Abstract
The lyso-phospholipid sphingosine 1-phosphate modulates lymphocyte trafficking, endothelial development and integrity, heart rate, and vascular tone and maturation by activating G-protein-coupled sphingosine 1-phosphate receptors. Here we present the crystal structure of the sphingosine 1-phosphate receptor 1 fused to T4-lysozyme (S1P1-T4L) in complex with an antagonist sphingolipid mimic. Access to the binding pocket is completely occluded by the N-terminus and extracellular loops of the receptor. Access is gained by ligands entering laterally between helices I and VII within the transmembrane region of the receptor. This structure, along with mutagenesis, agonist structure-activity relationship data and modeling, provides a detailed view of the molecular recognition and hydrophobic volume triggering that activates S1P1 resulting in the modulation of immune and stromal cell responses.
G protein-coupled receptors (GPCRs) convert exogenous signals into a cellular response by initiating a variety of intracellular signal cascades. The sphingosine 1-phosphate receptor subtype 1 (S1P1) (1) belongs to a sub-class of the GPCR family originally termed the endothelial differentiation gene (EDG) family of lipid receptors. The family was later renamed to reflect activation by two distinct lipids, S1P and lysophosphatidic acid (2). The S1P receptor family comprises five members (S1P1–5) with significant sequence identity within the ligand binding region including the conserved sphingolipid binding pocket. Activation of the S1P1 receptor through exogenous ligands, both physiological and pharmacological, results in significant inhibition of lymphocyte recirculation (3). This physiological effect was leveraged in the development of the nonselective S1P agonist pro-drug FTY720 (fingolimod) recently approved for the clinical treatment of relapsing remitting multiple sclerosis (4).
The structures of multiple members of the GPCR family have been determined at atomic resolution in both agonist and antagonist conformation as well as in complex with a cognate G protein (5–16). Together, these structures help define the repertoire of structural changes associated with the class A GPCR family that are required to recognize ligands of very different physico-chemical properties, yet signal through common mechanisms. We report here the structural characterization of a lipid-sensing GPCR, the S1P1 receptor fused to T4-lysozyme, in complex with the selective antagonist sphingolipid mimic (R)-3-Amino-(3-hexylphenylamino)-4-oxobutylphosphonic acid (ML056) (17) to 3.35 Å using traditional x-ray diffraction data processing methods and to 2.8 Å resolution using an experimental microdiffraction data assembly method to help process data of rapidly decaying microcrystals (Table S1) (18). Analysis of the structure along with structure-activity and mutagenesis data, reveals key interactions associated with the binding of physiological phospho-sphingolipids and related molecules (class I ligands). In addition, we report interactions within the binding pocket that are not required for class I ligand binding or function, but which directly affect binding of class II ligands, small-molecule S1P1 agonists that are not dependent on polar lipid head group interactions (19).
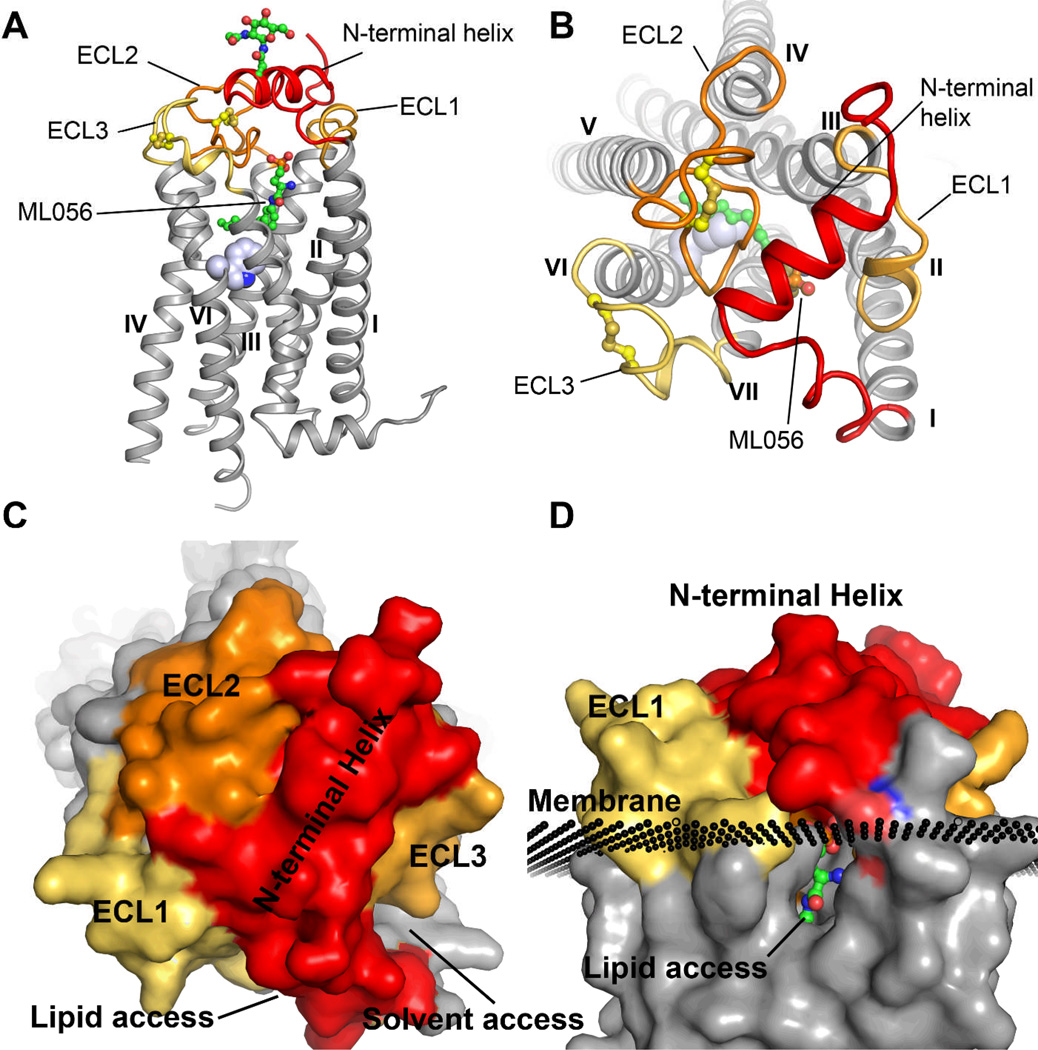
Overall the S1P1-T4L receptor structure shares many common features with previously determined receptors, including seven trans-membrane helices arranged in a structurally conserved bundle. The extracellular region for all GPCRs is composed of three loops: ECL1 between helices II and III, ECL2 between helices IV and V, and ECL3 between helices VI and VII. However, a number of distinguishing characteristics are associated with the ligand binding pocket in the S1P1 receptor (Fig. 1A). The N-terminus folds over the top of the receptor and contributes binding interactions, while at the same time forming a helical cap, which limits access to the amphipathic binding pocket (Fig. 1B). Both ECL1 and ECL2 pack tightly against the N-terminal helix further occluding ligand access to the receptor from the extracellular milieu while contributing significant surface area to the binding pocket. The limited access to the ligand binding site from the extracellular region explains why S1P1 ligands, including S1P, show slow saturation of receptor binding in the presence of excess ligand (1) (Fig. 1C). The ligands may gain access to the binding pocket from within the membrane bilayer, possibly through a gap between helices I and VII (Fig. 1D). This region of the receptor structure is more open than in other receptors due to a repositioning of helices I and II toward helix III in S1P1 relative to other receptors which is apparent after alignment using GPCR fold core residues (Fig. S4) (20). A similar mode of entry has been postulated for retinal loading into opsin, as well as for the entry of anandamide into the closely related cannabinoid receptors (21–23).
Fig. 1.
Overview of the structural features unique to the S1P1 receptor. A. Ribbon trace of the receptor with relevant features highlighted. The receptor folds in a traditional seven transmembrane helical bundle similar to other class A GPCRs. However, the extracellular domain of the S1P1 receptor adopts a novel fold incorporating helical elements from the N-terminus (red ribbon), as well as ECL1 (gold ribbon), that occlude access to the ligand binding pocket. ECL2 (orange ribbon) and ECL3 (yellow ribbon) are both constrained by intraloop disulfide bonds (yellow ball-and-stick) and contribute important interactions within the binding pocket. The compound ML056 is shown in green carbon ball-and-stick rendering and W2696.48 is rendered as a space-filling atom in blue as reference point for the binding pocket. The T4L fusion protein was omitted from this figure for clarity. B. Top view of the receptor highlighting the critical role of the N-terminal helix in capping the ligand and limiting solvent access to the binding pocket. There is a sizable access point for ligand entry and exit between helices I and VII. C. Surface rendering of the receptor from the top view highlighting the occluded nature of the binding pocket with two points of access either for lipid or solvent. D. Side-view of the lipid access channel relative to the predicted position of the membrane bilayer.
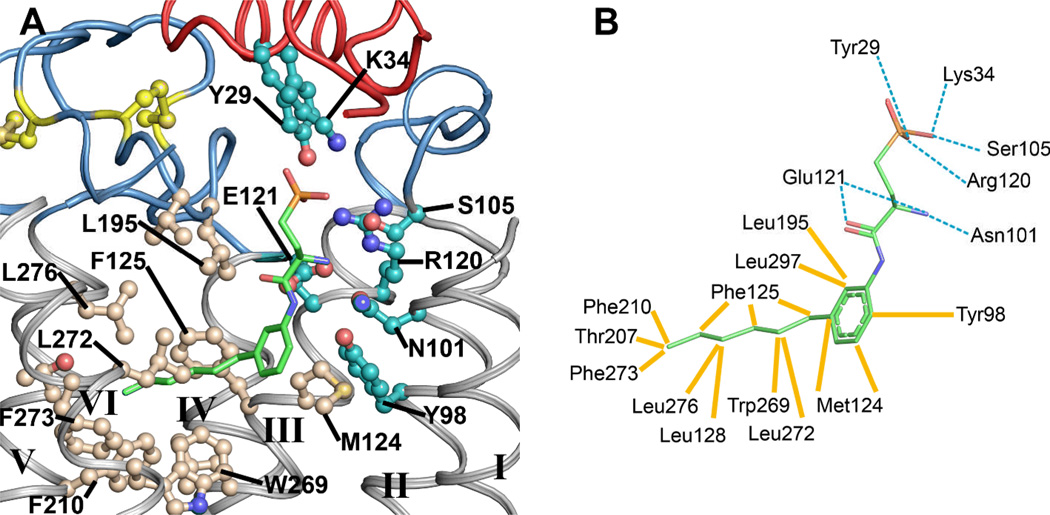
The binding pocket of the S1P1 receptor is highly amphipathic reflecting the nature of its hydrophobic-zwitterionic agonist. The established roles of R1203.28 and E1213.29 (24) that interact with the zwitterionic sphingosine head group (25, 26) are recapitulated in their interactions with the phosphonate and primary amine of ML056 (Fig. 2). However, a third predicted ionic interaction between R2927.35 and the phosphate head group is not apparent in the ML056 interactions. Instead, the phosphonate head group of ML056 is surrounded by a ring of positively charged and polar residues contributed by helices III and VII, ECL2 and the N-terminal capping helix, which form a positively charged pocket that provides charge complementarity and high affinity interactions to the phosphate group of the sphingolipids. The primary amine of ML056 is likely protonated at physiological pH, which results in a zwitterionic head group. This positive charge contributes to the binding affinity of sphingosine containing compounds through ionic interactions with the carboxylate side chain of E1213.29 as well as the amide carbonyl of N1012.60 (helix II). The involvement of N1012.60Y29 and K34 (27) in the binding of sphingosine-like head groups was not previously predicted. We, therefore, verified their involvement through mutagenesis coupled with signaling assays (Table S3).
Fig. 2.
Detailed structural representation of the interactions between ML056 and the S1P1 receptor. A. ML056 is colored as green carbons sticks. Polar residues within the binding pocket are colored with cyan carbons, whereas residues comprising the hydrophobic portion of the binding pocket are colored with tan carbons and the two disulfide bonds are shown in yellow ball-and-stick. The N-terminal helix and extracellular loops are shown as red and blue ribbons respectively. B. Two-dimensional residue interaction map illustrating the amino-acids within 4 Å of ML056. Polar interactions are represented by dotted cyan lines, whereas hydrophobic interactions are represented as solid yellow lines drawn to the closest atom of the ligand.
The phenyl acyl tail of ML056 inserts into a largely aromatic pocket consisting of residues from helices III, V, VI and VII, as well as ECL2. The majority of the hydrophobic portion of the binding pocket is lined with short aliphatic residues that define the shape of the pocket and four aromatic residues that provide the potential for specific interactions (Fig. 2). The involvement of some of these residues in the binding and signaling of the S1P receptor was determined previously through molecular modeling and mutagenesis. The acyl chain is precisely structured, which was not expected based on commonly held views of acyl chain flexibility (28). This reflects a role for selective hydrophobic interactions within the pocket that result in this preferred low energy minimum. The previously published mutagenesis data for the S1P1 receptor are, for the most part, consistent with the structure of the antagonist bound receptor.
