Abstract
In situ analysis of biomarkers is highly desirable in molecular pathology because it allows the examination of biomarker status within the histopathological context of clinical specimens. Immunohistochemistry and DNA in situ hybridization (ISH) are widely used in clinical settings to assess protein and DNA biomarkers, respectively, but clinical use of in situ RNA analysis is rare. This disparity is especially notable when considering the abundance of RNA biomarkers discovered through whole-genome expression profiling. This is largely due to the high degree of technical complexity and insufficient sensitivity and specificity of current RNA ISH techniques. Here, we describe RNAscope, a novel RNA ISH technology with a unique probe design strategy that allows simultaneous signal amplification and background suppression to achieve single-molecule visualization while preserving tissue morphology. RNAscope is compatible with routine formalin-fixed, paraffin-embedded tissue specimens and can use either conventional chromogenic dyes for bright-field microscopy or fluorescent dyes for multiplex analysis. Unlike grind-and-bind RNA analysis methods such as real-time RT-PCR, RNAscope brings the benefits of in situ analysis to RNA biomarkers and may enable rapid development of RNA ISH-based molecular diagnostic assays.
Biomarkers based on DNA, RNA, and proteins are increasingly used for cancer diagnosis, prognosis, and therapy guidance, heralding the era of personalized medicine.1 RNA biomarkers or gene expression signatures have emerged as a major class of cancer biomarkers, thanks to widespread use of genome-wide gene expression profiling technologies.2,3 To implement these genomic signatures in clinical diagnostic assays, the current platform of choice is real-time RT-PCR, which is considered the gold standard in gene expression analysis.4 However, this grind-and-bind approach has a serious drawback: the process of RNA extraction destroys the tissue context of gene expression measurements, making it impossible to map the observed signals to individual cells. Furthermore, these assays are prone to interference from unintended cell types (eg, noncancer cells) and from unwanted tissue elements (eg, fibrosis and necrosis). Microdissection techniques can alleviate this problem to some extent,5,6 but they are too cumbersome and laborious to be useful on a routine basis. By contrast, DNA in situ hybridization (ISH)7 and protein immunohistochemistry (IHC)8 are routinely used in clinical laboratories for DNA and protein biomarker analysis, allowing the integration of molecular information with histopathology for optimal clinical interpretation. To date, use of RNA ISH in clinical diagnostics has been limited to highly expressed genes such as Epstein-Barr virus (EBV)-derived transcripts EBER1/2 in EBV-related diseases.9,10 Conventional non-radioisotopic RNA in situ hybridization (ISH) techniques lack the sensitivity and specificity required to measure many low-abundance RNA biomarkers reliably.11
Here, we describe RNAscope, a novel RNA ISH method. Single-molecule visualization in individual cells is achieved through use of a novel probe design strategy and a hybridization-based signal amplification system to simultaneously amplify signals and suppress background. Many of the steps in RNAscope are similar to those in IHC. The RNAscope approach can be used with archival formalin-fixed, paraffin-embedded (FFPE) tissue samples on glass slides, and the stained slides can be visualized under either a standard bright-field microscope (with chromogenic labels) or an epifluorescent microscope (with fluorescent labels). The RNAscope approach allows multiplex detection for up to four target genes (limited by the number of spectrally discernible fluorescent dyes). The ability to analyze gene expression in situ in routine clinical specimen types, as well as high sensitivity and specificity, make RNAscope a promising platform for translating many RNA biomarkers into clinical use.
Materials and Methods
Cell Culture
SK-BR-3 breast adenocarcinoma cells (ATCC, Manassas, VA) were cultured in McCoy's medium. HuH-7 hepatocellular carcinoma cells (JCRB-Japanese Collection of Research Bioresources, Shinjuku, Japan) with or without hepatitis C virus (HCV) infection were cultured in modified Eagle's medium. MCF7 breast adenocarcinoma cells, SiHa cervical squamous cell carcinoma cells, HeLa cervical adenocarcinoma cells, and MS751 cervical epidermoid carcinoma cells (all from ATCC) were cultured in Dulbecco's modified Eagle's medium at 37°C in 5% CO2. MDA-MB-468 breast adenocarcinoma cells (ATCC) were cultured in L-15 (Leibovitz) medium in a CO2-free incubator at 37°C. All media were supplemented with 10% fetal bovine serum (Gibco; Invitrogen, Carlsbad, CA).
Tissue Specimens
Deidentified archival FFPE tumor tissues were purchased from Analytical Biological Services (Wilmington, DE). Tissue quality was assessed by performing RNAscope analysis for mRNA of the housekeeping gene ubiquitin C (UBC).
RNAscope Design Strategy
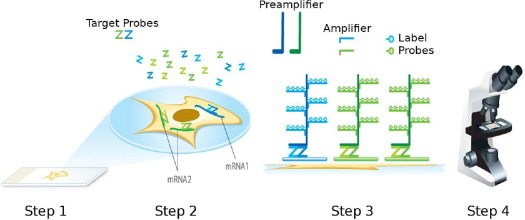
We sought to improve the signal-to-noise ratio of RNA ISH by amplifying target-specific signals but not background noise from nonspecific hybridization. We used a novel target probe design strategy (a double-Z design) (Figure 1). A series of target probes are designed to hybridize to the target RNA molecule. Each target probe contains an 18- to 25-base region complementary to the target RNA, a spacer sequence, and a 14-base tail sequence (conceptualized as Z). A pair of target probes (double Z), each possessing a different type of tail sequence, hybridize contiguously to a target region (∼50 bases). The two tail sequences together form a 28-base hybridization site for the preamplifier, which contains 20 binding sites for the amplifier, which, in turn, contains 20 binding sites for the label probe. Typically, a 1-kb region on the RNA molecule is targeted by 20 probe pairs; thus, sequential hybridizations with the preamplifier, amplifier, and label probe can theoretically yield up to 8000 labels for each target RNA molecule.
Figure 1.
Schematic of the RNAscope assay procedure. In step 1, cells or tissues are fixed and permeabilized to allow for target probe access. In step 2, target RNA-specific oligonucleotide probes (Z) are hybridized in pairs (ZZ) to multiple RNA targets. In step 3, multiple signal amplification molecules are hybridized, each recognizing a specific target probe, and each unique label probe is conjugated to a different fluorophore or enzyme. In step 4, signals are detected using an epifluorescent microscope (for fluorescent label) or standard bright-field microscope (for enzyme label).
This hybridization-mediated signal amplification scheme is similar to the branched DNA (bDNA) method described previously,12 but the double-Z probe design strategy should ensure superior background control because it is highly unlikely that a nonspecific hybridization event will juxtapose a pair of target probes along an off-target mRNA molecule to form the 28-base hybridization site for the preamplifier, and also because a single 14-base tail sequence will not bind the preamplifier with sufficient strength to result in successful signal amplification.
The label probe can be either fluorescently labeled for direct visualization under an epifluorescent microscope or conjugated to an alkaline phosphatase or horseradish peroxidase (HRP) molecule for chromogenic reactions [Fast Red with alkaline phosphatase and 3,3′-diaminobenzidine (DAB) with HRP]. The alkaline phosphatase or HRP-labeled probes have an added advantage, in that chromogen-stained slides can be viewed under a standard bright-field microscope similar to IHC procedures, making RNAscope assay results easier to read and archive in a clinical setting.
Multiple RNA species can be measured simultaneously in two ways: the target probes for different genes can have the same tail sequence recognized by the same signal amplification system, generating a pooled signal; alternatively, multiple signal amplification systems with different label probes can be used to detect each RNA species, allowing for multiplex detection of multiple target RNAs.
Custom software was written to automatically select target probe sequences with compatible melting temperature (Tm) and minimal cross-hybridization to off-target sequences.13 We determined that three probe pairs could generate readily visible signals with HRP/DAB detection (see Supplemental Figure S1 at http://jmd.amjpathol.org). We chose 10 to 20 pairs, for optimal signals and for added robustness against potentially variable target accessibility and partial RNA degradation.
RNAscope Assay Procedure for RNA Detection
The target genes and probed regions are listed in Supplemental Table S1 (available at http://jmd.amjpathol.org). Sequences of target probes, preamplifier, amplifier, and label probe are proprietary (Advanced Cell Diagnostics, Hayward, CA). For fluorescent detection, the label probe was conjugated to Alexa Fluor 488, 546, 647, or 750 (Molecular Probes; Invitrogen, Eugene, OR). For chromogenic detection using DAB, label probe was conjugated to HRP.
For cell lines, cells were placed on slides and fixed in 4% formaldehyde for 60 minutes, followed by protease digestion (2.5 μg/mL) at 23°C to 25°C. The cells were then incubated in order at 40°C with the following solutions: target probes in hybridization buffer A [6× SSC (1× SSC is 0.15 mol/L NaCl, 0.015 mol/L Na-citrate), 25% formamide, 0.2% lithium dodecyl sulfate, blocking reagents] for 3 hours; preamplifier (2 nmol/L) in hybridization buffer B (20% formamide, 5× SSC, 0.3% lithium dodecyl sulfate, 10% dextran sulfate, blocking reagents) for 30 minutes; amplifier (2 nmol/L) in hybridization buffer B at 40°C for 15 minutes; and label probe (2 nmol/L) in hybridization buffer C (5× SSC, 0.3% lithium dodecyl sulfate, blocking reagents) for 15 minutes. After each hybridization step, slides were washed with wash buffer (0.1× SSC, 0.03% lithium dodecyl sulfate) three times at room temperature. For multiplex detection, equimolar amounts of target probes, preamplifier, amplifier, and label probe of each amplification system were used. Chromogenic detection was performed using DAB followed by counterstaining with hematoxylin (American MasterTech Scientific, Lodi, CA).
For FFPE tissues, tissue sections in 5-μm thickness were deparaffinized in xylene, followed by dehydration in an ethanol series. Tissue sections were then incubated in citrate buffer (10 nmol/L, pH 6) maintained at a boiling temperature (100°C to 103°C) using a hot plate for 15 minutes, rinsed in deionized water, and immediately treated with10 μg/mL protease (Sigma-Aldrich, St. Louis, MO) at 40°C for 30 minutes in a HybEZ hybridization oven (Advanced Cell Diagnostics, Hayward, CA). Hybridization with target probes, preamplifier, amplifier, and label probe and chromogenic detection were as described above for cultured cells. A wide range of assay conditions were explored to optimize for FFPE samples prepared and fixed according to American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines14 (10% neutral buffered formalin for 6 to 72 hours at room temperature), including pretreatment conditions such as citrate buffer temperature, pH, incubation time, and protease concentrations.
Assays using archival FFPE specimens were typically performed in parallel with positive and negative controls, to ensure interpretable results. The endogenous housekeeping gene UBC was used as positive control to assess both tissue RNA integrity and assay procedure. Positive staining with signals easily visible under a 10× objective lens was considered to be adequate. The bacterial gene dapB was used as negative control to assess background signals; completely negative staining was routinely achieved using our standard protocol.
Conventional RNA ISH
A commercially available RISH kit (Biocare Medical, Concord, CA) was used to detect immunoglobulin κ chain mRNA in human tonsil tissue according to the manufacturer's protocol, in parallel with the RNAscope assay. No negative control was provided in the kit.
Estimation of Copy Number
RNAscope for DNA ISH
Trypsinized cells were treated in a hypotonic solution (0.075 mol/L KCl), washed, fixed in Carnoy's fixative (3:1 methanol/acetic acid), and air-dried on slides. The cells were then treated with RNase (Roche Diagnostics, Indianapolis, IN), followed by 0.005% pepsin (Sigma-Aldrich) treatment for 10 minutes at 37°C. DNA was denatured in denaturing solution (75% formamide, 2× SSC) for 5 minutes at 75°C and then was subjected to probe hybridization according to the RNAscope procedure described above.
QuantiGene 2.0
HeLa cells were cultured in triplicate: one culture was trypsinized, to get a cell count; another was lysed for QuantiGene 2.0 (Panomics; Affymetrix, Fremont, CA) analysis according to the manufacturer's protocol; the third was used for RNAscope analysis. The number of HER2 mRNA transcripts in the cell lysate was determined by using a standard curve constructed from known amounts of in vitro transcribed RNA. The per-cell copy number was determined by dividing the number of mRNA molecules in the lysate by the number of cells used in the assay. Confidence limits were estimated by linear regression based on triplicate measurements.
RNAscope
The number of HER2 mRNA transcripts determined by RNAscope was based on the fluorescent spot count of ∼100 randomly selected cells in microscopic images. The per-cell copy number was determined by dividing the total spot count by the number of cells counted.
Microscopic Imaging
Images were acquired using an Olympus IX71 fluorescent microscope (Olympus, Tokyo, Japan) and a PXL37 CCD camera (Photometrics, Tucson, AZ). For multiplex RNAscope staining and for studies with FFPE tissue specimens, images were acquired using a Zeiss Axioplan M1 microscope (Carl Zeiss MicroImaging, Göttingen, Germany) and a CRi Nuance multispectral imaging system (Caliper Life Sciences, Cambridge, MA). Overlapping signals from different fluorophores were separated by comparing composite signals against a reference spectral library generated with single-color stained samples.
Results
Validation of RNAscope Design Strategy
We hypothesized that selective amplification of target-specific signals without amplifying background signal should substantially improve signal-to-noise ratio, thereby also improving both sensitivity and specificity in RNA ISH. To achieve this goal, we developed a probe design concept much akin to fluorescence resonance energy transfer (FRET), in which two independent probes (double Z probes) have to hybridize to the target sequence in tandem in order for signal amplification to occur (Figure 1; see also under Materials and Methods). Because it is highly unlikely that two independent probes will hybridize to a nonspecific target right next to each other, this design concept should in theory ensure selective amplification of target-specific signals.
To validate this probe design strategy, we first determined whether the paired target probes are necessary to generate signals. The set of target probe pairs for 18S ribosomal RNA was split into two groups, one consisting of only the probes with the left tail and the other consisting of only the probes with the right tail (Figure 2A). The two groups of probes were hybridized either alone or in combination with HeLa cells. A ubiquitous fluorescent staining was observed when both groups were used together, but no fluorescent signal was detected when either group was used alone. The lack of visible signals (indistinguishable from the control with no target probe) from either half of the target probe pairs was remarkable, considering the high levels of 18S rRNA target molecules in the cells. This result confirmed that the individual 14-base tails were not sufficient for binding to the signal amplification system under the assay conditions.
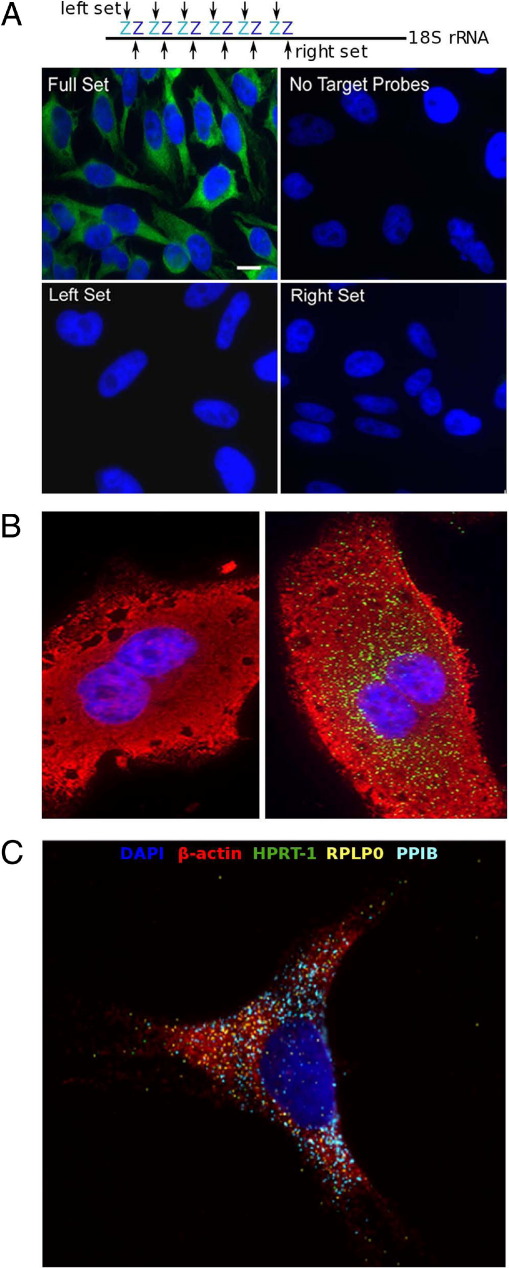
Figure 2.
Validation of RNAscope. A: HeLa cells were hybridized with either the full set of probes to 18S rRNA, the left half of the set, or the right half of the set (as shown in the schematic along the top). A no-probe control was performed in parallel as an indicator of background staining. Cells were counterstained with DAPI (blue), which masks nucleolar 18S RNA. Scale bar = 10 μm. B: HCV-uninfected (left) or HCV-infected (right) HuH-7 cells were hybridized with probe sets to HCV mRNA (green). Cells were costained with 18S rRNA target probes (red) as an internal control for assay success. Nuclei were counterstained with DAPI (blue). Original magnification, ×40. C: HeLa cells were hybridized with probes to β-actin, RPLP0 (60S acidic ribosomal protein P0), PPIB (peptidylprolyl isomerase B), and HPRT-1 (hypoxanthine phosphoribosyltransferase 1) in multiplex fluorescence format. Nuclei were counterstained with DAPI. Original magnification, ×40.
To demonstrate assay specificity, a probe set targeting HCV RNA was used to detect a viral transcript in HuH-7 cells stably infected with HCV. Positive punctate staining was detected in HCV-infected cells, but not in uninfected control cells (Figure 2B). To further determine the level of specificity achievable, we stained several cell lines containing different human papillomavirus (HPV) genotypes with as much as 85% overall sequence identity.15 Each target probe set gave positive staining only in the cell line known to harbor the corresponding HPV genotype (see Supplemental Figure S2 at http://jmd.amjpathol.org).
Multiplex Detection of mRNAs in Cell Lines
RNAscope, a probe hybridization-based assay, could have better multiplexing capability than an antibody-based IHC assay because the assay condition is universal to all RNA transcripts (which are located within the cytoplasm) and because cross-hybridization between target probes can be minimized by the probe design algorithm. To demonstrate the ability of RNAscope to detect multiple transcripts simultaneously, probe sets for the housekeeping genes HPRT1, PPIB, RPLP0, and ACTB (β-actin) were labeled with fluorescent dyes of different colors and hybridized to HeLa cells (Figure 2C). All four genes showed distinct expression patterns consistent with their known low, medium, and high expression levels. To determine whether multiplex analysis gave quantitative results similar to those of single-gene analysis, we detected mRNAs of HPRT1 and POLR2A in SK-BR-3 cells either singly or in a duplex format. The dot counts for each gene were similar for both single and duplex hybridization (see Supplemental Table S2 at http://jmd.amjpathol.org).
Single RNA Molecule Detection in Cell Lines
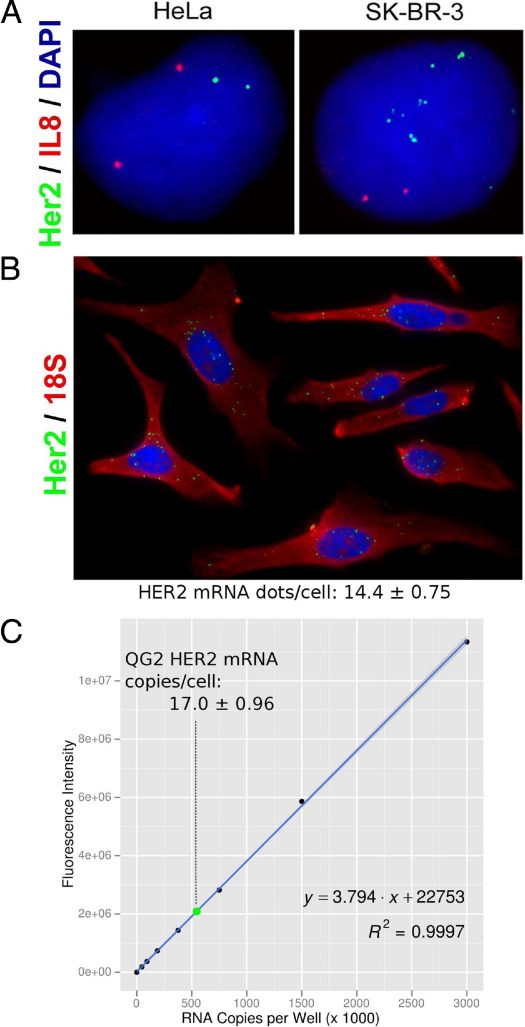
The punctate staining pattern seen for HCV, HPRT1, PPIB, POLR2A, and RPLP0 raised the possibility that each fluorescent dot in the RNAscope assay represents a single mRNA transcript. We used two different methods to test this. First, we used the same target probes to detect human epidermal growth factor receptor 2 (HER2) mRNA and genomic DNA in HeLa and SK-BR-3 cells under identical target probe hybridization and signal amplification conditions. For genomic DNA, two fluorescent dots were detected in HeLa cells, whereas many more dots were seen in SK-BR-3 cells, consistent with the diploid and amplified HER2 gene status in HeLa and SK-BR-3 cells, respectively (Figure 3A). These results demonstrate that the RNAscope assay is capable of single-molecule nucleic acid detection. The same probe set detected HER2 mRNA transcripts in HeLa and SK-BR-3 cells with dot numbers consistent with their gene amplification status (see Supplemental Figure S3A at http://jmd.amjpathol.org). We then compared the signal intensities of HER2 mRNA dots with those of HER2 genomic DNA dots. If the RNA staining represented more than one mRNA transcript, then the signal intensity of at least some of the mRNA dots would be expected to be greater than the genomic DNA dots. However, no mRNA signal had a fluorescent intensity greater than the maximum intensity observed for genomic DNA (see Supplemental Figure S3B at http://jmd.amjpathol.org). In fact, the signal distribution of the RNA dots was skewed toward the lower end, possibly because of the more restricted probe accessibility or spatial localization of RNA. In a second test, we counted the number of HER2 mRNA dots in HeLa cells and compared that with the number of mRNA transcripts determined in cell lysates by QuantiGene 2.016 (Figure 3, B and C). The dot number per cell determined by RNAscope (mean ± 95% confidence limit, 14.4 ± 0.75; Figure 3B) agreed well with the copy number per cell determined by QuantiGene (mean ± 95% confidence limit, 17.0 ± 0.96; Figure 3C). Taken together, these results are consistent with each RNA dot deriving from a single mRNA molecule.
Figure 3.
Single RNA molecule detection. A: HER2 genomic DNA in HeLa and SK-BR-3 cells was detected using the RNAscope probes and signal amplification system. Nuclei were costained for IL-8 for diploid genome Original magnification, ×40. B: HER2 mRNA detection in HeLa cells using the same probes and signal amplification system. A probe set to 18S rRNA was used as internal control for RNA detection. HER2 mRNA dots were counted; mean dots per cell (±95% confidence limit) are indicated. Nuclei were counterstained with DAPI (blue). Original magnification, ×40. C: HER2 mRNA copies per cell in HeLa cells determined by QuantiGene 2.0 (QG2) using a standard curve from in vitro transcribed RNA. Mean copies per cell (±95% confidence limit) are indicated (green dot), as calculated from triplicate measurements using linear regression.
In Situ Detection of mRNA in FFPE Tissues
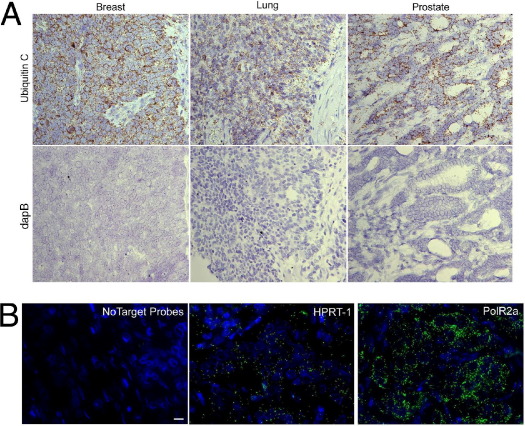
Because FFPE tissue is the most widely used clinical sample type in cancer diagnosis, we adapted the RNAscope assay for use on FFPE tissue and determined its general applicability for staining various tissue types. A probe set targeting the housekeeping gene UBC was hybridized to archival breast, lung, and prostate FFPE tissues, using standard tissue pretreatment in boiling citrate buffer and protease digestion. Using HRP-conjugated label probe, strong positive DAB staining was detected in all three tissue types, but no signal was present when the negative control dapB probes were used (Figure 4A).
Figure 4.
RNAscope detection of RNA in FFPE tumor tissues. A: Chromogenic staining (DAB) of primary tumor tissues (breast, lung, and prostate) hybridized with either probes to ubiquitin C (UBC) or probes against the bacterial gene dapB as negative control. Nuclei were counterstained with hematoxylin. Original magnification, ×40. B: Fluorescent detection of low-copy transcripts in FFPE samples. Breast tumor tissue section was hybridized with either no probes or with Alexa Fluor 488-labeled probe sets (green) to HPRT1 or POLR2A. Nuclei were counterstained with DAPI (blue). Scale bar = 10 μm.
To demonstrate the detection sensitivity of RNAscope in FFPE tissue, we hybridized probe sets for two low-expressing housekeeping genes, POLR2A and HPRT1, to breast tumor tissues. Positive signals were observed for both genes as discrete fluorescent dots (Figure 4B), which are almost identical to those seen in cell lines, suggesting the possibility of single-molecule detection.
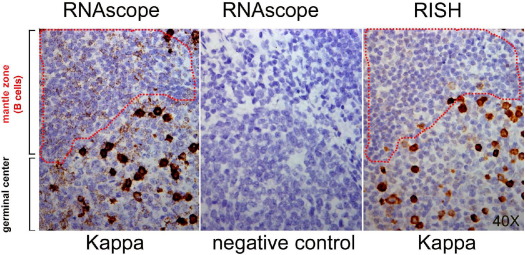
We next compared RNAscope with a commercial non-radioisotopic RNA ISH kit (RISH, Biocare, CA), used according to the manufacturer's protocol, for detecting immunoglobulin κ chain expression in B lymphocytes in human FFPE tonsil tissue. Plasma cells in the germinal center express high levels of κ light chain, whereas B cells in the mantle zone express much lower levels of κ light chain, levels that cannot be reliably detected by conventional non-radioisotopic ISH methods.17,18 Indeed, both RNAscope and RISH detected strong staining in plasma cells, although RNAscope staining was more intense. Importantly, whereas RNAscope demonstrated clear punctate staining in B cells in the mantle zone, RISH generated only a faint diffuse pattern in these cells (Figure 5), a pattern that can be difficult to distinguish from background.18
Figure 5.
Detection of Ig κ chain expression in B lymphocytes in FFPE human tonsil tissue. κ light chain mRNA transcripts were stained using RNAscope or a commercial non-radioisotopic RNA ISH kit. For RNAscope, a negative control (bacterial gene dapB) was also included. The dotted line outlines the mantle zone. Original magnification, ×40.
Multiplex Detection of mRNAs in FFPE Tissues
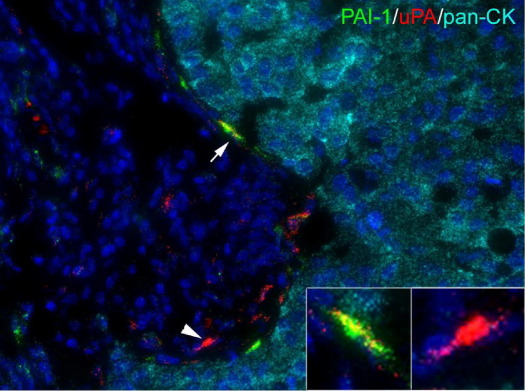
To demonstrate the multiplex ability of RNAscope in FFPE tissues, we analyzed the expression of urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in FFPE breast cancer tissues, together with cytokeratin markers (pooled probes for CK-8, CK-18, and CK-19), using a triplex fluorescence assay (Figure 6). uPA and PAI-1 are among the most established prognostic markers in breast cancer.19,20 Consistent with previous studies,21,22 strong uPA and PAI-1 mRNA staining was seen in tumor-associated stromal cells, but not in the tumor epithelium. Furthermore, our results indicate that uPA and PAI-1 mRNAs are rarely coexpressed in the same cell, suggesting that these two proteins may be produced by different stromal cell types.22
Figure 6.
Multiplex fluorescence detection of uPA and PAI mRNAs in breast cancer. Merged pseudo-colored image of a metastatic breast cancer tissue section stained with probes specific to cytokeratins [PanCK (CK-8, CK-18, and CK-19), labeled with Alexa Fluor 647], uPA (labeled with Alexa Fluor 546), and PAI-1 (labeled with Alexa Fluor 488). Both uPA expression (arrowhead and right inset) and coexpression with PAI-1 (arrow and left inset) were detected. Nuclei were counterstained with DAPI (blue). Original magnification, ×40.
Discussion
Here, we have described the development and validation of RNAscope, a novel non-radioisotopic RNA ISH technology that has the potential to be applied to routine clinical samples for biomarker analysis. ISH, which has a history of more than 40 years, has been continuously improved over time.7,23–25 Nonetheless, limitations in sensitivity and specificity and the time-consuming, complex procedures involved have impeded its adoption in clinical research and diagnostics.
Previous efforts at improving RNA ISH performance have focused largely on improving sensitivity,7,23 either by amplifying the mRNA targets before hybridization (eg, in situ PCR26) or amplifying the signals after target hybridization (eg, bDNA12 or tyramide signal amplification27). Target amplification before hybridization makes quantification problematic, because of amplification bias, and signal amplification can also amplify noise, limiting the achievable improvement in signal-to-noise ratio. In contrast, RNAscope was designed to amplify target-specific signals without also amplifying the background, resulting in marked improvement in signal-to-noise ratio. To date, we have developed RNAscope assays for more than 100 genes with varying expression levels (unpublished data).
In analyzing biomarkers in FFPE tissues, preanalytical variables such as time from tissue acquisition to fixation, fixatives used, and fixation time can negatively affect the resulting biomarker measurements.14 We found that reliable results could be obtained if tissues were prepared and fixed according to ASCO/CAP guidelines.14 The use of up to 20 target probe pairs, each spanning 40 to 50 nucleotides along the target RNA molecule, should provide additional robustness against the partial RNA degradation characteristic of FFPE samples.
Secreted proteins such as growth factors, cytokines, and extracellular matrix proteins are increasingly being used as therapeutic targets and diagnostic biomarkers. In cases in which detecting these proteins by IHC may lack sensitivity or cellular resolution (see, for example, Beck et al18), detecting their mRNAs by RNAscope may be an effective alternative, because mRNAs are localized intracellularly.
In summary, RNAscope represents a significant improvement in RNA ISH methodology and is compatible with clinical sample types and laboratory workflows. Our research group has begun to test this assay in routine clinical samples from clinical laboratories.28 Because the target probes are unmodified short oligonucleotides and can be uniformly designed and manufactured, RNAscope assays should be scalable to whole transcriptomes with universal reagents and assay conditions. Like other in situ methods, such as DNA ISH and protein IHC, RNAscope holds significant promise as a new platform for developing and implementing RNA-based molecular diagnostics.
Acknowledgments
We thank Steve Chen, Frank Witney, and Mickey Urdea for their support and encouragement; Tony Hartman, Catherine Wolf, Yunqing Ma, Quan Nguyen, Steve Lai, and Takuro Yaoi for their advice and contribution in assay development; and Dr. Peter Sarnow (Stanford University, Palo Alto, CA) for supplying HCV replicon-infected HuH-7 and parental cell lines.
Footnotes
Supported in part by grants from the NIH (R43/44CA122444 to Y.L.) and the Department of Defense (Breast Cancer Research Program grant W81XWH-06-1-0682 to Y.L.).
Disclosure: All authors are employed by and own stock in Advanced Cell Diagnostics, Inc.
Supplemental material for this article can be found at http://jmd.amjpathol.org or at doi: 10.1016/j.jmoldx.2011.08.002.
Supplementary Data
Number of probe pairs required for generating visible signal in chromogenic RNAscope assay. HeLa cell FFPE pellet was used as a model system for testing the number of probe pairs needed to detect POLR2A mRNAs in FFPE tissue specimens. A: Chromogenic staining was performed with the full PolR2A probe set consisting of 20 target probe pairs or with dapB probe set as negative control. Nuclei were counterstained with hematoxylin. B: The PolR2A probe set was divided into 20 individual target probe pairs, numbered 1 to 20 based on their location along the target (5′ to 3′). Chromogenic staining was then performed using one target pair (TP 10; left) or a combination of two target pairs (TP 10 and TP 11; middle) or a combination of three consecutive target pairs located at the middle of the full probe set (TP 9, TP 10, and TP 11; right). Insets: Higher magnification views. Similar experiments were also performed using one, two, or three target pairs located at the 5′ or 3′ end of the probe set; similar results were observed.
Detection of HPV E6/E7 mRNA. Cells from three cervical cancer cell lines known to contain different HPV subtypes [ie, SiHa (HPV-16), HeLa (HPV-18), and MS751 (HPV-45)] were stained by RNAscope with probe sets targeting HPV-16, HPV-18, and HPV-45. Positive staining was detected with an alkaline phosphatase label probe followed by reaction with Fast Red substrate (red), which was visualized under a fluorescence microscope. Cells were counterstained with DAPI (blue).
Single-copy HER2 mRNA detection in HeLa cells. A: The same fluorescently labeled HER2 probes and signal amplification system were used to detect HER2 genomic DNA (see Figure 3A) and HER2 mRNA (green) in HeLa and SK-BR-3 cells. B: Relative signal intensities of individual fluorescent dots in HeLa cells were quantified using CellProfiler image analysis software for identifying and quantifying cell phenotypes (Broad Institute, Cambridge, MA; http://www.cellprofiler.org) and were plotted as histograms for RNA and DNA.
References
- 1.Hamburg M.A., Collins F.S. The path to personalized medicine [Erratum appeared in N Engl J Med 2010;363:1092] N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C., Piccart M.J. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 3.van 't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Schreiber G.J., Kerkhoven R.M., Roberts C., Linsley P.S., Bernards R., Friend S.H. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 4.Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 5.Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuaqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., Liotta L.A. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 6.Bonner R.F., Emmert-Buck M., Cole K., Pohida T., Chuaqui R., Goldstein S., Liotta L.A. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278 doi: 10.1126/science.278.5342.1481. 1481, 1483. [DOI] [PubMed] [Google Scholar]
- 7.Levsky J.M., Singer R.H. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 8.Matos LL de, Trufelli D.C., de Matos M.G.L., da Silva Pinhal M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:9–20. doi: 10.4137/bmi.s2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulley M.L. Molecular diagnosis of Epstein-Barr virus-related diseases. J Mol Diagn. 2001;3:1–10. doi: 10.1016/S1525-1578(10)60642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambinder R.F., Mann R.B. Detection and characterization of Epstein-Barr virus in clinical specimens. Am J Pathol. 1994;145:239–252. [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmood R., Mason I. In-situ hybridization of radioactive riboprobes to RNA in tissue sections. Methods Mol Biol. 2008;461:675–686. doi: 10.1007/978-1-60327-483-8_45. [DOI] [PubMed] [Google Scholar]
- 12.Player A.N., Shen L.P., Kenny D., Antao V.P., Kolberg J.A. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2001;49:603–612. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 13.Bushnell S., Budde J., Catino T., Cole J., Derti A., Kelso R., Collins M.L., Molino G., Sheridan P., Monahan J., Urdea M. ProbeDesigner: for the design of probesets for branched DNA (bDNA) signal amplification assays. Bioinformatics. 1999;15:348–355. doi: 10.1093/bioinformatics/15.5.348. [DOI] [PubMed] [Google Scholar]
- 14.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., Hicks D.G., Lester S., Love R., Mangu P.B., McShane L., Miller K., Osborne C.K., Paik S., Perlmutter J., Rhodes A., Sasano H., Schwartz J.N., Sweep F.C., Taube S., Torlakovic E.E., Valenstein P., Viale G., Visscher D., Wheeler T., Williams R.B., Wittliff J.L., Wolff A.C. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–922. doi: 10.5858/134.6.907. [Erratum appeared in Arch Pathol Lab Med 2010;134:1101] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 16.Canales R.D., Luo Y., Willey J.C., Austermiller B., Barbacioru C.C., Boysen C., Hunkapiller K., Jensen R.V., Knight C.R., Lee K.Y., Ma Y., Maqsodi B., Papallo A., Peters E.H., Poulter K., Ruppel P.L., Samaha R.R., Shi L., Yang W., Zhang L., Goodsaid F.M. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 17.Segal G.H., Shick H.E., Tubbs R.R., Fishleder A.J., Stoler M.H. In situ hybridization analysis of lymphoproliferative disorders: Assessment of clonality by immunoglobulin light-chain messenger RNA expression. Diagn Mol Pathol. 1994;3:170–177. [PubMed] [Google Scholar]
- 18.Beck R.C., