Summary
We consider penalized linear regression, especially for “large p, small n” problems, for which the relationships among predictors are described a priori by a network. A class of motivating examples includes modeling a phenotype through gene expression profiles while accounting for coordinated functioning of genes in the form of biological pathways or networks. To incorporate the prior knowledge of the similar effect sizes of neighboring predictors in a network, we propose a grouped penalty based on the Lγ-norm that smoothes the regression coefficients of the predictors over the network. The main feature of the proposed method is its ability to automatically realize grouped variable selection and exploit grouping effects. We also discuss effects of the choices of the γ and some weights inside the Lγ-norm. Simulation studies demonstrate the superior finite sample performance of the proposed method as compared to Lasso, elastic net and a recently proposed network-based method. The new method performs best in variable selection across all simulation set-ups considered. For illustration, the method is applied to a microarray dataset to predict survival times for some glioblastoma patients using a gene expression dataset and a gene network compiled from some KEGG pathways.
Keywords: Elastic net, Generalized boosted Lasso, L1 penalization, Laplacian, Lasso, Microarray gene expression, Penalized likelihood
1. Introduction
Consider linear regression, especially for “large p, small n” problems, as arising in genomic and proteomic studies. In our motivating example, we wish to use gene expression profiles to predict survival times for glioblastoma patients after surgery, where p ≈ 1500 genes are available as predictors with only n < 100 samples For this type of problems it is well known that some regularization on parameters is necessary. In addition to predictive performance, it is also biologically important to select genes relevant to the outcome. Hence, both variable selection and parameter estimation are targeted. Many penalized methods have emerged, mostly within the last few years, such as Lasso (Tibshirani 1996), SCAD (Fan and Li 2001), elastic net (Zou and Hastie 2005) and LARS (Efron et al 2005). Although these methods have proven useful in various applications, they may not be efficient due to their generic nature in failing to account for specific relationships among the genes (or more generally, predictors). In particular, as pointed out by Li and Li (2008), the above generic methods (and other commonly used variable selection methods) treat all the genes equally a priori, thus ignoring individual features of the genes. It is known that the genes do not work in isolation or independently with each other; they function coordinately in pathways or networks. A large body of biological knowledge on gene functions and pathways is available through the Gene Ontology (GO) (Ashburner et al 2000) and KEGG (Kanehisa and Goto 2000) databases. In our example we will utilize a gene network compiled from the KEGG.
With gene expression data, after standardizing the expression levels of each gene to have mean zero and variance one across samples, due to co-expressions of neighboring or interacting genes in a network, one may assume a priori that the magnitudes of the effects of the neighboring genes are similar, though their directions may differ. Of course this assumption may or may not hold in practice, but under this assumption, Li and Li (2008) proposed a new penalty that utilizes the structure of a given gene network. There are two potential drawbacks with Li and Li’s method. First, it is computationally more challenging due to the two tuning parameters in their penalty function, and that a straightforward fitting procedure as suggested therein involves (n + p) observations for p variables, leading to high or even infeasible computational demand for large p. Second, their penalty function encourages the smoothness of (weighted) coefficients βi’s rather than of (weighted) |βi|’s as intended. The above two points motivate our proposed penalty, which is related to grouped penalties (Yuan and Lin 2007; Zhao et al 2006), but differs from the existing ones in its specific reference to a network. The main advantages of our method include a simpler computational task with only one tuning parameter, e.g., in developing fast algorithms for solution paths, and its ability of automatically realizing grouped variable selection and exploiting grouping effects. Li and Li’s method is not capable of grouped variable selection which partially explains why our method outperforms theirs (and Lasso and elastic net) in variable selection when grouping is reasonable. In addition, we discuss the choice of the group penalty and its associated weights.
The remainder of this paper is organized as follows. In Section 2 we first review several commonly used penalized regression approaches, including Lasso, elastic net and the method of Li and Li (2008). We then propose our new method and study its theoretical properties. Section 3 reports on simulation studies comparing the finite-sample performance of our new method with its competitors. Section 4 analyzes the motivating example. Section 5 discusses some possible modifications and extensions, followed by a short discussion in Section 6.
2. Methods
2.1 Penalized Regression
Consider a linear regression model: with E(εk) = 0. We assume throughout that training data (yk, xk1, …, xkp) for k = 1, …, n, have been standardized such that the sample means of y and of each xi are 0 and the sample variance of each xi is 1. One often estimates β = (β1, …, βp)′ by minimizing the squared error loss
leading to the ordinary least square estimator (OLSE) β̃ = arg minβ L(β). However, in some situations, e.g., if p ≈ n or p > n, or for the purpose of variable selection, it may be desirable to regularize β through a penalized least square estimator (PLSE):
where pλ(β) is a penalty function. Two popular choices are the ridge penalty (Hoerl and Kennard 1970): , and the L1 or Lasso (Tibshirani 1996) penalty: . Compared to the ridge, a nice feature of the Lasso penalty is its capability in variable selection: with a λ large enough, some β̂i will be exactly 0, effectively excluding the corresponding predictor xi from a model. A downside of the Lasso is that it can have no more than n non-zero β̂i’s, which limitis its application with p ≫ n as for typical microarray data. To overcome the problem, Zou and Hastie (2005) proposed the elastic net (Enet) that combines the ridge and Lasso penalties:
where the first term is used for variable selection, while the second is to exploit grouping effects (see section 2.5 for more details).
In spite of the success of the above methods, they are generic, possibly failing to take full advantage of prior knowledge of existing structures among predictors. For example, for microarray data as considered here, it is known that the genes work coordinately as dictated by a gene network. To incorporate biological knowledge of gene networks, Li and Li (2008) proposed a new penalty that is similar to Enet but also uses the normalized Laplacian matrix M of a network. Specifically, given a network that describes relationships among the predictors, denote di as the degree of predictor (or exchangeably, node) i in the network; that is di is the number of direct neighbors of node i in the network. Li and Li’s network-based penalty is
where i ~ j means that nodes i and j are direct neighbors in the network; alternatively, if the combinatorial Laplacian matrix Mu is used, the second term of pλ(β) becomes λ2Σi~j(βi − βj)2, which has a Bayesian interpretation in that the prior distribution of β follows a Gaussian conditional autoregressive model (Gelfand and Vounatsou 2003) with its neighboring structure induced by the network. Smoothing (weighted) βi’s over the network is motivated by the prior assumption that the (weighted) effects of the neighboring genes are similar; a possible justification for the use of di in pλ is to acknowledge the biological importance of “hub” genes with large di. Although some smoothness over a network is expected, depending on the specific type of the network and application, the exact relationships or the effects of di may still be debatable. Similar to Enet, the first term is used for variable selection while the second smoothes the parameters over the network. Two possible limitations with Li and Li’s penalty are: first, determining two tuning parameters (λ1, λ2) is computationally more intensive than choosing just one, and the presence of two tuning parameters poses a challenge in developing efficient algorithms, such as in identifying solution paths; second, because the second term enforces prior (or βi ≈ βj), it may fail even if (or |βi| = |βj|) but with opposite signs; the latter case is biologically reasonable, such as when one of two neighboring genes is up-regulated while the other is down-regulated in expression.
2.2 New Method
Here we propose a novel penalty, which is a sum of grouped penalties, each in the form of the Lγ-norm of the two coefficients for a pair of neighboring nodes in a given network. This penalty also allows the user to choose a weight for each node, which is to be shown under special cases to realize different types of shrinkages, enforcing various prior relationships among βi’s. For each node i with degree di, define wi = g(di, γ) as its weight, possibly depending on di and γ; for example, we will consider three specific choices: i) , ii) wi = di, and iii) , which lead to three different types of smoothing on the parameters as shown in Corollary 2. Our proposed penalty is
| (1) |
where γ′ satisfies 1/γ′ + 1/γ = 1 with γ > 1. Each term p(βi, βj) is essentially a weighted Lγ-norm of vector (βi, βj)′, and hence pλ(β; λ, w) is convex in β. Note that the constant 21/γ′ can be dropped, but its presence reduces 21/γ′p(βi, βj) to the L1-norm of (βi, βj)′ if |βi| = |βj|.
Some main motivations for pλ are the following. First, each term of pλ is a (weighted) grouped penalty, encouraging both βi and βj to be equal to zero simultaneously (Yuan and Lin 2006; Zhao et al 2006), which is in agreement with our assumption that two neighboring genes in a network should be more likely to (or not to) participate in the same biological process simultaneously; its theory is provided in Theorem 1 below. Second, the weight wi, is adopted to encourage , or |βi| ≈ |βj| for two neighboring genes i ~ j, similar to (but different from) that targeted by Li and Li (2008); this is supported by Corollary 2 below for some special cases, though a general theory still lacks. In addition, if γ = 1, pλ reduces to the L1-penalty in the Lasso. Third, a larger γ is chosen to more strongly smooth , |βi| or |βi|/di over the network; a special case is that, with wi = di, as γ → ∞, , which most strongly encourages |βi| = |βj| (Zhao et al 2006).
2.3 Shrinkage Effects
In general, there is no closed form for our proposed PLSE, as for most other PLSEs. Below we first characterize a relationship between any PLSE β̂ and OLSE β̃ in some special but yet illustrative situations.
Lemma 1
For the model E(Y) = Xβ, if pλ(β) is differentiable at β̂, we have
| (2) |
Below we consider a simple case with only two predictors, which are linked in a two-node network. We assume that the variables have been normalized such that Σkyk = 0, Σkxki = 0 and . Without loss of generality, we assume w1 = w2 = 1, ρ = corr(x1, x2) = Σkxk1xk2, and β̂1β̂2 ≠ 0. From Lemma 1, it is easy to derive our proposed PLSEs as
| (3) |
with λ′ = λ21/γ′. If |β̂1| ≠ |β̂2|, the shrinkage effects on the two parameters are unbalanced. For example, if |β̂1| > |β̂2|, β̂1 is scaled by a factor smaller than that for β̂2, and at the same time β̂1 is shifted by a factor smaller than that for β̂2. This unbalanced shrinkage is more severe for a larger γ: as γ → ∞, |β̂1|γ − 1/(|β̂1|γ + |β̂2|γ)1/γ′ → 1 while |β̂2|γ − 1/(|β̂1|γ + |β̂2|γ)1/γ′ → 0; hence, the scaling factor for the smaller β̂2 tends to 1 while that for β̂1 is always less than 1. In the case with ρ = 0 if |β̂1| > |β̂2|,
demonstrating an extreme case of unbalanced shrinkage; the above can be also directly derived from (2). In addition, if sign(β̂1) ≠ sign (β̂2) and ρ ≠ 0, there is a double shrinkage or penalization in that an PLSE is both shifted and scaled towards 0. For example, suppose that β̃1 > 0 and β̂1 > 0 while |β̃2 < 0 and β̂2 < 0. Then the second term in the numerator of each β̂j has an opposite sign to that of β̃j; that is, in addition to be scaled by a factor less than 1, each β̃j is shifted towards 0. On the other hand, if sign(β̂1) = sign (β̂2), the shrinkage effect by scaling is compensated by that of being shifted away from 0, because the second term in the numerator of each β̂j has the same sign as that of β̃j.
The double shrinkage is not unique to our proposed PLSE; in fact, the Enet estimate is also doubly shrunken:
| (4) |
Note that, even if ρ = 0, β̂j,E is still doubly penalized, whereas the double penalization on the network-based β̂j vanishes. Similarly, we conjecture that the Li and Li’s estimator is also doubly penalized. In contrast, the Lasso estimator is only shifted towards 0, at least for the case of two predictors.
Zou and Hastie (2005) used a scaling factor 1 + λ2, corresponding to the scaling factor in (4) with ρ = 0, to alleviate the bias effect of double penalization for Enet; the same strategy was adopted by Li and Li (2008) for their estimator. It is not clear how to correct for our proposed estimator, partly because the scaling factor depends on the estimate itself in a complicating way, though we study a simple proposal in Section 5.
2.4 Grouped Variable Selection
To establish statistical properties of grouped variable selection for our proposed method, we first derive a result for a general design matrix X, then illustrate the effect through an orthonormal X. To simplify notations, denote by V(i,j) (or V−(i,j)) the vector containing (or excluding the ith and jth components of vector V; M(i,j;k,l) (or M(i,j;−k,−l)) the submatrix of M with the ith and jth rows and including (or excluding) columns k and l; similarly, we can define other forms of submatrices and vectors. The proof of the following theorem is given in Web Appendix A.
Theorem 1
For any edge i ~ j, a sufficient condition for β̂i = β̂j = 0 is
| (5) |
and a necessary condition is
| (6) |
It is most illustrative to consider a simple situation with X′X = I.
Corollary 1
Assume that X′X = I. For any edge i ~ j, a sufficient condition for β̂i = β̂j = 0 is
| (7) |
and a necessary condition is
| (8) |
Corollary 1 clearly demonstrates the effect of grouped variable selection: if the (weighted) average of the OLSEs β̃i and β̃j, in terms of their weighted Lγ′-norm, is small enough, as compared to the tuning parameter λ, then PLSEs β̂i and β̂j are forced to be exactly 0 simultaneously. This is in contrast to other non-grouped penalties. For example, in the orthonormal case, the Lasso estimate β̂i,L = sign(β̃i) (|β̃i| − λ)+; it is obvious that the two Lasso estimates are shrunken or thresholded individually: even if the (weighted) average of their OLSEs is small enough, it is possible that only one of the two PLSEs is exactly 0.
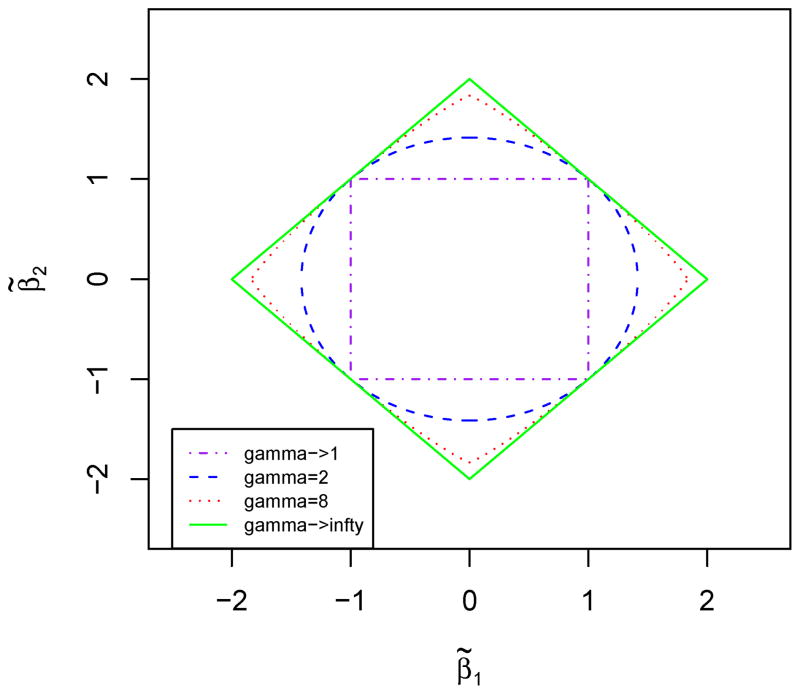
Corollary 1 also sheds light on the effect of the choice of γ in the Lγ-norm. For example, (7) becomes i) max(|β̃i|, |β̃j|) ≤ λ if γ → 1; ii) if γ = 2; iii) |β̃i|8/7 + |β̃j|8/7 ≤ 2λ8/7 if γ = 8; iv) |β̃i| + |β̃j| ≤ 2λ if γ → ∞. As shown in Fig 1, (7) is easier to be satisfied (i.e. covering a larger area) for a larger γ, leading to stronger grouped variable selection, more likely to result in β̂i = β̂j = 0, if the same λ is used.
Figure 1.
Constraint regions of (β̃1, β̃2) yielding β̂1 = β̂2 = 0 for various γ and λ = 1. This figure appears in color in the electronic version of this article.
2.5 Grouping Effects
We demonstrate the grouping effects of our proposed penalty: under some conditions, for two neighboring nodes, their non-zero regression coefficient estimates are shrunken to be closer to each other as the tuning parameter or their correlation increases. Web Appendix B shows some complicated shrinkage effects in a network, from which we have the below result for a simple case as used in the later simulation and in Li and Li (2008).
Corollary 2
Consider a subnetwork containing a transcription factor (TF, say gene 0) connected to each of its target genes i = 1, …, K; there is no connection between any two target genes and between this subnetwork and any other parts of the network. We further assume that the K target genes have the same β̂1 = … = β̂K, and that pλ(β) is differentiate at β̂0 and β̂1 with β̂0β̂1 > 0.
- If , then
(9) - if wi = di, then
(10) -
if , then
(11) where is the sample correlation between gene 1 and the TF, and ||Y||2 is the L2 norm of the vector of response values.
From the foregoing corollary, the grouping effect is evident for γ > 1 as , |β̂1 − β̂0|, or |β̂1 − β̂0/d0| is upper-bounded by a number that decreases as either γ or ρ1,0 increases. Note that d1 = 1. Although Corollary 2 is obtained under a simplified (but still meaningful) scenario, it is more general than Theorem 1 of Li and Li (2008); more importantly, it clearly suggests the necessity of choosing appropriate weights wi’s: different choices of weights realize different types of shrinkage on and smoothness among coefficients βi’s. The choice of weights for a penalty with a parameter appearing multiple times is important yet barely studied, as acknowledged but not elaborated by Zhao et al (2006). In addition, the corollary also suggests that a larger γ leads to a stronger grouping effect: , |β̂1 − β̂0|or |β̂1 − β̂0/d0| is forced to decrease more as γ increases. It is easy to see that, for instance, if , as γ → ∞, the left hand side of (9) tends to 1, while 1/γ′ decreases and tends to 1; because the right hand side tends to 0 as λ → ∞, we must have (or one of them is 0) for a λ large enough, which is the maximum grouping effect of using the L∞-norm (Zhao et al 2006; Bondell and Reich 2008).
2.6 Computation
We propose using a slightly modified generalized Boosted Lasso (GBL) algorithm of Zhao and Yu (2004) for implementation, which works like the stagewise regression (Efron et al 2004), involving only a coordinate-wise search and repeated calculations of the objective function; see Web Appendix C for details. The GBL algorithm yields an approximate solution path β̂(λ), a set of PLSEs β̂ at a finite number of tuning parameter values λ = λ(0) ≥ λ(1) ≥ … ≥ λ(r) ≥ 0. In particular β̂(λ) = β̂(λ(0)) ≈ 0 for any λ ≥ λ(0), and β̂(λ) = β̂(λ(r)) for any λ ≤ λ(r). For other λ(k) ≥ λ ≥ λ(k+1), we can linearly interpolate β̂(λ) between β̂(λ(k)) and β̂(λ(k+1)).
Although cross-validation and other model selection methods might be used, in this article we simply use an independent tuning dataset to calculate the prediction mean squared error (PMSE) for the response at each λ(k) for k = 0, …, r; if λ(k0) minimizes the PMSEs, then we choose β̂(λ(k0)) as the final parameter estimates, which in turn determines a subset of selected predictors (with non-zero estimates). Note that, rather than using λ, we can also parametrize the tuning parameter by fraction s = s(λ) = pλ{β̂(λ)}/pλ{β̂(λ(r))}, where β̂(λ(r)) is a minimally penalized estimate if λ(r) > 0; otherwise β̂(λ(r)) = β̂(0) is an OLSE (which may not be unique). In this way, the tuning parameter 0 ≤ s ≤ 1 facilitates comparison of various methods with different penalty functions.
3. Simulations
3.1 Simulation Set-ups
Our simulation set-ups closely followed that of Li and Li (2008): simulated data were generated from a linear model with iid noises ; each network consisted of nTF subnetworks, each with a TF and its 10 regulatory target genes. For each set-up, we considered two cases: one with nTF = 3 TFs and the other with nTF= 10 TFs, corresponding to a “small p, small n” situation with p = 33 < n and a “large p, small n” with p = 110 > n respectively. For each case with nTF = 3 TFs, two subnetworks were informative (p1 = 22) with non-zero βi’s while the other one with βi = 0 (p0 = 11); for nTF = 10, four subnetworks were informative (p1 = 44) while the other six were not (p0 = 66).
Each predictor was marginally distributed as N(0,1), and to mimic a regulatory relationship, the predictor of each target gene and the TF had a bivariate normal distribution with correlation ρ = 0.7; conditional on the TF, the target genes were independent. In set-up 1, we considered the case with the correct prior assumption: if i ~ j. Specifically, i) for p = 33, we had
and for p = 110, we had
The remaining set-ups perturbed the condition on the equality of the weighted coefficients for two neighboring genes. Set-up 2 was the same as set-up 1 except that the signs βi’s of the first three target genes in each subnetwork were flipped to their opposites; for example, for the first subnetwork, the first three target genes’ βi’s were changed from to . Set-up 3 was the same as set-up 1 except that in the second (for p = 33), or in the second and fourth (for p = 110) subnetworks, the in the target genes’ coefficients was replaced by 10. Set-up 4 was the same as set-up 3 except that the signs of the coefficients of the first three target genes in each informative subnetwork were flipped, as in set-up 2. Set-up 5 was similar to Set-up 1 except that five out of 10 targets of each informative TF had βi = 0, hence the prior belief of co-appearance of a TF and its targets was not correct.
There were n = 50, 50 and 200 cases in each training, tuning and test datasets respectively. The training data were used to fit the model, while the tuning data were used to select the tuning parameters. For Lasso Enet, Li and Li, a grid search over an equally spaced 100 points (between 0 and 1 as parametrized as fraction s) was used to determine λ or λ1, while another grid search over an equally spaced 100 points between 0 and 0.05 was used to find an optimal λ2 for Enet and Li and Li’s method; for our method, we searched over all λi as returned from GBL. The test data were used to calculate PMSE for the response; we also calculated the number of zero estimates of the βi for informative and non-informative genes, denoted as q1 and q0 respectively.
For each set-up, 100 independent datasets were generated, from which the means and standard deviations (SDs) were calculated for each PMSE, q1 and q0; note that the Monte Carlo standard error was simply SD/10.
3.2 Simulation Results
For the traditional situation with small p, first, in terms of PMSE, our method with γ = 8 was a consistent winner, closely followed by our method with γ = 2 (Table 1). Li and Li’s method, Enet and Lasso performed similarly. Second, in terms of variables selection, a similar conclusion holds: our method performed best, removing a comparable number of noise variables while keeping most of the informative variables as compared to the other three methods.
Table 1.
Means (SDs) of prediction mean squared error (PMSE), number of removed informative variables (q1) and number of removed non-informative variables (q0) from 100 simulated datasets. The minimal mean PMSEs for each set-up are boldfaced.
| Set-up | Methods |
P1 = 22, p0 = 11
|
P1 = 44, p0 = 66
|
||||
|---|---|---|---|---|---|---|---|
| PMSE | q1 | q2 | PMSE | q1 | q0 | ||
| 1 | Lasso | 54.0 (9.0) | 5.2 (2.1) | 6.9 (2.7) | 166.6 (32.9) | 20.1 (2.5) | 53.9 (6.4) |
| Enet | 54.8 (10.3) | 5.4 (2.3) | 7.1 (2.7) | 164.3 (29.3) | 10.6 (9.2) | 31.4 (24.0) | |
| Li&Li | 54.6 (9.7) | 5.4 (2.3) | 7.1 (2.7) | 154.6 (28.3) | 5.0 (7.6) | 15.1 (21.2) | |
| γ = 2 | 46.7 (7.6) | 0.2 (0.5) | 10.1 (1.2) | 138.1 (32.3) | 3.2 (3.7) | 60.0 (5.4) | |
| γ = 8 | 43.6 (6.8) | 0.0 (0.0) | 10.2 (1.0) | 132.0 (35.8) | 3.2 (4.3) | 60.0 (4.8) | |
| γ = ∞ | 46.3 (8.0) | 0.1 (0.8) | 9.8 (1.2) | 162.9 (46.6) | 7.3 (5.9) | 56.6 (6.8) | |
| 2 | Lasso | 59.2 (12.7) | 11.6 (3.1) | 9.6 (2.1) | 160.8 (39.0) | 30.2 (4.0) | 61.1 (4.2) |
| Enet | 58.2 (12.6) | 12.4 (3.5) | 9.8 (2.0) | 161.1 (45.5) | 29.0 (8.5) | 57.8 (15.1) | |
| Li&Li | 58.2 (12.8) | 12.3 (3.3) | 9.8 (2.0) | 161.7 (44.7) | 26.0 (11.7) | 52.1 (22.3) | |
| γ = 2 | 57.2 (11.9) | 2.8 (3.1) | 9.0 (2.7) | 161.2 (44.3) | 16.8 (8.2) | 61.3 (5.1) | |
| γ = 8 | 55.5 (11.4) | 2.1 (3.2) | 8.1 (3.2) | 169.9 (57.4) | 19.6 (10.1) | 60.2 (7.5) | |
| γ = ∞ | 59.1 (21.6) | 2.9 (4.1) | 7.3 (3.4) | 186.0 (67.6) | 23.6 (10.0) | 61.0 (7.4) | |
| 3 | Lasso | 45.4 (8.3) | 6.6 (2.4) | 7.1 (2.8) | 115.3 (24.2) | 23.2 (3.2) | 56.5 (6.1) |
| Enet | 45.7 (8.1) | 6.7 (2.5) | 7.3 (2.8) | 118.0 (27.8) | 18.2 (9.5) | 45.0 (22.2) | |
| Li&Li | 45.6 (7.8) | 6.7 (2.4) | 7.3 (2.8) | 116.5 (22.5) | 11.6 (11.1) | 29.6 (27.1) | |
| γ = 2 | 41.7 (7.2) | 1.8 (2.4) | 10.2 (1.1) | 107.5 (25.9) | 7.6 (5.3) | 61.0 (4.4) | |
| γ = 8 | 39.9 (7.4) | 1.0 (2.6) | 10.2 (1.2) | 107.5 (37.2) | 8.8 (6.6) | 61.4 (3.9) | |
| γ = ∞ | 42.6 (7.7) | 2.9 (3.5) | 9.9 (1.4) | 129.6 (41.9) | 13.2 (7.6) | 59.2 (6.3) | |
| 4 | Lasso | 50.6 (11.4) | 12.5 (3.7) | 9.5 (2.4) | 117.5 (31.6) | 31.7 (3.9) | 61.6 (4.2) |
| Enet | 49.2 (9.7) | 13.4 (3.7) | 9.7 (2.3) | 115.8 (30.9) | 31.5 (7.1) | 60.5 (12.0) | |
| Li&Li | 49.2 (9.9) | 13.3 (3.6) | 9.8 (2.3) | 113.6 (28.6) | 29.1 (10.0) | 56.8 (18.5) | |
| γ = 2 | 50.3 (12.0) | 4.7 (4.0) | 8.8 (2.9) | 122.4 (34.3) | 18.4 (8.6) | 61.9 (5.4) | |
| γ = 8 | 48.5 (10.1) | 3.6 (4.1) | 8.4 (3.0) | 135.6 (51.2) | 22.7 (11.1) | 61.7 (6.1) | |
| γ = ∞ | 51.2 (18.8) | 4.1 (4.5) | 7.4 (3.5) | 148.6 (56.1) | 26.7 (10.5) | 62.0 (6.3) | |
| Set-up | Methods |
p1 = 12, p0 = 21
|
p1 = 24, p0 = 86
|
||||
| PMSE | q1 | q0 | PMSE | q1 | q0 | ||
| 5 | Lasso | 38.8 (9.5) | 3.1 (1.5) | 15.5 (3.4) | 112.7 (29.8) | 11.4 (2.6) | 75.5 (5.8) |
| Enet | 38.0 (9.7) | 3.3 (1.7) | 15.7 (3.5) | 112.9 (29.0) | 10.8 (3.8) | 71.1 (17.4) | |
| Li&Li | 37.6 (8.4) | 3.2 (1.6) | 15.7 (3.5) | 111.8 (27.9) | 9.4 (5.0) | 61.7 (29.3) | |
| γ = 2 | 37.9 (7.4) | 0.2 (0.5) | 10.7 (1.8) | 110.3 (28.4) | 4.2 (3.1) | 66.6 (5.8) | |
| γ = 8 | 38.1 (6.8) | 0.1 (0.5) | 10.0 (2.1) | 113.3 (29.4) | 5.7 (3.6) | 67.1 (7.0) | |
| γ = ∞ | 40.7 (9.6) | 0.5 (1.1) | 9.3 (3.8) | 131.1 (38.1) | 7.8 (4.3) | 68.2 (9.2) | |
When p was large, in terms of PMSE, there were mixed results in terms of which method was the winner: for set-ups 1 and 3, in which βi’s in the same subnetwork shared the same sign, our method, especially with γ = 8, was the clear winner; on the other hand, for set-up 2, our method with γ = 2 performed similarly as the other three methods, whereas in set-up 4, Li and Li’s was the winner, followed by Enet and Lasso. It is noted that our method with γ = 8, and especially with γ = ∞, might not perform well. This was somewhat surprising, and could be related to the more severe double penalization and stronger grouped variable selection with a larger γ as analyzed earlier. This point was confirmed by observing larger biases of the resulting estimates found in Table 2.
Table 2.
Mean, variance and mean squared error (MSE) of regression coefficient estimate from 100 simulated datasets.
| Set-up | P | Methods |
β1 = 5
|
β2 = 1.58
|
β11 = 1.58
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Var | MSE | Mean | Var | MSE | Mean | Var | MSE | |||
| 1 | 33 | Lasso | 6.07 | 4.72 | 5.81 | 1.33 | 1.35 | 1.40 | 1.38 | 1.36 | 1.39 |
| Enet | 6.42 | 4.05 | 6.03 | 1.36 | 1.54 | 1.57 | 1.40 | 1.37 | 1.38 | ||
| Li&Li | 6.43 | 4.03 | 6.02 | 1.36 | 1.53 | 1.56 | 1.39 | 1.36 | 1.38 | ||
| γ = 2 | 4.76 | 0.68 | 0.51 | 1.03 | 0.72 | 0.78 | 1.63 | 0.86 | 0.58 | ||
| γ = 8 | 4.29 | 0.29 | 0.80 | 1.48 | 0.35 | 0.36 | 1.48 | 0.34 | 0.35 | ||
| γ = ∞ | 3.63 | 0.44 | 2.32 | 1.60 | 0.60 | 0.60 | 1.56 | 0.64 | 0.63 | ||
| 1 | 110 | Lasso | 5.28 | 8.69 | 8.69 | 1.43 | 2.43 | 2.42 | 1.26 | 2.53 | 2.61 |
| Enet | 3.79 | 4.76 | 6.18 | 1.82 | 1.86 | 1.90 | 1.47 | 1.71 | 1.71 | ||
| Li&Li | 5.00 | 1.69 | 1.67 | 1.74 | 1.33 | 1.34 | 1.51 | 1.31 | 1.31 | ||
| γ = 2 | 3.82 | 1.02 | 2.41 | 1.51 | 1.29 | 1.28 | 1.53 | 1.66 | 1.64 | ||
| γ = 8 | 3.47 | 0.79 | 3.12 | 1.50 | 1.02 | 1.02 | 1.60 | 1.24 | 1.23 | ||
| γ = ∞ | 2.13 | 1.33 | 9.57 | 1.64 | 2.08 | 2.06 | 1.75 | 2.34 | 2.35 | ||
| Set-up | P | Methods |
β1 = 5
|
β2 = −1.58
|
β11 = 1.58
|
||||||
| Mean | Var | MSE | Mean | Var | MSE | Mean | Var | MSE | |||
| 2 | 33 | Lasso | 3.65 | 2.39 | 4.19 | −.09 | 0.23 | 2.44 | 0.83 | 0.99 | 1.54 |
| Enet | 4.36 | 2.14 | 2.53 | −.08 | 0.22 | 2.47 | 0.84 | 1.18 | 1.72 | ||
| Li&Li | 4.23 | 2.19 | 2.75 | −.08 | 0.22 | 2.47 | 0.83 | 1.14 | 1.69 | ||
| γ = 2 | 2.56 | 1.38 | 7.30 | −.17 | 0.54 | 2.52 | 1.17 | 0.85 | 1.01 | ||
| γ = 8 | 2.47 | 1.37 | 7.75 | −.28 | 0.96 | 2.63 | 1.28 | 0.90 | 0.98 | ||
| γ = ∞ | 2.18 | 1.95 | 9.88 | −.43 | 1.07 | 2.38 | 1.44 | 1.15 | 1.16 | ||
| 2 | 110 | Lasso | 2.54 | 4.31 | 10.31 | 0.13 | 0.34 | 3.25 | 0.94 | 1.92 | 2.32 |
| Enet | 2.87 | 4.85 | 9.32 | 0.16 | 0.41 | 3.44 | 0.95 | 1.96 | 2.34 | ||
| Li&Li | 2.88 | 3.97 | 8.43 | 0.16 | 0.43 | 3.45 | 0.93 | 1.72 | 2.13 | ||
| γ = 2 | 1.37 | 0.79 | 14.00 | 0.22 | 0.28 | 3.53 | 1.12 | 1.56 | 1.76 | ||
| γ = 8 | 1.07 | 0.80 | 16.22 | 0.24 | 0.36 | 3.67 | 1.05 | 1.53 | 1.80 | ||
| γ = ∞ | 0.47 | 0.46 | 20.98 | 0.23 | 0.39 | 3.65 | 1.06 | 2.05 | 2.30 | ||
Nevertheless, in term of variable selection, even in the large p case, our method consistently won. For any of the first four set-ups and with any of the three choices of γ, no matter how it worked in terms of PMSE, our new method always retained a larger number of informative variables while removing more or a similar number of noise variables as compared to the other methods. For set-up 5, although our method tended to delete fewer variables, it removed a much smaller proportion of informative ones among the deleted ones. Overall, our method with γ = 2 performed most consistently. Somewhat surprisingly, in spite of the closeness between the two penalties (see e.g. Fig 1), our method with γ = 8 worked distinguishingly better than that with γ = ∞.
4. Example
4.1 Data
We applied the methods to a microarray gene expression dataset with glioblastoma patients (Horvath et al 2006). As a primary malignant brain tumor of adults, glioblastoma is one of the most lethal with a median survival time from diagnosis only at 15 months in spite of various treatments. The data consisted of two independent sets drawn from two studies, called Set 1 and Set 2 respectively; as in Li and Li (2008), we used 50 and 61 samples with observed survival times from the two sets, and took the log survival time (in years) as the response. The gene expression profiles were measured on Affymetrix HG-U133A arrays, and processed by the RMA method (Irizarry et al 2003).
Wei and Li (2007) compiled a network of 1668 genes from 33 KEGG pathways, which was used here. Using R Bioconductor library hgu133a, we identified a subset of 1523 genes among the 1668 genes that were present on HG-U133A arrays. In our analyses only these 1523 genes were used. In the resulting network, there were 6865 edges in total; the distribution of the node degrees ranged from 1 to 81, with the mean at 9 and the three quartiles at 2, 4 and 11 respectively.
4.2 Analysis
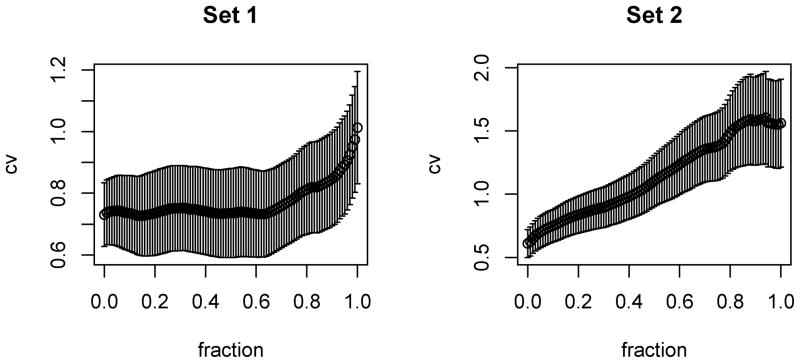
First, as in Li and Li (2008), we used Set 1 to build a model, then evaluated its predictive performance using Set 2. It turned out that the intercept-only model gave the smallest PMSE, as supported by Lasso, Enet and our method. We reasoned that perhaps the second set was somewhat different from the first one, and thus combined the two together before randomly splitting into training, tuning and test data; again it turned out that the intercept-only model was the best, as selected by Lasso and our method. As shown in Fig 2, it seems that the expression profiles were not predictive of survival time for Set 2, while they were more informative for Set 1. Hence, in the following we only used the data of Set 1.
Figure 2.
PMSEs (± SE) versus tuning parameter s based on ten-fold CV for Lasso for the two sets of the glioblastoma data.
For the small sample size n = 50, there would be a large variability associated with any PMSE for any method, suggesting limited utility in comparing PMSEs for various methods. Therefore, we focused on gene selection. We excluded one outlier with log survival time less than −3, while all other ones were between −2 and 2. We randomly split the data into training and tuning parts with n = 30 and 19 respectively.
We ran Lasso, Enet and our proposed method with γ = 2 and . While Li and Li (2008) were able to analyze the data based on a sophisticated and efficient implementation of their method, the straightforward implementation with data augmentation suggested therein failed because it required too large computer memory for a sample size of n + p and p predictors. With λ2 = 0 selected by the tuning data, Enet gave the same results as Lasso. Lasso and Enet selected 11 genes: ADCYAP1R1, ARRB1, CACNA1S, CTLA4, FOXO1, GLG1, IFT57, LAMB1, MPDZ, SDC2, and TBL1X; there was no edge linking any two of the 11 genes. By comparison, our method selected 17 genes: ADCYAP1, ADCYAP1R1, ARRB1, CCL4, CCS, CD46, CDK6, FBP1, FBP2, FLNC, FOXO1, GLG1, IFT57 MAP3K12, SSH1, TBL1X, and TUBB2C; there were three edges linking five of the 17 genes: FOXO1 was connected to FBP1 and FBP2, and ADCYAP1 connected to ADCYAP1R1. A literature search revealed that FOXO1, as a member of forkhead transcription factors, is linked to glioblastoma (Choe et al 2003; Seoane et al 2004). Another gene, CDK6, identified by our method, but missed by the Lasso and Enet, was also related to glioblastoma: it is a well-known oncogene, and in particular, glioblastoma multiforme is characterized by copy number changes (Ruano et al 2006) and elevated expression (Lam et al 2000) of CDK6. In addition, according to the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (Forbes et al 2006), among the selected genes by the three methods, IFT57, CDK6 and MAP3K12 have cancer-related mutations, of which, only IFT57 was detected by Lasso and Enet.
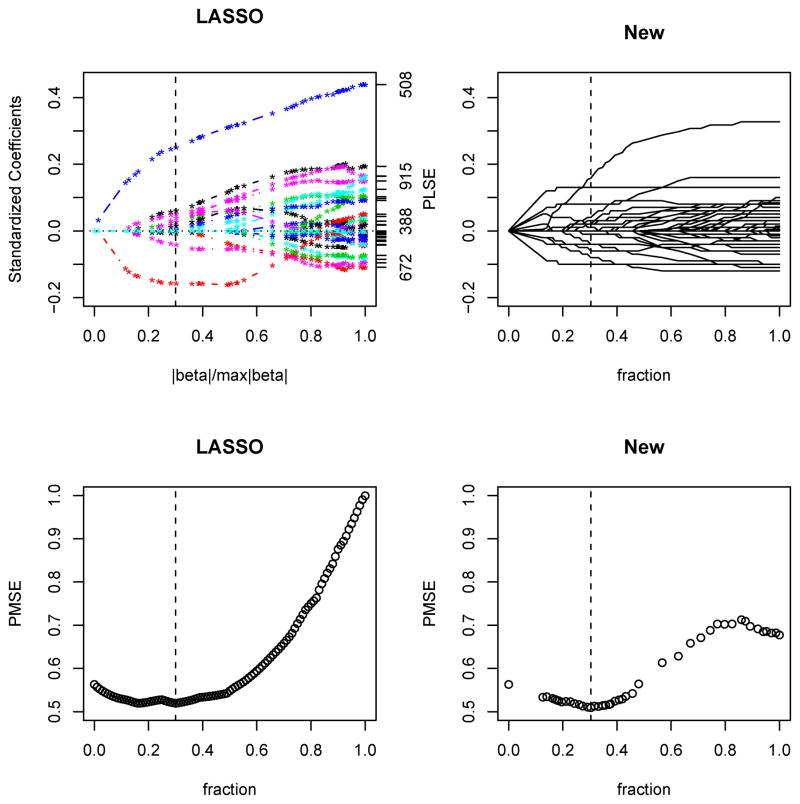
Figure 3 gives the solution pathways for the regression coefficient estimates by the two methods. Most of the genes had their estimates at or close to 0. FOXO1 had the largest coefficient estimate at 0.25 by Lasso and at 0.16 by our method. In the fitted model by Lasso, there was only one other gene, TBL1X (with a coefficient estimate at −0.16), that retained an estimate larger than 0.05 in absolute value. For our methods, in addition to FOXO1, other genes with the absolute values of their coefficient estimates larger than 0.05 were ADCYAP1 (at 0.08), CDK6 (−0.06) GLG1 (0.08) and ADCYAP1R1 (0.13). It is also clear that the regression coefficient estimates from our method tended to be smaller than the Lasso estimates, in agreement with the earlier observation in the simulation study that our PLSE seemed to be shrunken more than that of the Lasso. Another explanation is related to the penalty function used: for example, FOXO1 had 9 direct neighbors, most of which had zero coefficient estimates; our penalty would smooth that of FOXO1 more towards zero, the value shared by most of its neighbors; in contrast, the Lasso penalty would not have this kind of effects because there was no penalty term to link that of FOXO1 to its neighbors’. Finally, confirmed by the PMSEs estimated from the tuning data (Fig 3), by both Lasso (and Enet) and our method, parsimonious models with fewer and smaller coefficient estimates gave smaller PMSEs than the larger (and less penalized) ones; the minimum PMSEs selected by the Lasso and our method were 0.52 and 0.51 respectively, both obtained at the tuning parameter value s = 0.3.
Figure 3.
Solution paths or PMSE versus tuning parameter s based on tuning data for Lasso and our new method based on a linear model for the first set of the glioblastoma data. This figure appears in color in the electronic version of this article.
As an alternative, we also applied the three methods to the semi-parametric Cox proportional hazards model (PHM) by approximating a PHM as a linear model. They yielded results similar to their earlier ones (from the linear model) respectively, though our method seemed to be more stable in gene selection; see Web Appendix D for details.