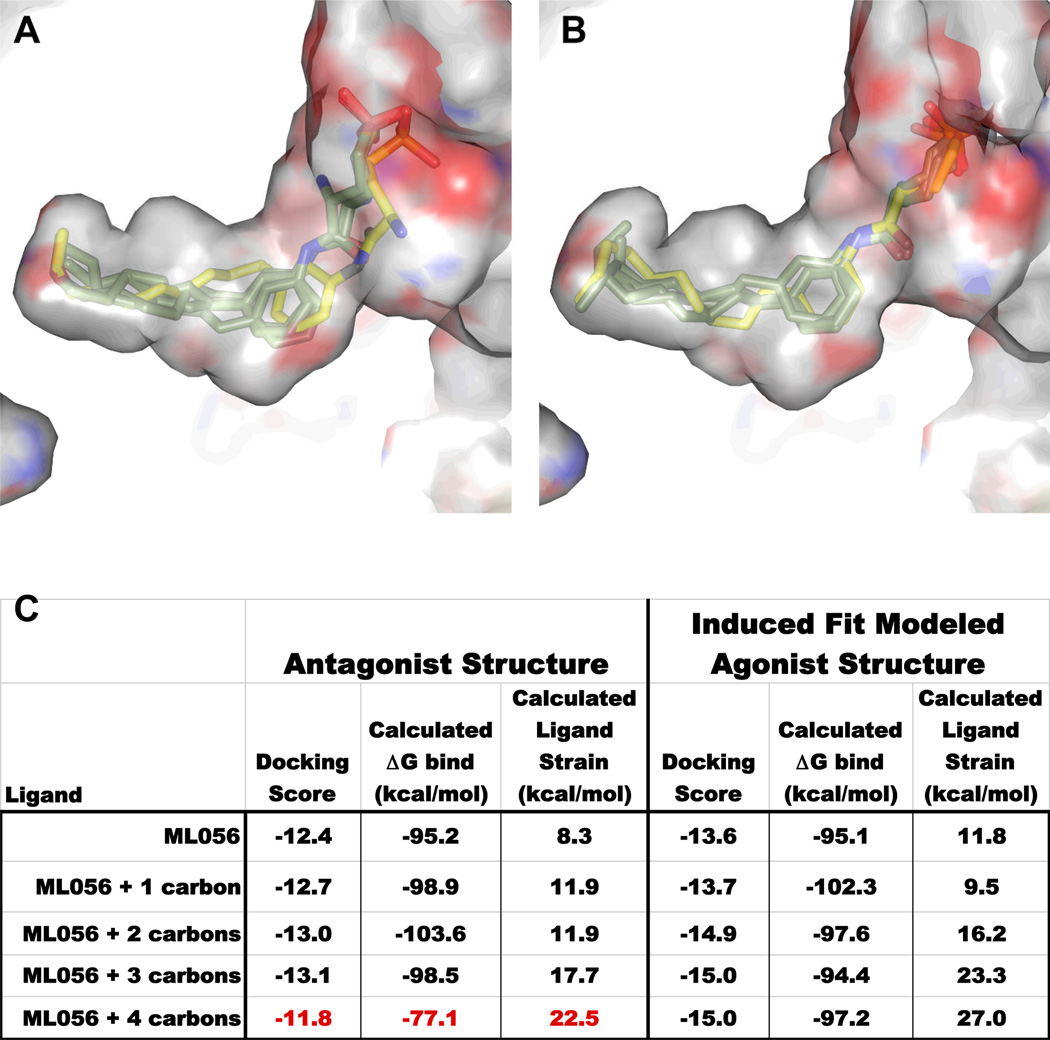
Structure-activity relationships from ML056-like compounds reveal an antagonist island that is easily converted to full agonism by small extensions of the acyl chain, suggesting that the volume occupied by the hydrophobic portion of the ligand plays a key role in triggering agonist signaling (29). In addition, changing the substitution pattern around the phenyl ring from para to meta switches the pharmacology of the compound series from agonist to antagonist, localizing the effect of hydrophobic volume changes within the binding pocket. Docking studies corroborate these trends in that the ML056 antagonist binding pocket of the S1P1 receptor could not accommodate an acyl carbon length greater than nine carbons long without significant ligand strain (Fig. 3A) (30, 31). The increased volume requirements for agonists were accommodated in the model by using an induced fit docking protocol that allowed the side-chains of the residues lining the hydrophobic portion of the binding pocket to move in response to a para-substituted regio-isomer of ML056 while holding the S1P head group interactions fixed (Fig. 3B) (32–34). The largest positional change occurred in F2736.52 whose movement along with slight positional changes in W2696.48 and F1253.33 accommodated binding of the endogenous S1P ligand as well as FTY720-P suggesting that these residues maybe be particularly important for signaling (Fig. 4A).
Fig. 3.
Docking study of ML056 analogs (green carbons) with progressively longer acyl chains. A ten carbon acyl chain (yellow carbons) switches the response from antagonism to agonism. A. Docking each of the five ligands into the antagonist binding pocket resulted in essentially superimposable docking poses for all of the ligands except ML056 + 4 (yellow carbons) which cannot maintain sphingolipid headgroup binding interactions due to high ligand strain. B. Docking each of the five ligands into the induced fit agonist modeled binding pocket reveals that all ligands can bind with reasonable docking scores while maintaining head group interactions. C. Table of docking and MM-GB/SA (Molecular Mechanics, Generalized Born and Solvent Accesibility) calculations for assessing the quality of the docking poses. The agonist compound ML056 + 4 docked into the antagonist binding pocket generates a steep drop in calculated ΔGbind along with an increase in calculated ligand strain.
