Abstract
The hedgehog (HH) signaling pathway is a highly regulated signaling pathway that is important not only for embryonic development, tissue patterning, and organogenesis but also for tissue repair and the maintenance of stem cells in adult tissues. In the adult hematopoietic system, HH signaling regulates intrathymic T-cell development, and it is one of the survival signals provided by follicular dendritic cells to prevent apoptosis in germinal center B cells. HH signaling is required for primitive hematopoiesis; however, conflicting data have been reported regarding the role of the HH pathway in adult hematopoiesis. Inappropriate activation of the HH signaling pathway occurs in several human cancers, including hematological neoplasms. Emerging data demonstrate abnormal HH pathway activation in chronic lymphocytic leukemia/small lymphocytic lymphoma, plasma cell myeloma, mantle cell lymphoma, diffuse large B-cell lymphoma, ALK-positive anaplastic large cell lymphoma, chronic myelogenous leukemia, and acute leukemias. In these neoplasms, HH signaling promotes proliferation and survival, contributes to the maintenance of cancer stem cells, and enhances tolerance or resistance to chemotherapeutic agents. Here, we review current understanding of HH signaling, its role in the pathobiology of hematological malignancies, and its potential as a therapeutic target to treat malignant hematological neoplasms.
The Hedgehog Signaling Pathway
Hedgehog (HH) proteins were first identified in Drosophila as secreted signaling proteins1 that have an essential role in embryonic development, determining the anteroposterior orientation of developing structures in the Drosophila embryo and larva.2 There is a single Hh gene in Drosophila, but three homologs have been found in vertebrates: sonic hedgehog (SHH), Indian hedgehog (IHH), and desert hedgehog (DHH).3 Notably, the three HH ligands activate the same signal transduction pathway, but they regulate different organ systems.3–5 The SHH gene is expressed in the central nervous system, lung, tooth, gut, and hair follicle.6–10 IHH is involved in endochondral bone formation,11 and DHH is expressed mostly in gonads.12
HH proteins undergo a maturation process before their active forms can be secreted from cells and activate HH signaling. The C-terminal domain has proteolytic activity and autocleaves the ligand molecule, generating an N-terminal signaling peptide. This ligand peptide is modified at both ends, esterified with cholesterol moiety at the C-terminal end, and palmitoylated with amide-linked palmitate at the N-terminal end.13,14 The bilipidated ligands form multimers, which leave the ligand-producing cell via dispatched (Disp), a 12-transmembrane protein, and are transported via glypicans and megalin to the major cell receptor, patched (Ptc in Drosophila; PTCH in humans).15
The two known human homologs, PTCH1 and PTCH2, are both composed of 12 transmembrane domains and two extracellular loops that have HH ligand binding sites. The PTCH1 gene is located at 9q22.1-q31 and encodes a 1447-amino-acid glycoprotein.16 The PTCH2 gene is located at 1p34.1 and encodes a 1203-amino-acid protein.17 PTCH2 and PTCH1 have different intracellular N- and C-terminal domains.18 PTCH1 and PTCH2 are considered to have different functions based on their different expression during development of the epidermis.17 PTCH1 mutations19 are associated with nevoid basal cell carcinoma syndrome (also known as Gorlin's syndrome, an autosomal dominant disorder characterized by multiple basal cell carcinomas and skeletal defects, among other findings),20 medulloblastoma, and meningioma; PTCH2 mutations, although rare, have been reported in medulloblastoma and basal cell carcinoma.17
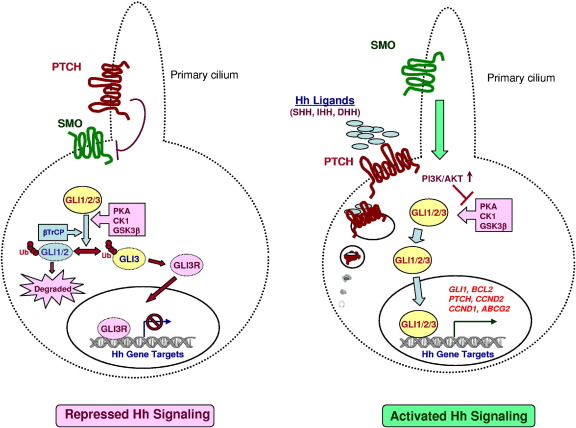
In vertebrates, primary cilium is the site where activation of HH signaling occurs. A schematic representation of HH signaling pathway activation is shown in Figure 1. In the absence of HH ligand, PTCH inhibits the smoothened protein (SMO); this protein has seven transmembrane domains, an extracellular N-terminus, and an intracellular C-terminus, with homology to G protein-coupled receptors (GPCRs).21,22 Binding of HH ligand to PTCH results in endocytosis of the PTCH-ligand complex, followed by migration of uninhibited SMO to the primary cilium. For localization of SMO to the cilia, a short motif C-terminal to the last transmembrane domain is required.23 β-Arrestin was reported to mediate this process.24 In the cilium, uninhibited SMO transduces the signal to the cytoplasm via intraflagellar transport proteins, with glioma-associated oncogene homolog (GLI) proteins as major targets. The GLI proteins regulate target gene expression by direct association with a consensus binding site located in the promoter region of the target genes.25 Activation of canonical HH signaling has profound effects on the stability of GLI family of zinc-finger transcription factors. In the absence of HH ligands, the three isoforms of GLI transcription factors (ie, GLI1, 118 kDa; GLI2, 175 kDa; and GLI3, 174 kDa) are ubiquitinated by β-transducin-repeat containing protein (β-TrCP), which is a phosphorylation-dependent E3 ubiquitin ligase. Ubiquitination of GLI proteins requires priming phosphorylations by protein kinase A (PKA), casein kinase 1 (CK1), and glycogen synthase kinase-3β (GSK-3β).26–28 After ubiquitination, GLI1 is completely degraded and cleared from the cell by the proteasomal system, whereas GLI2 and GLI3 are partially degraded to form low-molecular weight repressor forms (GLI2R or GLI3R), which migrate to the nucleus, bind to GLI-binding consensus sequences, and repress transcription of HH target genes. For GLI2, however, it was shown that degradation and clearance predominated over formation of the repressor form, which is unlike GLI3 and makes GLI2 more an activator than a repressor of HH signaling.26–28 Activation of canonical HH signaling leads to abrogation of phosphorylation-dependent proteolytic degradation of all GLI proteins, increasing their cytoplasmic and nuclear levels and so also increasing transcription of HH target genes: GLI1, PTCH1, PTCH2, BCL2, ABCG2, CCND2, FGF4, VEGFA (previously VEGF), PAX6, PAX7, PAX9, jagged 1 (JAG1), and MYCN, among others.
Figure 1.
Schematic diagram of repressed (left) and activated (right) canonical HH signaling pathways. In the absence of HH ligands, PTCH localizes to the primary cilium and prevents SMO from entering the cilium. This results in phosphorylation-dependent ubiquitination of GLI transcription factors, leading to either degradation and clearance from the cell (GLI1 and GLI2) or partial proteolysis of GLI3 to form a repressive form (GLI3R) lacking C-terminal activation domains. GLI3R enters into the nucleus, binds GLI binding consensus sequences, and represses transcription of HH target genes. In the presence of HH ligands (SHH, IHH, DHH), PTCH is internalized, allowing migration of uninhibited SMO to the primary cilium. This results in increased stability and nuclear accumulation of GLI transcription factors and transcription of HH target genes (GLI1, BCL2, PTCH, CCND2, CCND1, and ABCG2, among others).