5. Extensions
Here we consider a few possible modifications and extensions. First, if there are singletons that are not connected to any other genes in a network, to facilitate gene selection, we can add an L1-penalty for the coefficients of the singletons. Note that, partly due to constant 21/γ′ in the network-based penalty, each grouped Lγ-penalty is in the same scale of an L1-penalty, and hence only a single regularization parameter λ is needed for both types of the penalties. Second, if we do not have a network structure for a cluster of functionally related genes, we may treat them as fully connected to each other and apply the same network-based penalty. Alternatively, we can treat them as a separate group and apply an Lγ-norm of all the genes in the group with γ > 1 as a penalty; this strategy is effective if we believe a priori that the genes in the group are likely to be all relevant or irrelevant together. On the other hand, if we only have vague knowledge on the group, we can simply apply the L1-norm to the group.
Third, as is true in the first four simulation set-ups, if a TF is involved in a biological process, our penalty encourages simultaneous appearance of the TF and all its targets in a regression model; in practice, however, it may be that only a subset of the target genes are involved. To construct a penalty function to allow such a case, we can add an L1-penalty for the coefficients of the target genes. This is related to, but different from, the hierarchical penalty as proposed by Zhao et al (2006). Table 3 lists the results for the five simulation set-ups using the extended methods just described. It is somewhat surprising that the new penalty in general worked quite well; in particular, as compared with the previous grouped penalty, the new penalty gave smaller PMSEs, and retained a slightly larger number of informative genes while removing almost the same number of noise genes. However, the above new penalty depended on correctly selecting and thus further penalizing the target genes (because most of the target genes either were not informative or had much smaller coefficients than that of the TFs). In general, for any given network, it is unknown which subset of the genes should be imposed with an additional L1-penalty. When we simply applied an additional L1-penalty on each gene, the resulting performance was worse than using the network-based penalty alone: not only the PMSEs were larger, but more informative genes would be removed (Table 3).
Table 3.
Means (SDs) of prediction mean squared error (PMSE), number of removed informative variables (q1) and number of removed non-informative variables (q0) with an additional L1 penalty for only target genes (L1-T) or all the genes (L1-A), or with different weights in network-based regression from 100 simulated datasets. The minimal mean PMSEs for each set-up are boldfaced.
| Set-up | Methods |
p1 = 44, p0 = 66
|
||
|---|---|---|---|---|
| PMSE | q1 | q0 | ||
| 1 | γ = 2, L1-T | 127.2 (23.7) | 7.1 (2.8) | 56.0 (6.8) |
| γ = 8, L1-T | 122.9 (26.5) | 3.2 (2.8) | 60.4 (4.7) | |
| γ = ∞, L1-T | 128.3 (27.3) | 4.7 (2.9) | 59.3 (4.6) | |
| γ = 2, L1-A | 148.2 (32.5) | 7.6 (3.6) | 59.2 (5.6) | |
| γ = 8, L1-A | 144.8 (36.6) | 5.8 (4.2) | 59.1 (5.6) | |
| γ = ∞, L1-A | 161.8 (48.5) | 9.0 (4.8) | 57.5 (6.1) | |
| γ = 2, wi = di | 190.3 (56.9) | 11.6 (5.4) | 59.4 (6.4) | |
| γ = 8, wi = di | 188.2 (38.6) | 13.5 (4.4) | 57.8 (6.4) | |
| γ = ∞, | 114.1 (22.5) | 0.1 (0.8) | 55.3 (10.7) | |
| 2 | γ = 2, L1-T | 136.3 (28.4) | 15.7 (4.7) | 60.9 (4.3) |
| γ = 8, L1-T | 151.7 (36.4) | 16.0 (7.2) | 60.9 (5.0) | |
| γ = ∞, L1-T | 154.9 (45.4) | 16.9 (7.0) | 60.5 (5.2) | |
| γ = 2, L1-A | 163.2 (47.2) | 20.6 (7.0) | 60.9 (5.4) | |
| γ = 8, L1-A | 176.0 (56.2) | 22.0 (8.8) | 61.3 (6.0) | |
| γ = ∞, L1-A | 186.3 (60.6) | 25.1 (8.7) | 61.7 (6.3) | |
| γ = 2, wi = di | 199.5 (69.5) | 27.7 (9.0) | 62.4 (5.7) | |
| γ = 8, wi = di | 242.1 (96.4) | 31.6 (11.4) | 61.0 (6.5) | |
| γ = ∞, | 127.2 (24.3) | 3.0 (4.9) | 56.4 (8.5) | |
| 3 | γ = 2, L1-T | 92.6 (18.1) | 10.0 (2.7) | 58.7 (5.1) |
| γ = 8, L1-T | 98.2 (25.0) | 7.2 (4.8) | 61.4 (4.1) | |
| γ = ∞, L1-T | 99.7 (23.0) | 8.2 (4.2) | 60.3 (4.9) | |
| γ = 2, L1-A | 113.5 (31.6) | 11.9 (4.6) | 60.3 (4.9) | |
| γ = 8, L1-A | 115.5 (36.5) | 11.2 (5.8) | 60.9 (5.2) | |
| γ = ∞, L1-A | 129.2 (44.3) | 14.2 (6.3) | 59.9 (6.5) | |
| γ = 2, wi = di | 153.5 (43.8) | 18.3 (6.5) | 61.9 (6.4) | |
| γ = 8, wi = di | 137.7 (58.9) | 16.9 (5.3) | 58.7 (6.3) | |
| γ = ∞, | 86.2 (18.9) | 0.7 (2.2) | 56.7 (9.3) | |
| 4 | γ = 2, L1-T | 100.0 (20.0) | 17.5 (4.8) | 61.2 (4.0) |
| γ = 8, L1-T | 115.7 (31.7) | 17.7 (8.2) | 61.3 (6.1) | |
| γ = ∞, L1-T | 116.4 (31.6) | 18.7 (7.6) | 60.7 (6.3) | |
| γ = 2, L1-A | 125.3 (33.4) | 22.3 (7.3) | 61.7 (5.6) | |
| γ = 8, L1-A | 140.8 (45.5) | 25.3 (8.8) | 62.6 (5.4) | |
| γ = ∞, L1-A | 144.3 (45.7) | 26.9 (8.3) | 62.9 (5.0) | |
| γ = 2, wi = di | 163.2 (54.8) | 31.4 (8.6) | 63.8 (3.5) | |
| γ = 8, wi = di | 202.6 (65.7) | 36.2 (10.7) | 62.4 (6.3) | |
| γ = ∞, | 92.6 (18.5) | 4.8 (6.2) | 57.9 (7.2) | |
| Set-up | Methods |
p1 = 24, p0 = 86
|
||
| PMSE | q1 | q0 | ||
| 5 | γ = 2, L1-T | 90.3 (17.4) | 3.8 (1.9) | 68.9 (4.7) |
| γ = 8, L1-T | 100.6 (22.7) | 3.2 (2.7) | 66.3 (5.1) | |
| γ = ∞, L1-T | 102.1 (27.0) | 4.0 (2.7) | 65.7 (7.1) | |
| γ = 2, L1-A | 114.6 (34.1) | 5.8 (3.3) | 69.8 (5.6) | |
| γ = 8, L1-A | 120.1 (34.8) | 6.6 (3.8) | 68.7 (7.3) | |
| γ = ∞, L1-A | 133.4 (43.8) | 8.3 (4.1) | 69.7 (8.7) | |
| γ = 2, wi = di | 151.8 (50.2) | 10.4 (4.1) | 72.9 (8.8) | |
| γ = 8, wi = di | 173.8 (103.2) | 11.9 (6.6) | 71.1 (10.8) | |
| γ = ∞, | 83.2 (15.1) | 0.3 (1.1) | 59.7 (8.1) | |
Fourth, we investigated the robustness of the network-based regression with an incorrect choice of weights. In the simulation set-ups 1–4, the correct weights should be or ; instead, we used much smaller wi = di for all five set-ups. As shown in Table 3, it is interesting to note that the proposed method still performed better than or as well as Lasso, Enet and Li and Li’s method in terms of variable selection, but it often gave much larger PMSEs, presumably resulting from larger biases of the regression coefficients due to over-shrinkage (e.g., of β0 of a TF towards β1 of its targets as shown in Corollary 2).