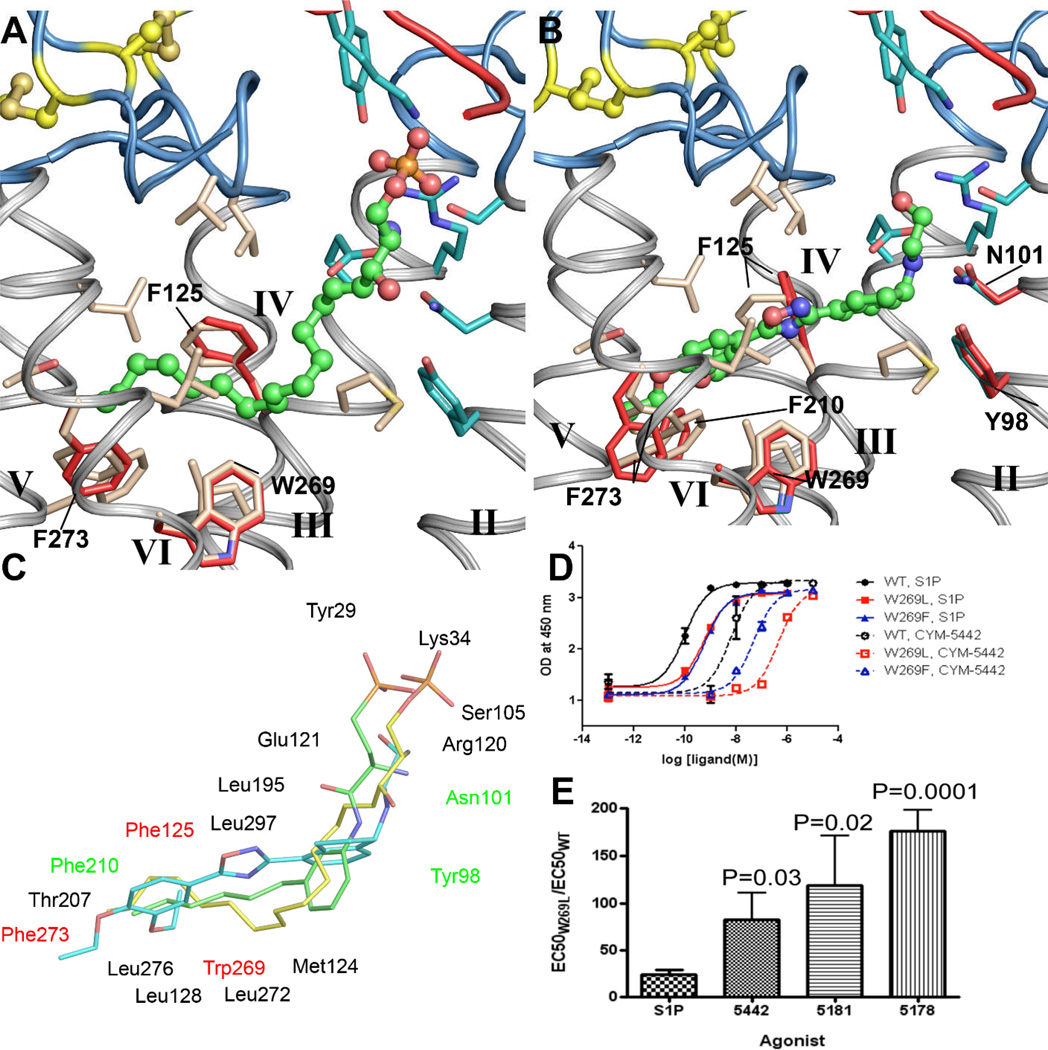
Fig. 4.
A comparison of putative binding interactions for class I and class II S1P1 agonists. A. The docked binding pose of S1P in the modeled S1P1 receptor agonist binding pocket. The red and blue ribbons are from N-terminal and extracellular loops respectively. Residues that deviate from their crystallographically determined positions (tan carbons) are rendered twice, the red atoms represent the modeled agonist position determined by induced fit docking. B. The docked binding pose of CYM-5442 in the modeled agonist binding pocket. Residues that deviate from their crystallographically determined positions (tan carbons) are rendered twice, the red atoms represent the modeled agonist position determined by induced fit docking. The binding pocket must accommodate slightly greater ligand volume in the hydrophobic portion. C. Overlay of the docked ligands S1P (yellow carbons) and CYM-5442 (cyan carbons) compared to the structurally determined conformation of ML056 (green carbons). Binding pocket residues are indicated with black font for those that do not change between the agonist and antagonist binding models, red font for those that change for the agonist induced fit docking model, and green font for those that change conformation induced by CYM-5442 docking. D. Ligand-induced ERK phosphorylation in WT, W2696.48L, and W2696.48F Jump-In stable cell lines stimulated with increasing concentrations of either S1P or CYM-5442 (mean ± S.D. of triplicate samples). The data are from one of three independent experiments showing a minimal effect on S1P signaling. The progressive loss in potency of CYM-5442 tracks with a loss of aromatic π-stacking interactions. E. A differential loss of agonist responsiveness of the mutant W269L S1P1 receptor compared to WT receptor was observed. For each experiment, the EC50 for agonist activation of ERK phosphorylation of W269L S1P1 receptor was divided by the EC50 of WT S1P1 receptor.