Several regulatory steps control HH pathway signaling. HH-interacting protein (HIP) competes with PTCH for HH binding and attenuates HH signaling.29 In mammalian cells, the suppressor of fused protein (SUFU) is a key regulator of the HH pathway, especially at the level of GLI. SUFU inhibits nuclear translocation of GLI proteins by sequestering them in the cytoplasm and thus inhibiting their transcriptional activity.30 In addition to cytoplasmic sequestration, SUFU also localizes to the nucleus and there suppresses GLI-driven transcription. Nuclear SUFU associates with GLI-binding sites on the DNA and represses gene transcription by recruiting the SAP18-mSin3 complex to GLI promoters.31 Additional modes of regulating HH signaling include PKA-mediated phosphorylation of GLI1 on residue Thr374, in the vicinity of the nuclear localization sequence, resulting in its cytoplasmic sequestration,32 and increased stability and nuclear accumulation of GLI1 induced by GTPase HRas and GTPase NRas via the RAS-RAF-MEK signaling axis.33
The HH Signaling Pathway in Hematopoiesis and Lymphopoiesis
Vertebrate hematopoiesis has two different phases: primitive (embryonic) hematopoiesis and definitive (adult) hematopoiesis. Primitive or embryonic hematopoiesis is characterized by the commitment of embryonic mesoderm to hematopoietic precursors such as embryonic (or primitive) erythrocytes and macrophages.34 The requirement of IHH in primitive hematopoiesis was established by Dyer et al,35 who showed that IHH ligand secreted by the endoderm was sufficient to induce formation of hematopoietic and endothelial cells. That IHH is a signal playing a key role in the development of the earliest hematovascular system was also supported by another study, in which IHH-deficient embryonic stem cell lines were unable to form blood islands and showed abnormal vascular morphology.36
Definitive hematopoiesis is autonomously initiated with the formation of multipotent hematopoietic stem cells in the aorta-gonad-mesonephros region.37 In contrast to primitive hematopoiesis, there is conflicting data regarding the role of HH pathway in definitive hematopoiesis. A study with zebrafish embryo with loss-of-function SHH mutation showed defects in adult hematopoietic stem cell formation.38 Several other studies have been conducted on the function of SMO in hematopoiesis, but their conclusions are fairly inconsistent. Gao et al39 showed that SMO-deficient hematopoietic stem cells were not affected in terms of differentiation, self-renewal, and regeneration of the immune system; they also demonstrated that SMO-deficient hematopoietic stem cells preserved the gene expression signature specific for hematopoietic stem cells. Similar results were observed by Hofmann et al40 and Dierks et al.41 Zhao et al,42 however, demonstrated that SMO-deficient mice clearly had a defect in long-term hematopoietic stem cell function in primary and secondary transplants and that therefore SMO is required for hematopoietic stem cell renewal in vivo. More recently, a Gli1null mouse study by Merchant et al43 showed decreased proliferation of hematopoietic stem cells and myeloid progenitors. Gli1null mice had more long-term hematopoietic stem cells that were more quiescent and showed increased engraftment after transplantation. In contrast, myeloid development was adversely affected, with decreased in vitro colony formation, decreased in vivo response to granulocyte colony-stimulating factor (G-CSF), and impaired leukocyte recovery after treatments with cytotoxic drugs. Results from the Merchant et al43 study support the notion that GLI1 is a regulator of self-renewal of hematopoietic stem cells and drives myeloid cell proliferation: loss of GLI1 impairs hematopoiesis in situations of stress and impairs the ability to recover after cytotoxic injury or respond to stimulatory cytokines.
The thymus is the organ of T-cell development and maturation. In the embryonic stage, progenitor cells are seeded to the thymus from fetal liver; after birth, progenitor cells migrate from the bone marrow to the thymus. In the adult, common lymphoid progenitors enter the blood vessels in the corticomedullary junction or medulla and undergo progressive differentiation of pre-T-cells (thymocytes) as double negative (DN; CD4−CD8−), double positive (DP; CD4+CD8+), and finally single positive (SP; either CD4+CD8− or CD4−CD8+) mature T-cell subsets. The DN stage is further divided into four smaller subsets, based on the expression status of CD25 and CD44: DN1 cells are CD25−CD44+, DN2 cells are CD25+CD44+, DN3 cells are CD25+CD44−, and DN4 cells are CD25−CD44−.44 T-cell lineage specification and T-cell receptor β (TCR-β) rearrangement occur in the DN2 and DN3 stages.45 Pre-TCR signaling is required for survival, proliferation, and differentiation to the DP stage, and is negatively regulated by SHH and IHH signaling.46–50 During transition from DP to SP, both positive and negative selections occur. This developmental program is regulated by the thymic stroma, and one way in which the stroma signals to developing thymocytes is by production of HH ligands.
The different components of the HH signaling pathway have been reported to be expressed in the thymus and to play an important role regulating the proliferation and differentiation of thymocytes. HH ligands are produced by stromal, thymic, epithelial, and dendritic cells.46,47,51–53 The fact that thymocytes also express HH ligands (especially IHH in DP-stage thymocytes) was demonstrated by Outram et al.46 PTCH and SMO are expressed mainly by immature thymocytes. SMO shows a peak expression at DN2, with a decrease in subsequent stages.47 GLI1 shows the highest expression at DN2 and DN3, then decreases to its minimal level at DP. GLI1 is required for normal differentiation of DN3.54 GLI2 is expressed at DN1 and DN2 at the highest level, down-regulated at DN3, and up-regulated at DN4.48 The expression of GLI3 is highest at DN1 and DN4 in fetal thymocytes, but is not detected in adult thymocytes. GLI3 is necessary for DN to DP differentiation, especially after pre-TCR signaling.55 HH signaling is necessary for proliferation and survival of DN1 and differentiation to DN2 (early stages of thymocyte development).46,53,56 As differentiation progresses, immature thymocytes move away from the cortical-medullary junction (which has high expression of HH proteins), resulting in a gradual loss of HH stimulation. These events coincide with the induction of TCR-β rearrangement. Generation of a productive TCR-β rearrangement results in pre-TCR assembly, surface expression, and signaling.53 HH signaling also influences late stages of T-cell development and T-cell activation.57–60
Sacedón et al61 reported that SHH ligand is produced by follicular dendritic cells within germinal centers of lymphoid follicles, and that SHH ligand protects germinal center B cells from apoptosis. They also showed that germinal center B cells express the HH receptors PTCH and SMO, and that their survival is altered after inhibition of the HH signaling pathway. These data indicate that HH signaling has a role in the biology of lymphoid follicles and is involved in rescuing or protecting germinal center cells from apoptosis.
HH proteins are expressed in all lymphoid organs and are secreted as soluble ligands by stromal cells; IHH is secreted in bone marrow and SHH in lymph nodes and spleen.61,62 The expression levels of HH-related proteins (HH ligands and the transcription factors GLI1, GLI2, and GLI3) were explored in reactive lymph nodes using immunohistochemistry (for HH ligands, a polyclonal antibody that recognizes all three HH proteins was used).63 HH ligands were detected in the cytoplasm of follicular dendritic cells, endothelial cells, and macrophages, but not in germinal center lymphocytes. GLI1 expression was not seen, but GLI2 expression was detected in some centroblasts and centrocytes surrounding follicular dendritic cells within germinal centers supporting the existence of an HH ligand-mediated stroma-lymphocyte interaction, as shown by Sacedón et al.61 Germinal center lymphocytes, follicular dendritic cells and endothelial cells were negative for GLI1. Expression of GLI3 was detected in follicular dendritic cells and endothelial cells, but not in germinal center lymphocytes. In the thymus, GLI3 was expressed by cortical and medullary stromal cells (including epithelial cells, endothelial cells, and histiocytes), but thymocytes were negative.64
Aberrant Activation of HH Signaling in Cancer
The initial observation of GLI1 involvement in glioma development was the first indication of the role of HH signaling in tumorigenesis.65 Later, a definitive link between the HH pathway and cancer was established by the identification of heterozygote mutations affecting PTCH and resulting in abnormal activation of the HH signaling pathway in sporadic basal cell carcinomas, rhabdomyosarcomas, and neural tumors.66 In addition, association of HH signaling with other tumors such as pancreatic adenocarcinoma and lung, gastric, prostate, and esophageal cancer was also identified.67–69
In cancer, several mechanisms by which HH signaling can be aberrantly activated have been described. The first is through mutation of members of the HH signaling leading to hyperactivation of the pathway (eg, PTCH1 mutations in Gorlin's syndrome, sporadic basal cell carcinomas, and a significant fraction of medulloblastomas; SUFU-inactivating mutations in medulloblastoma and rhabdomyosarcoma; and activating mutation of SMO in basal cell carcinomas).16,66,70–73 In these cases, HH signaling activation is ligand-independent, and HH deregulation is thought to be the initiating pathogenic event. The second model displays excessive and/or inappropriate expression of HH ligands (ligand-dependent signaling activation), resulting in autocrine and/or paracrine stimulation of cancer cells. An additional model in which the tumor cells secrete HH ligands activating HH signaling in the stroma has also been suggested. In this model, in response to the tumor-secreted HH ligands, stromal cells secrete growth factors contributing to proliferation and/or survival of the tumor cells. This model of HH activation has been shown in pancreatic and colorectal cancers.74 The autocrine pattern of HH activation has been described in some epithelial cancers (breast, pancreas, lung, prostate, and gastrointestinal tract).69,75–79 In contrast to cancers with mutational activation of HH signaling, in the ligand-dependent model HH deregulation is not the initiating pathogenic event but still plays a role in the pathobiology of the tumor by contributing to tumor maintenance and growth.
Aberrant Activation of the HH Signaling Pathway in Hematological Malignancies
Although inappropriate activation of the HH signaling pathway has been shown in some cancers, the contribution of the HH signaling pathway in hematological malignancies has not been thoroughly examined. Emerging data indicate that the HH pathway is active in some hematopoietic malignancies and that this activation contributes to the biology of these neoplasms. To date, hematopoietic malignancies have not been found to harbor activating or inhibiting mutations in HH signaling components, and current data suggest that in most hematopoietic neoplasms HH signaling does not play an independent role in tumor initiation but instead contributes to tumor maintenance, growth, and resistance to chemotherapy. There are also data suggesting that HH signaling can participate in cancer stem cell survival and/or expansion in some malignant hematopoietic neoplasms, such as plasma cell myeloma, chronic myelogenous leukemia and acute leukemias.
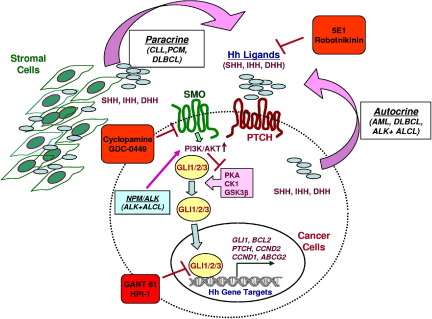
The mechanisms by which HH signaling is activated in hematological malignancies are not clear at this time. A schematic representation of mechanisms of activation of HH signaling in hematological cancers is shown in Figure 2.
Figure 2.
Schematic diagram of mechanisms of activation of HH signaling in hematological cancers. Both paracrine and autocrine activation of HH signaling has been observed in hematological cancers (paracrine mainly in CLL and plasma cell myeloma, and both paracrine and autocrine in DLBCL). An autocrine mechanism of activation has also been reported in acute myeloid leukemia and in ALK-positive anaplastic large cell lymphoma (ALKL+ALCL). Activation of PI3K-AKT (activation of other oncogenic pathways not shown) contributes to the activation of HH signaling. NPM-ALK in ALK-positive ALCL enhances the stability and consequently the transcriptional activity of GLI1 through activation of PI3K-AKT signaling. Examples of HH inhibitors, either natural products or synthetic compounds are shown (red boxes). 5E1 and robotnikinin bind and inactivate HH ligands, cyclopamine and GDC-0499 bind and inactivate SMO, and GANT-61 and HPI-1 interfere with GLI transcriptional activity.
A paracrine pattern in which stromal cells secrete HH ligands contributing to the activation of HH signaling in the tumor cells has been described in low-grade B-cell neoplasms, including chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL), an indolent form of B-cell lymphoma, as well as in plasma cell myeloma, and has been suggested in mantle cell lymphoma (MCL).80 An autocrine pattern has been suggested in acute myeloid leukemia and anaplastic lymphoma kinase (ALK)-positive anaplastic large cell lymphoma (ALCL). Both paracrine and autocrine patterns have been found in diffuse large B-cell lymphoma (DLBCL).
GLI1 is located at 12q13, and gains at this locus and/or extra copies of chromosome 12 have been reported in B-cell lymphomas, including splenic and nodal marginal zone lymphoma, DLBCL, and MCL.81–84 We have detected a few extra copies of GLI1, DHH, or SMO genes, caused by chromosomal aneuploidy rather than true gene amplification, in DLBCL cell lines and in patient samples (unpublished data), which may contribute to HH activation but does not fully explain activation of the pathway. Mutations of HH signaling-related genes have not been reported, and it is not known whether specific microRNAs are involved in the activation of HH signaling in malignant lymphomas and leukemias. It has been shown that the microRNA-17/92 cluster synergizes with HH signaling in cerebellar granule cell precursors and in medulloblastoma.85
Recent evidence suggests that activation of HH signaling in cancer in general, but also in hematological neoplasms in particular, results at least in part from integration of multiple deregulated oncogenic signaling inputs in the final signaling effectors of the HH pathway, the GLI transcription factors (see review by Stecca and Ruiz I Altaba).86 Growth factors such as platelet-derived growth factor, epidermal growth factor, and insulin-like growth factor 1 increase activation of HH signaling that is dependent of the activation of PI3K-AKT pathway.87,88 Interplay among NF-κB, RAS-RAF-MEK, TGF-β, notch, and HH signaling is also supported by multiple lines of evidence.86,89 This view of the interconnectivity of numerous oncogenic pathways has important implications for understanding how the major oncogenic pathways interact and also for the development of more effective and focused anticancer therapies.
HH Signaling in B-Cell Neoplasms
Using a c-Myc-driven mouse model that generated a wide range of B-cell malignancies including 40% slow-growing (low-grade) plasmacytomas, Dierks et al80 showed that lymphoma cells undergo apoptosis in the absence of their stromal microenvironment and that, to some extent, they are protected from apoptosis in the presence of HH ligands or SMO agonists. They also showed that the suppression of IHH secretion by the stromal cells using an IHH short hairpin RNA construct substantially decreased lymphoma cell growth; in the same study, similar results were obtained using human malignant lymphomas.80 These findings suggest that HH ligands secreted by the tumor microenvironment (stromal cells from bone marrow, lymph node, and spleen) are survival factors for low-grade B-cell lymphomas and plasma cell myeloma and support a paracrine role for HH signaling in low-grade B-cell neoplasms. This is of interest because inhibition of HH signaling could provide a novel strategy in these neoplasms.
Two additional studies have documented the role of HH signaling in CLL.90,91 Hegde et al90 found that the PTCH-SMO-GLI axis is functional in CLL cells, and that inhibition of HH signaling by cyclopamine abrogated bone marrow stroma-induced survival of CLL cells. These authors also reported a correlation between expression levels of GLI1 and GLI2 in tumor samples with disease progression and clinical outcome.90 In the other study, Desch et al91 found that activation of HH signaling via SMO-independent GLI1 is more relevant in CLL than the classical PTCH-SMO-GLI axis, and that the observed CLL sensitivity to cyclopamine (reported by Hegde et al90) is in part due to nonspecific off-target effects.91 They based these conclusions on observations that primary CLL cells were highly sensitive to GLI inhibition (as determined by using the GLI antagonist GANT61 or by silencing GLI1 gene expression using shRNA) and that silencing SMO did not influence CLL viability. Although the two sets of findings seem discordant, both of these studies support the functionality of HH signaling and the possibility of targeting this pathway in CLL. Studies with more potent and more specific SMO inhibitors than cyclopamine, such as vismodegib (GDC-0449), may help to further elucidate the role of SMO in the activation of HH signaling in this neoplasm.
More recently, HH signaling has been found to be active and functional in DLBCL, the most frequent aggressive subtype of B-cell lymphoma.92 High expression of the HH ligand receptors SMO and PTCH, and also the transcription factors GLI1 and GLI2, but not GLI3, have been detected in tumor samples and in DLBCL cell lines.63,92 Inhibition of HH signaling using the SMO inhibitor KAAD-cyclopamine resulted in decrease of cell number and colony formation in DLBCL cells, in particular in those DLBCL cell lines with high expression of SMO.92 Similar results in cell viability have been obtained using other SMO inhibitors as well as gene-silencing approaches (GLI1 and GLI2). It was shown that inhibition of HH signaling predominantly induced cell cycle arrest in DLBCL cell lines of germinal center type [cells carrying the t(14;18) rearrangements] and mainly apoptosis in those of activated B-cell type (ABC type). The resistance to apoptosis observed in germinal center DLBCL cells can be overcome using BCL2 functional inhibitors, suggesting that the functionality and expression levels of BCL2 are important in determining susceptibility to apoptosis induced by SMO inhibitors.92
Variable cytoplasmic expression levels of HH ligands (SHH, IHH, and DHH), including expression of the HH precursor (full length) and the N-terminal HH peptide, have been detected in DLBCL tumors and cell lines.63,92,93 Moreover, the secreted HH N-terminal peptide was detected in culture medium collected from DLBCL cell lines, and blocking secreted HH ligands with the monoclonal antibody 5E1 or silencing the expression of SHH and IHH genes by siRNA resulted in decreased expression levels of GLI1 associated with decreased expression of its downstream target genes.92 These data indicate that HH ligands are synthesized, processed, and secreted by DLBCL cells and that the canonical HH ligand-PTCH-SMO-GLI axis is functional in DLBCL cells. These data provide evidence of the existence of an autocrine HH signaling loop in this lymphoma type. In the same study, however, expression of HH ligands or secreted HH N-terminal peptide were not detected in cell lysates or in the culture medium collected from primary CLL cells, respectively.92 The gain of cell-autonomous activation of the HH pathway that is seen in DLBCL cells (in contrast to CLL cells, in which HH ligands are provided by stromal cells) likely represents a survival and/or proliferative advantage for the lymphoma cells and is an indication, at least to some degree, of stromal independence.
In addition to the above-mentioned autocrine model, a paracrine pattern of activation of HH signaling in DLBCL has also been demonstrated.94 Coculturing DLBCL cell lines with the human bone marrow stroma cell line HS5 in transwell experiments (without direct contact between DLBCL and stromal cells) resulted in activation of HH signaling in the tumor cells, that can be blocked with SMO inhibitors. In these experiments, it was also shown that stroma-induced activation of HH signaling contributes to the increase of chemotolerance of DLBCL cells. This stroma-induced chemotolerance induced by activation of HH signaling in the tumor cells was mediated by up-regulation of the expression of antiapoptotic proteins such as BCL2, BCL2A1, and BCL-XL, as well as by up-regulation of the ATP binding cassette sub-family G member 2 (ABCG2). Functional inhibition of SMO, ABCG2, and BCL2 abrogated stroma-induced chemotolerance in DLBCL cells, highlighting the potential adjuvant therapeutic value of these strategies in DLBCL and other cancers with abnormal activation of HH signaling. In the same study, ABCG2 was found to be a direct transcriptional target of HH signaling.94
In another report, Hegde et al95 documented the role of HH signaling in MCL, an aggressive subtype of B-cell lymphoma, showing that PTCH, SMO, GLI1, and GLI2 are expressed both in MCL cell lines and in primary MCL cells. Furthermore, they also demonstrated that treatment with exogenous HH ligands increased cell proliferation of one of the MCL cell lines (JVM2); inhibition of HH signaling in that same cell line resulted in decrease in cell proliferation.95
Recently, expression of GLI transcription factors was studied in classic Hodgkin's lymphoma.64 GLI3 was found universally expressed in Hodgkin-Reed Sternberg cells, but was not expressed or was significantly less frequently expressed in other lymphomas. Hodgkin-Reed Sternberg cells express relatively low levels of other HH-related molecules, including HH ligands and the transcription factors GLI1 and GLI2. GLI3 is also expressed by stromal cells in lymph node (including follicular dendritic cells, histiocytes, and endothelial cells) and in thymus (histiocytes, endothelial cells, and epithelial cells), but not by mature or immature lymphocytes.63,64 Hager-Theodorides et al55,96 showed that GLI3 in thymic stromal cells regulates T-cell selection and differentiation of thymocytes. Whether GLI3 plays a similar regulatory role in the modulation of the T-cell response and microenvironment in classic Hodgkin's lymphoma remains to be determined.