Finally, due to the grouped penalty, our proposed network-based regression performs well in variable selection, but may suffer from a large bias of PLSE β̂ (see Table 2). To reduce the possible bias of PLSE, a simple strategy is to choose a larger weight for a hub or more important gene, for example, , even when the correct as in simulation set-ups 1–2. We used in the simulations with γ = ∞; i.e., p(βi, βj) = max(|βi|/di, |βj|/dj). By comparing Tables 1 and 3, we can see that, although the method tended to keep slightly more genes (both informative and non-informative ones), it gave consistently smaller PMSEs than that from using the other weight in the simulations.
6. Discussion
We have proposed a penalized regression method to incorporate network structures of predictor variables, motivated by applications arising from analyzing genomic and proteomic data to account for gene networks. As biological data on gene networks and pathways have been rapidly accumulating, e.g. fueled by high-throughput DNA-protein and protein-protein interaction experiments, there is an increasingly rich source of network information available. On the other hand, there is always the issue of high-noise levels and small sample sizes associated with most genomic and proteomic studies, prompting the use of biological knowledge, such as embedded in gene networks, to improve analysis efficiency. Hence, in spite of its fairly recent developments, we expect to see more uses of gene networks in other domains, such as classification and clustering. In particular, as shown in our example for Cox regression, it seems straightforward, at least in principle, to apply our proposed network-based penalty to generalized linear models (e.g. Zhu and Hastie 2004) and other classification models (Zhu et al 2009), though more work, especially in developing fast and accurate computational algorithms, is needed.
Our proposed method seemed to work best in terms of variable selection: as compared to its competitors, it removed more or an equal number of noise variables while retaining more informative variables across a range of simulation scenarios; this good performance can be explained by its capability of grouped variable selection. Meanwhile, the message for its predictive performance is somewhat mixed: sometimes it did not work as well as Li and Li’s method. As our analysis suggested, it could be due to overly biased parameter estimates, possibly resulting from double penalization, especially when the L∞-norm was used. This is one of the weaknesses of the proposed penalty, though we have observed that using larger weights to reduce bias might be productive. Finally, although the prior assumption on the similarity of (weighted regression coefficients is reasonable for some applications, e.g. in eQTL mapping (Pan 2009), it is in general unclear how the parameters should be smoothed over a network and any such specific prior assumption needs to be validated by experimental data. Nevertheless, even if the prior assumption does not hold, by the bias-variance tradeoff, it may still gain by penalization (or smoothing), and the tuning parameters will balance the tradeoff; this point was supported by our simulation results. Furthermore, the introduction of the weights in the penalty offers some flexibility to realize various types of smoothing and shrinkage over a network. In practice, to deal with the unknown smoothness structure in a network and to optimize performance, we can adaptively choose the weights and Lγ-norm by treating them as tuning parameters based on cross-validation or independent tuning data. Certainly more studies are warranted in these directions.
Supplementary Material
Acknowledgments
WP would like to thank Hongzhe Li and Zhi Wei for kindly providing the KEGG network data, Feng Tai for help with the expression data, and Hongzhe Li and Melanie Wall for helpful discussions. The authors thank the two referees, an AE and the Editor (N. Wang) for constructive and helpful comments that led to a substantial improvement; in particular, section 5 was added under the suggestions of the reviewers. This research was partially supported by NIH grant GM081535; in addition, WP and BX by NIH grant HL65462, and XS by NSF grants IIS-0328802 and DMS-0604394.
Footnotes
Web Appendices A–D referenced in Sections 2.4–2.6 and 4.2 are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- Allen C, Vongpunsawad S, Nakamura T, James CD, Schroeder M, Cattaneo R, Giannini C, Krempski J, Peng KW, Goble JM, Uhm JH, Russell SJ, Galanis E. Retargeted oncolytic measles strains entering via the EGFRvIII receptor maintain significant antitumor activity against gliomas with increased tumor specificity. Cancer Res. 2006;66:11840–11850. doi: 10.1158/0008-5472.CAN-06-1200. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondell HD, Reich BJ. Simultaneous regression shrinkage, variable selection, and supervised clustering of predictors with OSCAR. Biometrics. 2008 doi: 10.1111/j.1541-0420.2007.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, Inge L, Smith BL, Sawyers CL, Mischel PS. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2004;32:407–499. [Google Scholar]
- Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, Stratton MR. Cosmic 2005. Br J Cancer. 2006;94:318–22. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand AE, Vounatsou P. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics. 2003;4:11–25. doi: 10.1093/biostatistics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Hoerl AE, Kennard RW. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Shu Q, Lee Y, Scheck AC, Liau LM, Wu H, Geschwind DH, Febbo PG, Kornblum HI, Cloughesy TF, Nelson SF, Mischel PS. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proceedings of National Academy of Sciences. 2006;103:17402–1707. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Di Tomaso E, Ng HK, Pang JC, Roussel MF, Hjelm NM. Expression of p19INK4d, CDK4, CDK6 in glioblastoma multiforme. Br J Neurosurg. 2000;14:28–32. doi: 10.1080/02688690042870. [DOI] [PubMed] [Google Scholar]
- Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24:1175–1118. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- Pan W. Network-based multiple locus linkage analysis of expression traits. Division of Biostatistics, University of Minnesota; 2009. Available at http://www.biostat.umn.edu/rrs.php as Research Report 2009-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano Y, Mollejo M, Ribalta T, Fiaqo C, Camacho FI, Gsmez E, de Lope AR, Hernandez-Moneo JL, Martmnez P, Melindez B. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massagui J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the LASSO. Journal of the Royal Statistical Society, B. 1996;58:267–288. [Google Scholar]
- Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J Royal Statist Soc B. 2006;68:49–67. [Google Scholar]
- Zhao P, Yu B. Tech Rept. Dept of Statistics; UC-Berkeley: 2004. Boosted Lasso. [Google Scholar]
- Zhao P, Rocha G, Yu B. Grouped and hierarchical model selection through composite absolute penalties. Annals of Statistics. 2006 (to appear) [Google Scholar]
- Zhu J, Hastie T. Classification of gene microarrays by penalized logistic regression. Biostatistics. 2004;5:427–443. doi: 10.1093/biostatistics/5.3.427. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Shen X, Pan W. Network-based support vector machine for classification of microarray samples. BMC Bioinformatics. 2009;10(Suppl 1):S21. doi: 10.1186/1471-2105-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. JRSS-B. 2005;67:301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.