Inspection of known agonists of the S1P1 receptor allows their classification into two basic groups based upon receptor interactions: Class I) ligands that can be either lipid-like compounds mimicking S1P and its sphingophospholipid head group interactions exemplified by FTY720-P, or non-lipid like small molecules possessing polar groups that interact with the S1P head group region, sharing similar bond lengths and volume to drive affinity and efficacy as exemplified by SEW2871, and Class II) small molecule agonists that do not require interactions with the polar head group interacting region of the receptor, exemplified by CYM-5442 (19, 35), and instead make specific variant binding interactions. The binding of the endogenous ligand S1P, as well as other head group-containing class I agonists to the S1P1 receptor, involves strong ionic interactions between the zwitterionic head group and a cluster of charged and polar residues from helices II, III and VII and the N-terminal capping helix of the receptor (19, 25, 36). The interactions within the S1P binding region are likely similar to those exhibited by the head group of ML056. The length and position of the acyl chain relative to the amino-phosphate or aminophosphonate groups are, in general, what dictate the pharmacology of these compounds (37). The class II agonists to the S1P1 receptor were developed in response to the observation that certain library screening hits do not interact with the conserved polar residues, R1203.28 and E1213.29, which was hypothesized to increase their potential for tuning receptor subtype selectivity while also providing greater variability in physico-chemical properties (19, 35). Recently, we reported development of such a ligand, CYM-5442, which does not require interactions with either R1203.28 or E1213.29 of the S1P1 receptor, yet induces ERK phosphorylation in vitro and potent lymphopenia in vivo with nanomolar potency, and showed non-competitive inhibition of ERK signaling with ML056 (19). In conjunction with structural efforts, we have investigated the mechanism of binding of CYM-5442 to the S1P1 receptor through mutagenesis of the ligand-interacting portion of the binding pocket.
A series of conservative point mutations along the hydrophobic ligand-binding pocket were made to probe for defective binding or signaling in response to either S1P or CYM-5442 (38)and complement previous studies (39, 40). Three such S1P1 receptor mutants, F2105.47L, F2656.44L, and W2696.48L (Table S3), decreased or abolished CYM-5442-induced ERK phosphorylation and binding while only the W2696.48L mutation minimally affected either binding or signaling induced by S1P and other orthosteric ligands (Fig. 4D). This contrasts with the negative effect of W2696.48A on ligand induced [35S]GTPγS Emax (28), indicating that S1P binding is highly dependent on hydrophobicity, but not aromaticity at this position. The class II agonist, CYM-5442, was dependent on the presence of an aromatic residue, displaying a complete disruption of signaling and binding by the W2696.48L mutation (Fig. 4B & Table S3). The hierarchy of loss of agonist responsiveness for the W2696.48L S1P1 mutant receptor relative to WT (CYM-5442 >> S1P) was preserved for structural analogues of CYM-5442, such as CYM-5181 and CYM-5178 (Fig. 4E & Table S2), suggesting an interaction between the 3,4-diethyloxy-phenyl ring common to these compounds and the aromatic ring of W2696.48. Results from a radioligand binding competition assay also indicate that W2696.48 plays a more critical role in binding CYM-5442 than in binding S1P (Fig. S6) (41). These mutagenesis data, together with structure activity relationship (SAR) data and the structural information, allowed us to model the likely binding orientation of CYM-5442 and rationalize the high selectivity exhibited by this series of ligands (Fig. 4B). These data also suggest that while hydrophobic packing interactions are the basis of both retinal and S1P interactions with the conserved tryptophan, the altered aromatic interactions with this highly conserved residue provides differences in ligand orientation, function and, thus, a foundation for specific anchoring in the pocket.