Activation of the HH pathway has also been documented in plasma cell myeloma. As noted above, HH ligands secreted by tumor microenvironment are survival factors for plasma cell myeloma.80 PTCH, SMO, and GLI1 were found to be overexpressed in both human plasma cell myeloma cell lines and primary tumor samples, compared with normal plasma cells and B-cells.97 Peacock et al97 found that HH signaling is required for self-renewal and maintenance of the tumor stem cell compartment (CD138−, CD19+) in plasma cell myeloma. Moreover, inhibition of HH pathway activity induced the loss of tumor stem cell compartment, accompanied by the induction of plasma cell differentiation and inhibition of clonogenic growth potential.97
HH Signaling in T-Cell Neoplasms
Dysregulated HH signaling may also be important in mature T-cell malignancies. ALK-positive ALCL is an aggressive type of non-Hodgkin's lymphoma of T-cell-null lineage. This lymphoma is characterized by chromosomal aberrations that lead to constitutive activation of ALK. The most common is translocation t(2;5)(p23;q35), which produces a fusion between the nucleophosmin gene (NPM1) and ALK, leading to expression of the NPM-ALK fusion protein.98 HH ligands and GLI1 have been found to be highly expressed in ALK-positive ALCL tumors and cell lines.87 NPM-ALK, through activation of PI3K-AKT, contributes to activation of HH signaling, which in turn contributes to cell proliferation and survival in ALK-positive ALCL and to the oncogenic effect of NPM-ALK.87 An autocrine role for HH signaling in ALK-positive ALCL has also been suggested. Extra copies of the SHH gene, but not of GLI1, have been found In ALK-positive ALCL tumors and cell lines.87
HH Signaling in Chronic Myelogenous Leukemia and Acute Leukemias
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative neoplasm characterized by the presence of the Philadelphia chromosome [t(9;22)] resulting in the expression of the BCR-ABL1 fusion gene. Dierks et al41 showed that HH signaling is activated in BCR-ABL1-positive leukemia stem cells through up-regulation of SMO. Pharmacological inhibition of SMO reduced CML leukemia stem cells in vivo and enhanced time to relapse. They also showed that the development of retransplantable BCR-ABL1-positive leukemias was abolished in the absence of SMO expression. Zhao et al42 reported similar findings. They showed that loss of SMO causes depletion of CML stem cells, whereas constitutive active SMO augments CML stem cell number and accelerates disease.42
Activation of HH signaling was also reported in acute myeloid leukemias by Kobune et al,99 who showed that, in acute myeloid leukemia cell lines, the degree of HH activation was greater in primary CD34+ blasts and in CD34+ cell lines, compared with CD34− blasts.99 HH inhibition (achieved using cyclopamine or by using the monoclonal antibody 5E1 to neutralize HH ligands) induced apoptosis in CD34+ cell lines and sensitized them against the chemotherapeutic agent cytarabine. In that study, cyclopamine treatments failed to affect growth or survival of acute myeloid leukemia cell lines without SMO, supporting the specificity of cyclopamine.99 Lin et al100 reported that expression of HH signaling components is also frequent in human B-cell acute lymphoblastic leukemias, and that the level of pathway activity can be modulated by HH ligand or SMO inhibitors. They also showed, both in vitro and in vivo, that inhibition of HH signaling affects highly clonogenic B-cell acute lymphoblastic leukemia cells primarily by limiting their self-renewal potential. Some reports have also suggested that HH signaling promotes the growth of T-cell acute lymphoblastic leukemia.101,102
Small-Molecule Modulators of HH Signaling
More than 50 compounds have been identified that inhibit HH signaling. Although the mechanisms of action of some of the HH signaling inhibitors are not fully understood, these molecules target several key steps in the process of HH activation (Figure 2). These small-molecule compounds can be categorized as follows: i) HH ligand inhibitors [HH neutralizing antibodies (5E1), and robotnikinin]; ii) SMO antagonists (cyclopamine, the first inhibitor discovered, and its derivatives, as well as synthetic compounds such as GDC-0449, Cur61414, and statins); iii) intraflagellar transport protein inhibitors (HH pathway inhibitors HPI-2, HPI-3, and HPI-4); and iv) direct or indirect GLI transcriptional inhibitors, including GLI antagonists (GANTs). GANT61 interferes with GLI binding to the gene promoters. HPI-1 targets an unclear primary cilium-independent process such as a post-translational modification of the GLI protein and/or interaction between GLIs with a cofactor103 and physalins (physalin F) that indirectly antagonize GLI function.104,105
Conclusions
In summary, emerging data demonstrate abnormal activation of the HH signaling pathway in lymphoid and myeloid neoplasms. Despite the inability of HH signaling to start tumorigenesis in hematological malignancies, HH signaling has been shown to have proliferative and survival roles, contributing to maintenance of the cancer stem-cell component and enhancing tolerance or resistance to chemotherapeutic agents. For these reasons, it will be important to further understand the biology of HH signaling in these neoplasms, and in particular to identify the causative factors for its aberrant activation and deregulation, its crosstalk with other oncogenic signaling pathways, and how HH target genes contribute to maintenance of the malignant phenotype. Although HH signaling pathway blockade alone might be insufficient for the treatment of lymphomas or leukemias, the current available data provide a rationale for using HH pathway antagonists as adjuvant therapeutic agents to increase susceptibility to current chemotherapeutic agents or to potentiate other targeted therapies for hematological neoplasms.
Footnotes
Supported by translational grant funds from the Leukemia & Lymphoma Society (to R.R.S. and F.V.), an NIH Physician-Scientist award (1 K08 CA143151-01 to F.V.), an NIH SPORE lymphoma grant (UT M.D. Anderson Cancer Center Lymphoma SPORE 1P50CA136411-01A1, to F.V.), and a Lauri Strauss Leukemia Foundation grant award (to R.R.S. and F.V.).
C.Y.O. and R.R.S. contributed equally to the present work.
