Abstract
Heparin and heparan sulfate contain a rare 3-O-sulfoglucosamine residue critical for anticoagulation and virus recognition, respectively. The glycosidic linkage proximate to this 3-O-sulfoglucosamine is resistant to cleavage by all heparin lyases. Heparin lyase II has a broad specificity. The crystal structure of the wild type heparin lyase II identified its active site and showed a close spatial proximity between Asn405 and the 3-OH group of the bound glucosamine residue. In this study, we mutated Asn405 to the less sterically demanding Ala405 or Gly405, which broadened the substrate specificity of heparin lyase II and caused it to cleave the resistant linkage proximate to the 3-O-sulfoglucosamine residue.
Keywords: heparin lyase II, heparinase, site-directed mutagenesis, protein engineering, heparin, 3-O-sulfo glucosamine, substrate specificity
1. Introduction
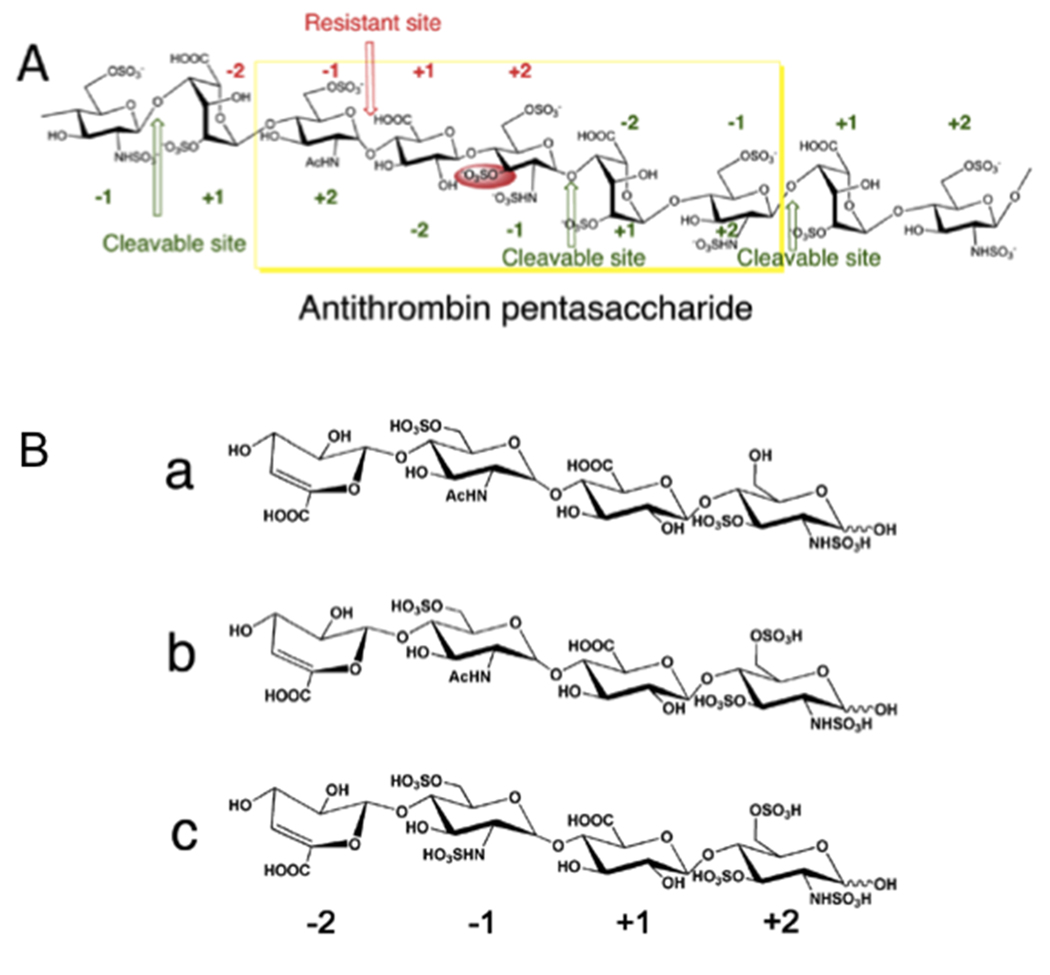
Heparin and heparan sulfate (HS) are negatively charged, linear polysaccharides consisting of repeating uronic acid-glucosamine disaccharide units. The major uronic acid components in heparin and HS are iduronic acid (IdoA) and glucuronic acid (GlcA), respectively. Heparin (Figure 1A) is more highly sulfated than heparan sulfate, with its uronic acid residues commonly modified with 2-O-sulfo groups and its glucosamine residues predominantly substituted with N-sulfo and 6-O-sulfo groups and in rare cases with 3-O-sulfo groups. The signature 3-O-sulfo group containing glucosamine residue found within the antithrombin (AT) binding sequence of heparin plays an essential role in binding and activating antithrombin [1–3] (Figure 1A). In HS a 3-O-sulfo group containing glucosamine residue is also contained in the glycoprotein (gD) binding site required for Herpes simplex virus type 1 (HSV-1) entry to cells [4–6].
Figure 1.
HepII action on 3-O-sulfo glucosamine containing substrates. A. Structure of a portion of the heparin chain containing an antithrombin binding site. Four sites are shown that should be HepII cleavable based on the residues at the −1+1 sites. Three of these are HepII cleavable (shown in green) and one, containing a 3-O-sulfo glucosamine residue at subsite +2 is resistant (shown in red). B. Chemical structures of used tetrasaccharide substrates. Tetrasaccharides a, b and c were prepared by exhaustively digesting heparin with HepI. The four monosaccharides in each structure are labeled as −2 to +2 from non-reducing end to reducing end. The 3-O-sulfo glucosamine residue is at subsite +2 and the putative HepII cleavage site is −1 +1.
Heparin and HS can be depolymerized through a β-elimination mechanism by bacterial heparin lyases (or heparinases), a group of polysaccharide cleaving enzymes commonly expressed and serving nutritional purposes in various microorganisms [7]. Among the identified three heparin lyases, heparin lyase II (HepII, EC number 4.2.2.7) from Pedobacter heparinus (previously known as Flavobacterium heparinum) displays a uniquely wide range of substrate specificity, cleaving both heparin and HS at the linkage between glucosamine residue and either iduronic or glucuronic acid residue having various sulfation patterns [8,9,10]. Despite this broad specificity, neither HepII nor any of the other currently known heparin lyases are capable of cleaving the glycosidic linkage (−1,+1) when the proximate glucosamine residue at the reducing end of the linkage (+2 subsite) contains a 3-O-sulfo group (Figure 1A) [11]. Recent studies on HepII in our laboratory have determined the structure of HepII with and without complexed HS oligosaccharide substrate or a disaccharide product [12,13]. Cleavage of substrates containing both uronic acid epimers IdoA and GlcA occurred within the same catalytic site in HepII. Potential active site residues participating in the various mechanistic steps of β-elimination have been identified along with their suggested roles. Specifically, His406 was found to be responsible for charge neutralization of the carboxyl group of uronic acid at the cleavage site. Tyr257 and His202 serve as the catalytic bases in the subsequent step by abstracting the proton of the C5 carbon from GlcA-containing (HS) and IdoA-containing (heparin) substrate, respectively [12]. In the crystal structure of HepII complexed with oligosaccharides occupying +1 and +2 sites the 3-O-hydroxy group of the glucosamine unit at the +2 subsite is buried and making contacts with the protein sidechains. The present study uses site-directed mutagenesis to perform protein engineering on HepII to test our hypothesis that expanding the size of the substrate-binding site near glucosamine 3-O-hydroxy group will make this mutant able to cleave substrates containing a 3-O-sulfo glucosamine residues at the +2 subsite. Several residues were mutated and the ability of the resulting enzymes to act on resistant substrates containing a 3-O-sulfo glucosamine residue was examined.
2. Materials and Methods
2.1 Chemicals
Heparin and HS sodium salts were purchased from Celsus Laboratories, Cincinnati, OH. Disaccharide standards of heparin and HS were obtained from Iduron Co. Other chemicals including chromatography and gel preparation reagents used in this work were the same as previously reported [12,13].
2.2 Preparation of recombinant wild type and mutant HepII
The mutants were prepared using the previously described expression plasmid containing HepII gene [13]. Site-directed mutagenesis was performed by the QuickChange method using Site-directed Mutagenesis Kit (Stratagene), according to the manufacturer’s protocol. Mutations were confirmed by DNA sequencing. All mutants were expressed in E. coli BL21(DE3).
2.3 Preparation of the heparin-derived tetrasaccharide substrates
Heparin-derived tetrasaccharides were prepared as described in previous literature [14]. Briefly, heparin was exhaustively digested by heparin lyase I in 50 mM sodium phosphate buffer, pH 7.4. The resulting disaccharide components were removed by chromatography on a 100 × 5.0 cm Bio-Gel P10 column eluted with 0.2 M NaCl and the oligosaccharide fractions were desalted on a 100 × 2.0 cm P2 column and lyophilized. The product mixture was fractionated on a 20 × 250 mm semipreparative strong anion exchange (SAX)-high performance liquid chromatography (HPLC) column eluted with a salt gradient with absorbance detected at 232 nm. Individual tetrasaccharide peaks were desalted and further purified by repeated separation on the same HPLC column. Purity of each tetrasaccharide was determined to be >95% by analytical HPLC. Structures of these tetrasaccharides were characterized by LC-MS and 1D and 2D NMR [14–16].
2.4 Activity analysis of mutant HepII on polysaccharide substrates by polyacrylamide gel electrophoresis (PAGE)
Heparin and HS were prepared to a final concentration of 2 mg/ml in 50 mM sodium phosphate buffer, pH 7.0, and was individually treated with 0.5 mg/ml WT or mutant HepII at 37°C overnight. Polyacrylamide gel was prepared in 22% and 5% of total acrylamide for resolving gel and stacking gel, respectively. The gel was loaded with 5 µg of each polysaccharide digestion sample and subjected to electrophoresis for 80 min at 200 V. The gel was visualized with 0.5% (w/v) Alcian blue in 2% (v/v) acetic acid solution.
2.5 Activity analysis of mutant HepII on defined tetrasaccharide substrates by liquid chromatography-mass spectroscopy (LC-MS)
Tetrasaccharide assays were performed each using 2 mg/ml of WT or mutant HepII reacting with 0.15 mg/ml of tetrasaccharide substrate in 50 mM sodium phosphate buffer, pH 7.0, at 37°C for 12 h. Control reactions were prepared using the same amount of substrate with no enzyme. Inactivated protein, generated by heating the sample to 100°C, was removed by centrifugation at 13,000 × g for 30 min at the end of reaction. The supernatants containing the reaction products were analyzed by LC-MS using an Agilent 1200 LC/MSD instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with an ion trap. An Acquity UPLC BEH C18 column (2.1 × 150 mm, 1.7 µm, Waters, Milford, MA, USA) was used. Eluent A and B were water/acetonitrile (85:15) v/v, and water/acetonitrile (35:65) v/v, respectively. Both eluents contained 12 mM tributyl amine (TrBA) and 38 mM NH4OAc with pH adjusted to 6.5 with HOAc. A gradient of solution A for 10 min followed by a linear gradient from 10 to 40 min (0–50% solution B) was used at flow rate of 100 µL/min. The column effluent entered the source of the ESI-MS for continuous detection by MS. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V, and a source of temperature of 350 °C, to obtain the maximum abundance of the ions in a full scan spectrum (200–1500 Da). Nitrogen was used as a drying (8 L/min) and nebulizing gas (40 psi).
3. Results and Discussion
3.1 Structure of WT and modeled structure of Asn405 mutant HepII bound with an HS disaccharide
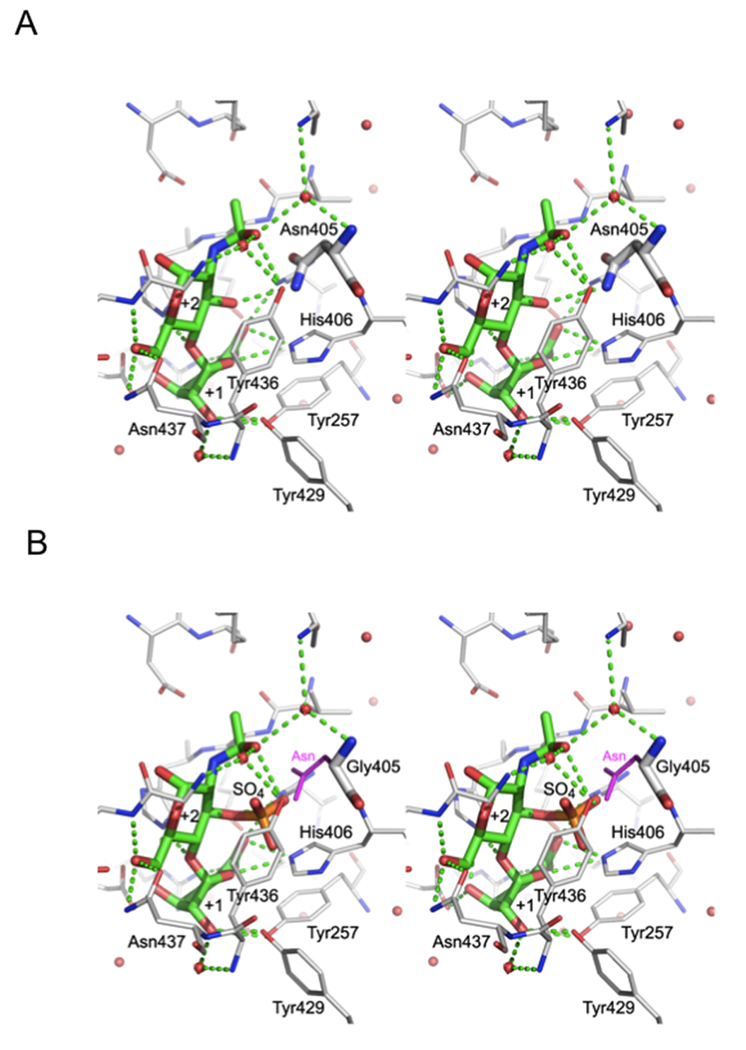
The crystal structure of the catalytic site of WT HepII (Figure 2A) suggests a possible explanation for the inability of HepII to cleave the polysaccharide or oligosaccharide substrate between −1 and +1 subsites when a 3-O-sulfo glucosamine residue is located at the +2 subsite (Figure 1A) [13]. In the crystal structure, the 3-OH group of the glucosamine at the +2 subsite was found to be pointing toward the bottom of the substrate-binding cavity in very close proximity to Asn405, His406, Tyr436 and Arg261. We speculated that if the glucosamine residue at the +2 subsite contained a 3-O-sulfo group, collision between this 3-O-sulfo group and the side chains of the above amino acid residues would sterically prevent proper positioning of the uronic acid reside at the +1 subsite, thus, preventing catalysis (Figure 2B). Of these, Arg261 is involved in hydrogen bonding to the carboxylic group of +1 sugar while Tyr436 stacks against the hydrophobic face of +2 sugar; their replacement is likely to decrease significantly substrate binding and adversely affect activity. His406 is also implicated in the enzymatic activity of HepII and the most likely residue to mutagenize was Asn405. Replacement by glycine without a sidechain opens up space for the additional sulfate group attached to O3 (Figure 2B). To test our hypothesis we performed site-directed mutagenesis of Asn405 and also attempted to replace His406 by a smaller sidechain. We prepared two single mutants N405A and N405G and two double mutants, N405A/H406A N405G/H406A. These mutants were expressed in E. coli with a His tag and purified to homogeneity.
Figure 2.
Vicinity of the +2 sugar. A. The stereo view of crystal structure of WT HepII with bound disaccharide [13] occupying subsites +1 and +2. The disaccharide and Asn405 are shown in thick lines while the neighboring sidechains are in thinner lines. B. The N405G mutant was modeled with a disaccharide containing a 3-O-sulfo glucosamine residue at subsite +2. The asparagine sidechain of the wild type enzyme is painted in magenta to show that it would collide with the 3-O-sulfo group.
3.2 Activity of HepII mutant enzymes on polysaccharide substrates
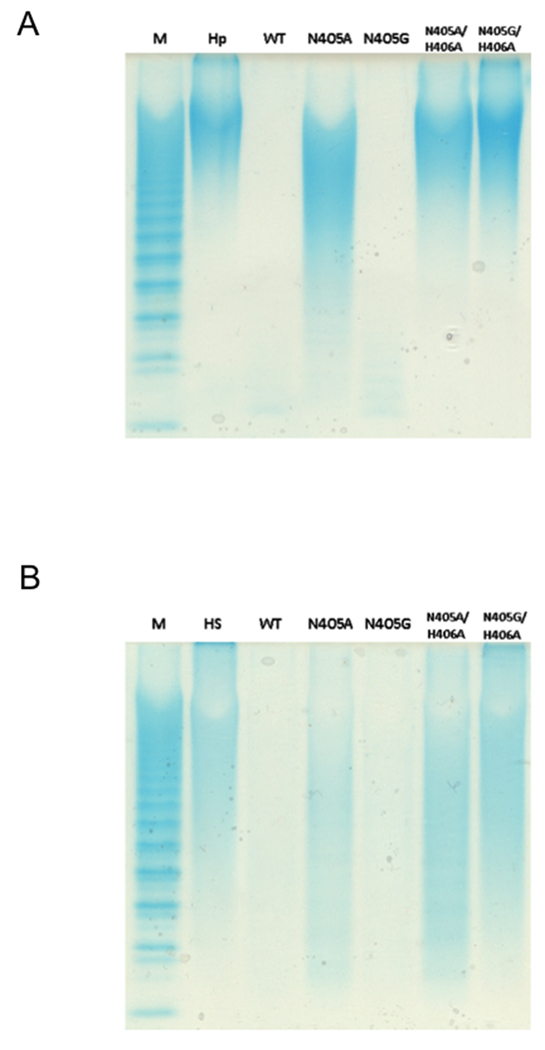
The activity of these mutants was first examined using heparin and HS polysaccharides as substrates. Exhaustive treatment with each enzyme (1–2 h) was monitored by PAGE (Figure 3). Wild type (WT) HepII served as a positive control completely digesting both heparin and HS to a barely detectable or undetectable mixture of disaccharide and tetrasaccharide products [16]. The N405G mutant enzyme afforded complete conversion on heparin and HS and N405A enzyme showed a substantial decrease in the molecular weight of both polysaccharides. The double mutants, N405A/H406A and N405G/H406A, showed little or no activity on both substrates with a barely observable activity towards HS. These results are in agreement with our previous findings on the key role of His406 in hydrogen-bonding the carboxylate oxygen of the cleavage site uronic acid for charge neutralization. Since His406 is responsible for this step in the elimination of both GlcA-containing (HS) and IdoA-containing (heparin) substrates, loss of enzymatic activity is not unexpected for mutations at this site [13].
Figure 3.
PAGE (22%) analysis with alcian blue staining of polysaccharides digested using HepII and HepII mutants. Panel A. Markers (M) with degree of polymerization (dp) of major bands labeled, untreated heparin, heparin treated with WT HepII, with HepII (N405A), with HepII (N405G), with HepII (N405A/H406A), with HepII (N405G/H405A). Panel B. Markers (M), untreated heparan sulfate, heparan sulfate treated with WT HepII, with HepII (N405A), with HepII (N405G) with HepII (N405A/H405A), with Hep (N405G/H406A), M, ladder of heparin oligosaccharide standards [20].
3.3 Activity of HepII mutant enzymes on defined tetrasaccharide substrates
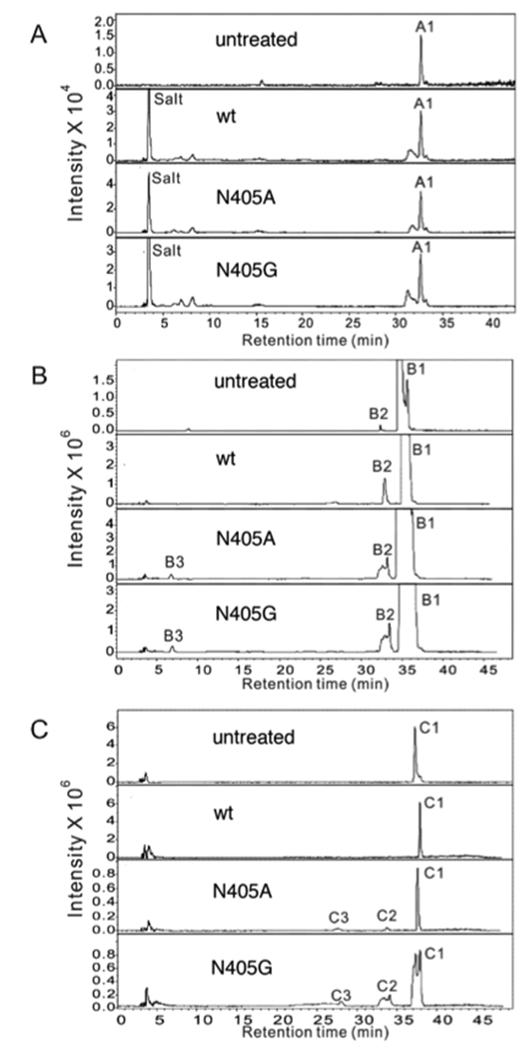
Three structurally defined tetrasaccharide substrates (a, b and c, Figure 1B) were next used to evaluate the effect of the Asn405 mutation on catalysis. The tetrasaccharides were prepared through the complete HepI depolymerization of heparin [16]. Each tetrasaccharide contains a 3-O-sulfo glucosamine residue (+2 subsite) next to the glycosidic uronic acid at the reducing end of the cleavage site (−1,+1) making these substrates resistant to WT HepII and all other known heparin lyases. Each tetrasaccharide contains a GlcA residue at subsite +1 and either a GlcNS6S or GlcNAc6S residue at subsite −1. The tetrasaccharides were individually treated with WT or mutant HepII under exhaustive reaction conditions, and any products resulting from cleavage between −1 and +1 were identified by LC-MS analysis. The observed and theoretical mass of the saccharide substrates and products, their identity and the substrate conversion percentage are summarized in Table 1.
Table 1.
Observed and theoretical mass of the saccharide substrates and products.
| Fractions | Found ions |
Observed Mol Mass |
Theoretical Mol Mass |
Assignment | Conversion %b | |||
|---|---|---|---|---|---|---|---|---|
| DPa | Sequences | WT | N405A | N405G | ||||
| A1 | [476.8]2- | 955.6 | 956.0 | 4 | ΔUA-GlcNAc6S-GlcA-GlcNS3S | 0 | 0 | 0 |
| B1 | [516.3]2- | 1034.6 | 1035.0 | 4 | ΔUA-GlcNAc6S-GlcA-GlcNS3S6S | 0 | 5 | 5 |
| B2 | [575.3]- | 576.6 | 576.9 | 2 | ΔUA-GlcNS3S6S | |||
| B3 | [457.4]- | 458.4 | 459.0 | 2 | ΔUA-GlcNAc6S | |||
| C1 | [535.5]2- | 1073.0 | 1073.9 | 4 | ΔUA-GlcNS6S-GlcA-GlcNS3S6S | 0 | 16 | 22 |
| C2 | [575.3]- | 576.6 | 576.9 | 2 | ΔUA-GlcNS3S6S | |||
| C3 | [495.3]- | 496.3 | 497.0 | 2 | ΔUA-GlcNS6S | |||
Degree of polymerization.
Conversion percentage is calculated by [product peak area/ (product peak area + substrate peak area)] × 100 (Fig. 4).
WT HepII, showed no activity on any of the three tetrasaccharide substrates tested and in this experiment served as a negative control. None of the Asn405 mutants were able to cleave the tetrasaccharide a substrate into two disaccharide products. In contrast, tetrasaccharide b and tetrasaccharide c served as substrates for both the N405A and N405G mutants, being partially cleaved into the two expected disaccharide products. The disaccharide product, derived from the reducing end of both tetrasaccharides b and c, had a molecular mass 576.6, corresponding to the 3-O-sulfo disaccharide, ΔUA-GlcNS3S6S. The disaccharide products originating from the non-reducing end of tetrasaccharide b and tetrasaccharide c had masses of 458.4 (ΔUA-GlcNAc6S) and 496.3 (ΔUA-GlcNS6S), respectively (Figure 1B, Table 1). The two double mutants, N405A/H406A and N405G/H406A, failed to cleave any of the tetrasaccharide substrates (data not shown). Even after extensive HepII treatment only 5% of tetrasaccharide b substrate and 20% of tetrasaccharide c substrate were converted to disaccharide products (Figure 4, Table 1).
Figure 4.
Extracted ion chromatography of tetrasaccharide a, b and c on treatment with WT and mutant HepII by LC-MS analysis. A. tetrasaccharide a; B. tetrasaccharide b; C. tetrasaccharide c. The four panels shown in each set of chromatograms (from top to bottom) correspond to untreated tetrasaccharide, WT treated tetrasaccharide, N405A tetrasaccharide, and N405G tetrasaccharide. Fractions from each treatment are listed in Table 1.
Extensive prior studies by others [17,18,19] on short oligosaccharides, including tetrasaccharides, have demonstrated that HepII is unable to cleave linkages (−1+1 subsite) with a proximate 3-O-sulfated glucosamine at the +2 subsite. Specifically, tetrasaccharide b and tetrasaccharide c have been reported to be resistant to cleavage by all known heparin lyases due to the presence of the 3-O sulfo group [11,19]. The crystal structure of the active site of WT HepII clearly shows a close proximity between Asn405 and the 3-OH group on the +2 subsite glucosamine [12]. The results of the current study clearly support the hypothesis that Asn405 sterically prevents appropriate binding of substrates having a 3-O-sulfo glucosamine residue at subsite +2 preventing cleavage of the −1+1 subsite linkage in these resistant substrates. The mutation of Asn405 to Ala with a smaller side chain or Gly without a sidechain serves to further reduce the substrate specificity of HepII. It is interesting to note that the more highly sulfated substrate tetrasaccharide c is a better HepII (N405A or N405G) substrate than tetrasaccharide b presumably due to enhanced binding to these mutant enzymes. In contrast, tetrasaccharide a is not a substrate possibly owing to the decreased sulfation at its reducing end +2 glucosamine residue adversely impacting its binding to the mutant HepII enzymes. Tetrasaccharide b and tetrasaccharide c comprise a large proportion of the tetrasaccharides produced in heparin lyase-catalyzed depolymerization of heparin, consistent with the high frequency of these sequences within the AT binding site of heparin. Thus, the present work demonstrates the potential of protein engineering to create new enzymes for use as tools for sequence analysis. Additional work will be required to enhance the activity and stability of such mutant enzymes before they can be routinely used in heparin analysis.
Highlights.
Heparin and heparan sulfate contain a rare 3-O-sulfo glucosamine residue
3-O-sulfo glucosamine is critical for anticoagulation and virus recognition
The glycosidic linkage proximate to this 3-O-sulfo glucosamine is resistant to heparin lyase cleavage
Protein engineering of heparin lyase II broadened its specificity causing it to cleave the resistant linkage proximate to the 3-O-sulfo glucosamine residue.
Acknowledgements
The authors thank the NIH for partially supporting this work through grants GM38060, HL096972, HL101721 (to RJL), GM93131 (to JDE) and CIHR for support through grant GSP-48370 (to MC).
Abbreviations
- HS
heparan sulfate
- GAG, GlcA
glucuronic acid
- IdoA
iduronic acid
- GlcNAc
N–acetylglucosamine
- GlcNS6S
6-O-sulfo-N-sulfoglucosamine
- GlcNAc6S
6-O-sulfo-N–acetylglucosamine
- GlcNS3S
3-O-sulfo-N-sulfoglucosamine
- GlcNS3S6S
3,6-O-sulfo-N-sulfoglucosamine
- AT
antithrombin
- Hep
heparin lyase
- PAGE
polyacrylamide gel electrophoresis
- LC
liquid chromatography
- MS
mass spectrometry
- dp
degree of polymerization.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petitou M, Duchaussoy P, Lederman I, Choay J, Sinay P. Binding of heparin to antithrombin-III - A chemical proof of the critical role played by a 3-sulfated 2-amino-2-deoxy-D-glucose residue. Carbohydrate Research. 1988;179:163–172. doi: 10.1016/0008-6215(88)84116-8. [DOI] [PubMed] [Google Scholar]
- 2.Walenga JM, Petitou M, Samama M, Fareed J, Choay J. Importance of a 3-O-sulfate group in a heparin pentasaccharide for antithrombotic activity. Thrombosis Research. 1988;52:553–563. doi: 10.1016/0049-3848(88)90128-4. [DOI] [PubMed] [Google Scholar]
- 3.Richard B, Swanson R, Olson ST. The signature 3-O-sulfo group of the anticoagulant heparin sequence is critical for heparin binding to antithrombin but is not required for allosteric activation. J Biol Chem. 2009;284:27054–27064. doi: 10.1074/jbc.M109.029892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shukla D, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 5.Tiwari V, Clement C, Duncan MB, Chen JH, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1. Journal of General Virology. 2004;85:805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 6.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue B, Liu J, Shukla D. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. Journal of Virology. 2006;80:8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linhardt RJ, Galliher PM, Cooney CL. Polysaccharide lyases. Applied Biochemistry and Biotechnology. 1986;12:135–176. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- 8.Lohse DL, Linhardt RJ. Purification and Characterization of Heparin Lyases from Flavobacterium heparinum. Journal of Biological Chemistry. 1992;267:24347–24355. [PubMed] [Google Scholar]
- 9.Desai UR, Wang HM, Linhardt RJ. Specificity Studies on the Heparin Lyases from Flavobacterium heparinum. Biochemistry. 1993;32:8140–8145. doi: 10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- 10.Desai UR, Wang HM, Linhardt RJ. Substrate Specificity of the Heparin Lyases from Flavobacterium heparinum. Archives of Biochemistry and Biophysics. 1993;306:461–468. doi: 10.1006/abbi.1993.1538. [DOI] [PubMed] [Google Scholar]
- 11.Shriver Z, Sundaram M, Venkataraman G, Fareed J, Linhardt R, Biemann K, Sasisekharan R. Cleavage of the antithrombin III binding site in heparin by heparinases and its implication in the generation of low molecular weight heparin. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10365–10370. doi: 10.1073/pnas.97.19.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaya D, et al. Catalytic Mechanism of Heparinase II Investigated by Site-directed Mutagenesis and the Crystal Structure with Its Substrate. Journal of Biological Chemistry. 2010;285:20051–20061. doi: 10.1074/jbc.M110.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaya D, Tocilj A, Li YG, Myette J, Venkataraman G, Sasisekharan R, Cygler M. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. Journal of Biological Chemistry. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- 14.Xiao ZP, Tappen BR, Ly M, Zhao WJ, Canova LP, Guan HS, Linhardt RJ. Heparin Mapping Using Heparin Lyases and the Generation of a Novel Low Molecular Weight Heparin. Journal of Medicinal Chemistry. 2011;54:603–610. doi: 10.1021/jm101381k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laremore TN, Ly M, Solakyildirim K, Zagorevski DV, Linhardt RJ. High-resolution preparative separation of glycosaminoglycan oligosaccharides by polyacrylamide gel electrophoresis. Analytical Biochemistry. 2010;401:236–241. doi: 10.1016/j.ab.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao ZP, Zhao WJ, Yang B, Zhang ZQ, Guan HS, Linhardt RJ. Heparinase 1 selectivity for the 3,6-di-O-sulfo-2-deoxy-2-sulfamido-alpha-D-glucopyranose (1,4) 2-O-sulfo-alpha-L-idopyranosyluronic acid (GlcNS3S6S-IdoA2S) linkages. Glycobiology. 2011;21:13–22. doi: 10.1093/glycob/cwq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugahara K, Tohnooka R, Yamada SH, Khoo KH, Morris HR, Dell A. Structural studies on the oligosaccharides isolated from bovine kidney heparan-sulfate and characterization of bacterial heparitinases used as substrates. Glycobiology. 1994;4:535–544. doi: 10.1093/glycob/4.4.535. [DOI] [PubMed] [Google Scholar]
- 18.Rhomberg AJ, Shriver Z, Biemann K, Sasisekharan R. Mass spectrometric evidence for the enzymatic mechanism of the depolymerization of heparin-like glycosaminoglycans by heparinase II. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12232–12237. doi: 10.1073/pnas.95.21.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada SH, Yoshida K, Sugiura M, Sugahara K, Khoo KH, Morris HR, Dell A. Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin-III-binding site of porcine intestinal heparin. Journal of Biological Chemistry. 1993;268:4780–4787. [PubMed] [Google Scholar]
- 20.Edens RE, Al-Hakim A, Weiler JM, Rethwisch DG, Fareed J, Linhardt RJ. Gradient polyacrylamide gel electrophoresis for determination of the molecular weights of heparin preparations and low-molecular-weight heparin sderivatives. Journal of Pharmaceutical Sciences. 1992;81:823–827. doi: 10.1002/jps.2600810821. [DOI] [PubMed] [Google Scholar]