Designing selectivity is an important component of the development process for S1P1 receptor modulators given that signaling along the other S1P receptor subtypes is associated with a number of clinically adverse events such as bronchoconstriction (42). The recently approved therapeutic, Gilenya™, is a first-in-class S1P1 pro-drug modulator that once phosphorylated by sphingosine kinase is a potent agonist for the S1P3, S1P4 and S1P5 receptors. Second generation therapeutics, derivatives of CYM-5442, currently under development are able to selectively agonize the S1P1 pathway without loss of potency. The structural basis for this enhanced selectivity can be rationalized directly based on discrete binding pocket substitutions as compared to S1P3 and S1P4. The S1P3 receptor has one significant substitution F2636.55 relative to the S1P1 receptor’s L2766.55, which occurs further down in the hydrophobic binding pocket introducing steric clashes with the aromatic ring system of class II agonists. The S1P4 receptor also has only one significant change in the binding pocket L1253.32 relative to the S1P1 receptor’s M1243.32, which may result in a slight constriction of the binding pocket at a critical location where the phenyl ring of FTY720-P and the indole ring of CYM-5442 are predicted to bind. The binding pocket of the S1P5 receptor is very similar to the S1P1 receptor with no apparent substitutions to inform selectivity, which is consistent with the lack of selectivity exhibited by most, if not all, of the S1P1 agonists. In addition to the S1Px receptors, this structure will be particularly useful for modeling additional members of the EDG family as well as increasing the repertoire of structural knowledge being collected on class A GPCRs.
By taking advantage of specific aromatic interactions within the hydrophobic binding pocket class II agonists avoid features that mimic the S1P head group relying more on shape complementarity to the agonist pocket allowing more selectivity between receptor subtypes. The ability of the GPCR fold to adapt to multiple diverse chemotypes appears to be driven by topological differences in the extracellular domain. Sphingolipid binding receptors channel access to the binding pocket a small region between helices I and VII adjacent to the plasma membrane, where sphingolipid molecules gain access from within the confines of the membrane rather than the extracellular milieu. Based on recent work on retinal loading in opsin, activation of the receptor may impact pocket accessibility through slight rearrangement of the transmembrane helices (43). Persistent signaling of S1P1 in the presence of certain ligands may be explained by occlusion of ligand egress from the binding pocket after activation.(44)
Supplementary Material
References and Notes
- 1.Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Ann Rev Biochem. 2009;78:743. doi: 10.1146/annurev.biochem.78.072407.103733. [DOI] [PubMed] [Google Scholar]
- 2.Chun J, et al. Pharmacol Rev. 2002;54:265. doi: 10.1124/pr.54.2.265. [DOI] [PubMed] [Google Scholar]
- 3.Mandala S, et al. Science. 2002;296:346. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann V, et al. Nat Rev Drug Disc. 2010;9:883. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen SG, et al. Nature. 2011;469:175. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Nature. 2007;450:383. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 8.Warne T, et al. Nature. 2008;454:486. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherezov V, et al. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien EY, et al. Science. 2010;330:1091. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaakola VP, et al. Science. 2008;322:1211. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palczewski K, et al. Science. 2000;289:739. [Google Scholar]
- 13.Wu B, et al. Science. 2010;330:1066. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F, et al. Science. 2011;332:322. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen SG, et al. Nature. 2011;477:549. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebon G, et al. Nature. 2011;474:521. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanna MG, et al. Nat Chem Biol. 2006;2:434. doi: 10.1038/nchembio804. [DOI] [PubMed] [Google Scholar]
- 18.Diffraction data were processed using two different methods – yielding data sets at 3.35 Å and 2.8 Å resolution. A lower resolution dataset using traditional integration and scaling methods was derived from selecting reflection groups based on resolution shell. A higher resolution dataset was derived from an experimental micro-diffraction data assembly method based on assembling data based on peak profile correlations. Further details can be found in the Supporting Online Material section online at Science
- 19.Gonzalez-Cabrera PJ, et al. Mol Pharmacol. 2008 [Google Scholar]
- 20.Hanson MA, Stevens RC. Structure. 2009;17:8. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst DP, et al. Journal of Biological Chemistry. 2010;285:17954. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schadel SA, et al. J Biol Chem. 2003;278:24896. doi: 10.1074/jbc.M302115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipek S, Stenkamp RE, Teller DC, Palczewski K. Annual review of physiology. 2003;65:851. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.In addition to numbering residue positions in the primary amino acid sequence, Ballesteros-Weistein numbering is shown as a superscript. The most conserved residue in each transmembrane helix is designated as x.50, where x is the number of the transmembrane helix, and the second number (50) indicates the relative position of the specific residue. The second number decreases towards the N terminus and vice versa. Only residues on transmembrane helices are noted.
- 25.Parrill AL, et al. J Biol Chem. 2000;275:39379. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 26.Jo E, et al. Chem Biol. 2005;12:703. doi: 10.1016/j.chembiol.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 27.K34 does not have strong electron density and placing its side-chain unambiguously in a position interacting with the phosphonate of W146 relies on omit maps rather than standard 2|Fo|-|Fc| maps. Its position on the N-terminal capping helix coupled with modeling and docking studies as well as subsequent mutagenesis have establish a role for this residue in sphingosine ligand binding interactions in support of its placement in the weak electron density maps.