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.J., von Kessler D.P., Parks S., Beachy P.A. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 3.Echelard Y., Epstein D.J., St-Jacques B., Shen L., Mohler J., McMahon J.A., McMahon A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 4.Marigo V., Tabin C.J. Regulation of patched by sonic hedgehog in the developing neural tube. Proc Natl Acad Sci USA. 1996;93:9346–9351. doi: 10.1073/pnas.93.18.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone D.M., Hynes M., Armanini M., Swanson T.A., Gu Q., Johnson R.L., Scott M.P., Pennica D., Goddard A., Phillips H., Noll M., Hooper J.E., de Sauvage F., Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich L.V., Johnson R.L., Milenkovic L., McMahon J.A., Scott M.P. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 7.Bellusci S., Furuta Y., Rush M.G., Henderson R., Winnier G., Hogan B.L. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- 8.Hardcastle Z., Mo R., Hui C.C., Sharpe P.T. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- 9.Litingtung Y., Lei L., Westphal H., Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 10.St-Jacques B., Dassule H.R., Karavanova I., Botchkarev V.A., Li J., Danielian P.S., McMahon J.A., Lewis P.M., Paus R., McMahon A.P. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 11.Vortkamp A., Lee K., Lanske B., Segre G.V., Kronenberg H.M., Tabin C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 12.Bitgood M.J., Shen L., McMahon A.P. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 13.Porter J.A., Ekker S.C., Park W.J., von Kessler D.P., Young K.E., Chen C.H., Ma Y., Woods A.S., Cotter R.J., Koonin E.V., Beachy P.A. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 14.Pepinsky R.B., Zeng C., Wen D., Rayhorn P., Baker D.P., Williams K.P., Bixler S.A., Ambrose C.M., Garber E.A., Miatkowski K., Taylor F.R., Wang E.A., Galdes A. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy R.A., Barth J.L., Chintalapudi M.R., Knaak C., Argraves W.S. Megalin functions as an endocytic sonic hedgehog receptor. J Biol Chem. 2002;277:25660–25667. doi: 10.1074/jbc.M201933200. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R.L., Rothman A.L., Xie J., Goodrich L.V., Bare J.W., Bonifas J.M., Quinn A.G., Myers R.M., Cox D.R., Epstein E.H., Jr, Scott M.P. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 17.Smyth I., Narang M.A., Evans T., Heimann C., Nakamura Y., Chenevix-Trench G., Pietsch T., Wicking C., Wainwright B.J. Isolation and characterization of human patched 2 (PTCH2), a putative tumour suppressor gene inbasal cell carcinoma and medulloblastoma on chromosome 1p32. Hum Mol Genet. 1999;8:291–297. doi: 10.1093/hmg/8.2.291. [DOI] [PubMed] [Google Scholar]
- 18.Motoyama J., Takabatake T., Takeshima K., Hui C. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet. 1998;18:104–106. doi: 10.1038/ng0298-104. [DOI] [PubMed] [Google Scholar]
- 19.Shimkets R., Gailani M.R., Siu V.M., Yang-Feng T., Pressman C.L., Levanat S., Goldstein A., Dean M., Bale A.E. Molecular analysis of chromosome 9q deletions in two Gorlin syndrome patients. Am J Hum Genet. 1996;59:417–422. [PMC free article] [PubMed] [Google Scholar]
- 20.Gorlin R.J., Goltz R.W. Multiple nevoid basal-cell epithelioma, jaw cysts and bifid rib: A syndrome. N Engl J Med. 1960;262:908–912. doi: 10.1056/NEJM196005052621803. [DOI] [PubMed] [Google Scholar]
- 21.Ogden S.K., Fei D.L., Schilling N.S., Ahmed Y.F., Hwa J., Robbins D.J. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456:967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philipp M., Caron M.G. Hedgehog signaling: is Smo a G protein-coupled receptor? Curr Biol. 2009;19:R125–R127. doi: 10.1016/j.cub.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs J.J., Whalen E.J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., Lefkowitz R.J. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki H., Hui C., Nakafuku M., Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 26.Tempe D., Casas M., Karaz S., Blanchet-Tournier M.F., Concordet J.P. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol Cell Biol. 2006;26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y., Bai C.B., Joyner A.L., Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntzicker E.G., Estay I.S., Zhen H., Lokteva L.A., Jackson P.K., Oro A.E. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuang P.T., McMahon A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 30.Monnier V., Dussillol F., Alves G., Lamour-Isnard C., Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- 31.Cheng S.Y., Bishop J.M. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng T., Chi S., Zhang X., Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. J Biol Chem. 2006;281:9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- 33.Stecca B., Mas C., Clement V., Zbinden M., Correa R., Piguet V., Beermann F., Ruiz I Altaba A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller G., Lacaud G., Robertson S. Development of the hematopoietic system in the mouse. Exp Hematol. 1999;27:777–787. doi: 10.1016/s0301-472x(99)00024-7. [DOI] [PubMed] [Google Scholar]
- 35.Dyer M.A., Farrington S.M., Mohn D., Munday J.R., Baron M.H. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development. 2001;128:1717–1730. doi: 10.1242/dev.128.10.1717. [DOI] [PubMed] [Google Scholar]
- 36.Byrd N., Becker S., Maye P., Narasimhaiah R., St-Jacques B., Zhang X., McMahon J., McMahon A., Grabel L. Hedgehog is required for murine yolk sac angiogenesis. Development. 2002;129:361–372. doi: 10.1242/dev.129.2.361. [DOI] [PubMed] [Google Scholar]
- 37.Medvinsky A., Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 38.Gering M., Patient R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Gao J., Graves S., Koch U., Liu S., Jankovic V., Buonamici S., El Andaloussi A., Nimer S.D., Kee B.L., Taichman R., Radtke F., Aifantis I. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell. 2009;4:548–558. doi: 10.1016/j.stem.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann I., Stover E.H., Cullen D.E., Mao J., Morgan K.J., Lee B.H., Kharas M.G., Miller P.G., Cornejo M.G., Okabe R., Armstrong S.A., Ghilardi N., Gould S., DE Sauvage F.J., McMahon A.P., Gilliland D.G. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell. 2009;4:559–567. doi: 10.1016/j.stem.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dierks C., Beigi R., Guo G.R., Zirlik K., Stegert M.R., Manley P., Trussell C., Schmitt-Graeff A., Landwerlin K., Veelken H., Warmuth M. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C., Chen A., Jamieson C.H., Fereshteh M., Abrahamsson A., Blum J., Kwon H.Y., Kim J., Chute J.P., Rizzieri D., Munchhof M., VanArsdale T., Beachy P.A., Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia [Erratum appeared in Nature 2009, 460:652] Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merchant A., Joseph G., Wang Q., Brennan S., Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood. 2010;115:2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godfrey D.I., Zlotnik A. Control points in early T-cell development. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 45.Crompton T., Outram S.V., Hager-Theodorides A.L. Sonic hedgehog signalling in T-cell development and activation. Nat Rev Immunol. 2007;7:726–735. doi: 10.1038/nri2151. [DOI] [PubMed] [Google Scholar]
- 46.Outram S.V., Hager-Theodorides A.L., Shah D.K., Rowbotham N.J., Drakopoulou E., Ross S.E., Lanske B., Dessens J.T., Crompton T. Indian hedgehog (Ihh) both promotes and restricts thymocyte differentiation. Blood. 2009;113:2217–2228. doi: 10.1182/blood-2008-03-144840. [DOI] [PubMed] [Google Scholar]
- 47.Outram S.V., Varas A., Pepicelli C.V., Crompton T. Hedgehog signaling regulates differentiation from double-negative to double-positive thymocyte. Immunity. 2000;13:187–197. doi: 10.1016/s1074-7613(00)00019-4. [Erratum appeared in Immunity 2000, 13:585] [DOI] [PubMed] [Google Scholar]
- 48.Rowbotham N.J., Hager-Theodorides A.L., Furmanski A.L., Ross S.E., Outram S.V., Dessens J.T., Crompton T. Sonic hedgehog negatively regulates pre-TCR-induced differentiation by a Gli2-dependent mechanism. Blood. 2009;113:5144–5156. doi: 10.1182/blood-2008-10-185751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varas A., Sacedón R., Hidalgo L., Martínez V.G., Valencia J., Cejalvo T., Zapata A., Hernández-López C., Vicente A. Interplay between BMP4 and IL-7 in human intrathymic precursor cells. Cell Cycle. 2009;8:4119–4126. doi: 10.4161/cc.8.24.10149. [DOI] [PubMed] [Google Scholar]
- 50.Gutiérrez-Frías C., Sacedón R., Hernández-López C., Cejalvo T., Crompton T., Zapata A.G., Varas A., Vicente A. Sonic hedgehog regulates early human thymocyte differentiation by counteracting the IL-7-induced development of CD34+ precursor cells. J Immunol. 2004;173:5046–5053. doi: 10.4049/jimmunol.173.8.5046. [DOI] [PubMed] [Google Scholar]
- 51.Sacedón R., Varas A., Hernández-López C., Gutiérrez-deFrías C., Crompton T., Zapata A.G., Vicente A. Expression of hedgehog proteins in the human thymus. J Histochem Cytochem. 2003;51:1557–1566. doi: 10.1177/002215540305101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varas A., Hernández-López C., Valencia J., Mattavelli S., Martínez V.G., Hidalgo L., Gutiérrez-Frías C., Zapata A.G., Sacedón R., Vicente A. Survival and function of human thymic dendritic cells are dependent on autocrine Hedgehog signaling. J Leukoc Biol. 2008;83:1476–1483. doi: 10.1189/jlb.1107792. [DOI] [PubMed] [Google Scholar]
- 53.El Andaloussi A., Graves S., Meng F., Mandal M., Mashayekhi M., Aifantis I. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nat Immunol. 2006;7:418–426. doi: 10.1038/ni1313. [DOI] [PubMed] [Google Scholar]
- 54.Drakopoulou E., Outram S.V., Rowbotham N.J., Ross S.E., Furmanski A.L., Saldana J.I., Hager-Theodorides A.L., Crompton T. Non-redundant role for the transcription factor Gli1 at multiple stages of thymocyte development. Cell Cycle. 2010;9:4144–4152. doi: 10.4161/cc.9.20.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hager-Theodorides A.L., Dessens J.T., Outram S.V., Crompton T. The transcription factor Gli3 regulates differentiation of fetal CD4- CD8- double-negative thymocytes. Blood. 2005;106:1296–1304. doi: 10.1182/blood-2005-03-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah D.K., Hager-Theodorides A.L., Outram S.V., Ross S.E., Varas A., Crompton T. Reduced thymocyte development in sonic hedgehog knockout embryos. J Immunol. 2004;172:2296–2306. doi: 10.4049/jimmunol.172.4.2296. [DOI] [PubMed] [Google Scholar]
- 57.Rowbotham N.J., Hager-Theodorides A.L., Cebecauer M., Shah D.K., Drakopoulou E., Dyson J., Outram S.V., Crompton T. Activation of the Hedgehog signaling pathway in T-lineage cells inhibits TCR repertoire selection in the thymus and peripheral T-cell activation. Blood. 2007;109:3757–3766. doi: 10.1182/blood-2006-07-037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowrey J.A., Stewart G.A., Lindey S., Hoyne G.F., Dallman M.J., Howie S.E., Lamb J.R. Sonic hedgehog promotes cell cycle progression in activated peripheral CD4(+) T lymphocytes. J Immunol. 2002;169:1869–1875. doi: 10.4049/jimmunol.169.4.1869. [DOI] [PubMed] [Google Scholar]
- 59.Stewart G.A., Lowrey J.A., Wakelin S.J., Fitch P.M., Lindey S., Dallman M.J., Lamb J.R., Howie S.E. Sonic hedgehog signaling modulates activation of and cytokine production by human peripheral CD4+ T cells. J Immunol. 2002;169:5451–5457. doi: 10.4049/jimmunol.169.10.5451. [DOI] [PubMed] [Google Scholar]
- 60.Rowbotham N.J., Furmanski A.L., Hager-Theodorides A.L., Ross S.E., Drakopoulou E., Koufaris C., Outram S.V., Crompton T. Repression of hedgehog signal transduction in T-lineage cells increases TCR-induced activation and proliferation. Cell Cycle. 2008;7:904–908. doi: 10.4161/cc.7.7.5628. [DOI] [PubMed] [Google Scholar]
- 61.Sacedón R., Díez B., Nuñez V., Hernández-López C., Gutierrez-Frías C., Cejalvo T., Outram S.V., Crompton T., Zapata A.G., Vicente A., Varas A. Sonic hedgehog is produced by follicular dendritic cells and protects germinal center B cells from apoptosis. J Immunol. 2005;174:1456–1461. doi: 10.4049/jimmunol.174.3.1456. [DOI] [PubMed] [Google Scholar]
- 62.Kobune M., Ito Y., Kawano Y., Sasaki K., Uchida H., Nakamura K., Dehari H., Chiba H., Takimoto R., Matsunaga T., Terui T., Kato J., Niitsu Y., Hamada H. Indian hedgehog gene transfer augments hematopoietic support of human stromal cells including NOD/SCID-beta2m-/- repopulating cells. Blood. 2004;104:1002–1009. doi: 10.1182/blood-2003-09-3347. [DOI] [PubMed] [Google Scholar]
- 63.Kim J.E., Singh R.R., Cho-Vega J.H., Drakos E., Davuluri Y., Khokhar F.A., Fayad L., Medeiros L.J., Vega F. Sonic hedgehog signaling proteins and ATP-binding cassette G2 are aberrantly expressed in diffuse large B-cell lymphoma. Mod Pathol. 2009;22:1312–1320. doi: 10.1038/modpathol.2009.98. [DOI] [PubMed] [Google Scholar]
- 64.Greaves W.O., Kim J.E., Singh R.R., Drakos E., Kunkalla K., Sánchez-Espiridión B., Garcia J.F., Medeiros L.J., Vega F. Glioma-associated oncogene homologue 3, a hedgehog transcription factor, is highly expressed in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Hum Pathol. 2011;42:1643–1652. doi: 10.1016/j.humpath.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinzler K.W., Bigner S.H., Bigner D.D., Trent J.M., Law M.L., O'Brien S.J., Wong A.J., Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 66.Hahn H., Wicking C., Zaphiropoulous P.G., Gailani M.R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A.B., Gillies S., Negus K., Smyth I., Pressman C., Leffell D.J., Gerrard B., Goldstein A.M., Dean M., Toftgard R., Chenevix-Trench G., Wainwright B., Bale A.E. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 67.Guessous F., Li Y., Abounader R. Signaling pathways in medulloblastoma. J Cell Physiol. 2008;217:577–583. doi: 10.1002/jcp.21542. [DOI] [PubMed] [Google Scholar]
- 68.Pasca di Magliano M., Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 69.Karhadkar S.S., Bova G.S., Abdallah N., Dhara S., Gardner D., Maitra A., Isaacs J.T., Berman D.M., Beachy P.A. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 70.Pietsch T., Waha A., Koch A., Kraus J., Albrecht S., Tonn J., Sörensen N., Berthold F., Henk B., Schmandt N., Wolf H.K., von Deimling A., Wainwright B., Chenevix-Trench G., Wiestler O.D., Wicking C. Medulloblastomas of the desmoplastic variant carry mutations of the human homologue of Drosophila patched. Cancer Res. 1997;57:2085–2088. [PubMed] [Google Scholar]
- 71.Beddis I.R., Mott M.G., Bullimore J. Case report: nasopharyngeal rhabdomyosarcoma and Gorlin's naevoid basal cell carcinoma syndrome. Med Pediatr Oncol. 1983;11:178–179. doi: 10.1002/mpo.2950110309. [DOI] [PubMed] [Google Scholar]
- 72.Taylor M.D., Liu L., Raffel C., Hui C.C., Mainprize T.G., Zhang X., Agatep R., Chiappa S., Gao L., Lowrance A., Hao A., Goldstein A.M., Stavrou T., Scherer S.W., Dura W.T., Wainwright B., Squire J.A., Rutka J.T., Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 73.Tostar U., Malm C.J., Meis-Kindblom J.M., Kindblom L.G., Toftgård R., Undén A.B. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 74.Yauch R.L., Gould S.E., Scales S.J., Tang T., Tian H., Ahn C.P., Marshall D., Fu L., Januario T., Kallop D., Nannini-Pepe M., Kotkow K., Marsters J.C., Rubin L.L., de Sauvage F.J. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 75.Kubo M., Nakamura M., Tasaki A., Yamanaka N., Nakashima H., Nomura M., Kuroki S., Katano M. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 76.Thayer S.P., di Magliano M.P., Heiser P.W., Nielsen C.M., Roberts D.J., Lauwers G.Y., Qi Y.P., Gysin S., Fernández-del Castillo C., Yajnik V., Antoniu B., McMahon M., Warshaw A.L., Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watkins D.N., Berman D.M., Burkholder S.G., Wang B., Beachy P.A., Baylin S.B. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 78.Berman D.M., Karhadkar S.S., Maitra A., Montes De Oca R., Gerstenblith M.R., Briggs K., Parker A.R., Shimada Y., Eshleman J.R., Watkins D.N., Beachy P.A. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 79.Oniscu A., James R.M., Morris R.G., Bader S., Malcomson R.D., Harrison D.J. Expression of Sonic hedgehog pathway genes is altered in colonic neoplasia. J Pathol. 2004;203:909–917. doi: 10.1002/path.1591. [DOI] [PubMed] [Google Scholar]
- 80.Dierks C., Grbic J., Zirlik K., Beigi R., Englund N.P., Guo G.R., Veelken H., Engelhardt M., Mertelsmann R., Kelleher J.F., Schultz P., Warmuth M. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nat Med. 2007;13:944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 81.Vega F., Cho-Vega J.H., Lennon P.A., Luthra M.G., Bailey J., Breeden M., Jones D., Medeiros L.J., Luthra R. Splenic marginal zone lymphomas are characterized by loss of interstitial regions of chromosome 7q, 7q31.32 and 7q36.2 that include the protection of telomere 1 (POT1) and sonic hedgehog (SHH) genes. Br J Haematol. 2008;142:216–226. doi: 10.1111/j.1365-2141.2008.07176.x. [DOI] [PubMed] [Google Scholar]
- 82.Rao P.H., Houldsworth J., Dyomina K., Parsa N.Z., Cigudosa J.C., Louie D.C., Popplewell L., Offit K., Jhanwar S.C., Chaganti R.S. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234–240. [PubMed] [Google Scholar]
- 83.Beà S., Colomo L., López-Guillermo A., Salaverria I., Puig X., Pinyol M., Rives S., Montserrat E., Campo E. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 84.Sander S., Bullinger L., Leupolt E., Benner A., Kienle D., Katzenberger T., Kalla J., Ott G., Müller-Hermelink H.K., Barth T.F., Möller P., Lichter P., Döhner H., Stilgenbauer S. Genomic aberrations in mantle cell lymphoma detected by interphase fluorescence in situ hybridization: Incidence and clinicopathological correlations. Haematologica. 2008;93:680–687. doi: 10.3324/haematol.12330. [DOI] [PubMed] [Google Scholar]
- 85.Northcott P.A., Fernandez-L A., Hagan J.P., Ellison D.W., Grajkowska W., Gillespie Y., Grundy R., Van Meter T., Rutka J.T., Croce C.M., Kenney A.M., Taylor M.D. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stecca B., Ruiz I Altaba A. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh R.R., Cho-Vega J.H., Davuluri Y., Ma S., Kasbidi F., Milito C., Lennon P.A., Drakos E., Medeiros L.J., Luthra R., Vega F. Sonic hedgehog signaling pathway is activated in ALK-positive anaplastic large cell lymphoma. Cancer Res. 2009;69:2550–2558. doi: 10.1158/0008-5472.CAN-08-1808. [DOI] [PubMed] [Google Scholar]
- 88.Yang L., Wang Y., Mao H., Fleig S., Omenetti A., Brown K.D., Sicklick J.K., Li Y.X., Diehl A.M. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol. 2008;48:98–106. doi: 10.1016/j.jhep.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakashima H., Nakamura M., Yamaguchi H., Yamanaka N., Akiyoshi T., Koga K., Yamaguchi K., Tsuneyoshi M., Tanaka M., Katano M. Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res. 2006;66:7041–7049. doi: 10.1158/0008-5472.CAN-05-4588. [DOI] [PubMed] [Google Scholar]
- 90.Hegde G.V., Peterson K.J., Emanuel K., Mittal A.K., Joshi A.D., Dickinson J.D., Kollessery G.J., Bociek R.G., Bierman P., Vose J.M., Weisenburger D.D., Joshi S.S. Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: a potential new therapeutic target. Mol Cancer Res. 2008;6:1928–1936. doi: 10.1158/1541-7786.MCR-08-0142. [DOI] [PubMed] [Google Scholar]
- 91.Desch P., Asslaber D., Kern D., Schnidar H., Mangelberger D., Alinger B., Stoecher M., Hofbauer S.W., Neureiter D., Tinhofer I., Aberger F., Hartmann T.N., Greil R. Inhibition of GLI, but not Smoothened, induces apoptosis in chronic lymphocytic leukemia cells. Oncogene. 2010;29:4885–4895. doi: 10.1038/onc.2010.243. [DOI] [PubMed] [Google Scholar]
- 92.Singh R.R., Kim J.E., Davuluri Y., Drakos E., Cho-Vega J.H., Amin H.M., Vega F. Hedgehog signaling pathway is activated in diffuse large B-cell lymphoma and contributes to tumor cell survival and proliferation. Leukemia. 2010;24:1025–1036. doi: 10.1038/leu.2010.35. [DOI] [PubMed] [Google Scholar]
- 93.Greaves W.O., Kim J.E., Kunkalla K., Medeiros L.J., Singh R., Vega F. Hedgehog signaling pathway is aberrantly activated in a subset of high grade lymphomas. Mod Pathol. 2010;23(Suppl 1s):298A. (abstract) [Google Scholar]
- 94.Singh R.R., Kunkalla K., Qu C., Schlette E., Neelapu S.S., Samaniego F., Vega F. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene. 2011 doi: 10.1038/onc.2011.195. [Epub ahead of press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hegde G.V., Munger C.M., Emanuel K., Joshi A.D., Greiner T.C., Weisenburger D.D., Vose J.M., Joshi S.S. Targeting of sonic hedgehog-GLI signaling: a potential strategy to improve therapy for mantle cell lymphoma. Mol Cancer Ther. 2008;7:1450–1460. doi: 10.1158/1535-7163.MCT-07-2118. [DOI] [PubMed] [Google Scholar]
- 96.Hager-Theodorides A.L., Furmanski A.L., Ross S.E., Outram S.V., Rowbotham N.J., Crompton T. The Gli3 transcription factor expressed in the thymus stroma controls thymocyte negative selection via Hedgehog-dependent and -independent mechanisms. J Immunol. 2009;183:3023–3032. doi: 10.4049/jimmunol.0900152. [DOI] [PubMed] [Google Scholar]
- 97.Peacock C.D., Wang Q., Gesell G.S., Corcoran-Schwartz I.M., Jones E., Kim J., Devereux W.L., Rhodes J.T., Huff C.A., Beachy P.A., Watkins D.N., Matsui W. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duyster J., Bai R.Y., Morris S.W. Translocations involving anaplastic lymphoma kinase (ALK) Oncogene. 2001;20:5623–5637. doi: 10.1038/sj.onc.1204594. [DOI] [PubMed] [Google Scholar]
- 99.Kobune M., Takimoto R., Murase K., Iyama S., Sato T., Kikuchi S., Kawano Y., Miyanishi K., Sato Y., Niitsu Y., Kato J. Drug resistance is dramatically restored by hedgehog inhibitors in CD34+ leukemic cells. Cancer Sci. 2009;100:948–955. doi: 10.1111/j.1349-7006.2009.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin T.L., Wang Q.H., Brown P., Peacock C., Merchant A.A., Brennan S., Jones E., McGovern K., Watkins D.N., Sakamoto K.M., Matsui W. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS One. 2010;5:e15262. doi: 10.1371/journal.pone.0015262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawahara T., Kawaguchi-Ihara N., Okuhashi Y., Itoh M., Nara N., Tohda S. Cyclopamine and quercetin suppress the growth of leukemia and lymphoma cells. Anticancer Res. 2009;29:4629–4632. [PubMed] [Google Scholar]
- 102.Ji Z., Mei F.C., Johnson B.H., Thompson E.B., Cheng X. Protein kinase A, not Epac, suppresses hedgehog activity and regulates glucocorticoid sensitivity in acute lymphoblastic leukemia cells. J Biol Chem. 2007;282:37370–37377. doi: 10.1074/jbc.M703697200. [DOI] [PubMed] [Google Scholar]
- 103.Hyman J.M., Firestone A.J., Heine V.M., Zhao Y., Ocasio C.A., Han K., Sun M., Rack P.G., Sinha S., Wu J.J., Solow-Cordero D.E., Jiang J., Rowitch D.H., Chen J.K. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heretsch P., Tzagkaroulaki L., Giannis A. Modulators of the hedgehog signaling pathway. Bioorg Med Chem. 2010;18:6613–6624. doi: 10.1016/j.bmc.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 105.Merchant A.A., Matsui W. Targeting Hedgehog: a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]