- 28.Fujiwara Y, et al. J Biol Chem. 2007;282:2374. doi: 10.1074/jbc.M609648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis MD, Clemens JJ, Macdonald TL, Lynch KR. J Biol Chem. 2005;280:9833. doi: 10.1074/jbc.M412356200. [DOI] [PubMed] [Google Scholar]
- 30.Lyne PD, Lamb ML, Saeh JC. J Med Chem. 2006;49:4805. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 31.Prime, version 3.0. New York, NY: Schrödinger, LLC; 2011. [Google Scholar]
- 32.Schrodinger Suite 2011 Induced Fit Docking protocol; Glide version 5.7. New York, NY: Schrodinger, LLC; 2011. [Google Scholar]; Prime, version 3.0. New York, NY: Schrodinger, LLC; 2011. [Google Scholar]
- 33.The induced fit model derived by leveraging structure activity relationships of the VPC compound series was used for subsequent analysis of class I S1P1 agonists without further modifications and was used as the starting point for induced fit modeling of class II ligands.
- 34.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. J Med Chem. 2006;49:534. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 35.Schurer SC, et al. ACS Chem Biol. 2008;3:486. doi: 10.1021/cb800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang DA, et al. J Biol Chem. 2001;276:49213. doi: 10.1074/jbc.M107301200. [DOI] [PubMed] [Google Scholar]
- 37.SEW2871 also been shown to interact with the head group binding region of the receptor, likely through a dipole moment interaction, while presenting aromatic ring systems to the hydrophobic binding pocket, and shares the same 15–16 carbon bond length.
- 38.The Jump-in system was used to provide a quantitative comparison between cell lines with similar surface expression, G-protein coupling and background protein expression.
- 39.Fujiwara Y, et al. J Biol Chem. 2007;282:2374. doi: 10.1074/jbc.M609648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A conservative mutagenesis approach was employed where hydrophobic to hydrophobic substitutions were preferred over substitutions that may drastically change the nature of the binding pocket or the overall protein fold.
- 41.Given the highly conserved nature of W2696.48 in all Class A GPCRs and its position directly above the third intracellular loop, a site of G-protein binding, the effects of this mutation were rigorously quantified by assaying stable single cell clones of wild-type (WT) and the W2696.48L mutant using ERK phosphorylation upon stimulation with increasing concentrations of S1P and CYM-5442 (Table S3). The WT and W2696.48L EC50 values differed by 15-fold when tested using S1P stimulation. They differed by greater than 100-fold when ERK phosphorylation was measured in response to CYM-5442 stimulation (Table S3). The W2696.48L mutant has a 10-fold greater effect on CYM-5442 signaling than on S1P signaling, suggesting that W2696.48 in S1P1 plays a more critical role in CYM-5442-induced receptor activation. An additional stable mutant, W2696.48F, was used to examine whether aromaticity at this position is sufficient for ligand-induced ERK phosphorylation in response to the class II agonists (Fig. 4D) The ratio of EC50 shifts comparing CYM-5442 to S1P for each mutant (W2696.48L = 12.3 and W2696.48F = 1.3) confirm that the aromatic ring of W2696.48 supplies a key interaction for CYM-5442 and the class II agonists.
- 42.Forrest M, et al. J Pharmacol Exp Therap. 2004;309:758. doi: 10.1124/jpet.103.062828. [DOI] [PubMed] [Google Scholar]
- 43.Hildebrand PW, et al. PloS one. 2009;4:e4382. doi: 10.1371/journal.pone.0004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.This work was supported in part by the Protein Structure Initiative (PSI):Biology grant U54 GM094618 for structure production, NIH Roadmap grant P50 GM073197 for technology development, NIH Roadmap Molecular Libraries Probe Production Centers grant U54 MH084512 for antagonist and class II agonist identification, and AI074564 and AI055509 to HR and Michael B. Oldstone for studying the role of S1P in infection and immunity. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Science or the National Institutes of Health. The authors thank E. Chien for preliminary analysis, M. Mileni for critical evaluation of the structure, J. Johnson, A. Leslie, W. Kabsch and A. Perrakis for discussion and review of the experimental microdiffraction data assembly processing method, and A. Walker for assistance with manuscript preparation. The authors acknowledge J. Smith, R. Fischetti and N. Sanishvili at the GM/CA-CAT beamline at the Advanced Photon Source, for assistance in development and use of the minibeam and beamtime. The GM/CA-CAT beamline (23-ID) is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Atomic coordinates and structure factors have been deposited in the Protein Data Bank with identification code 3V2W and 3V3Y.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.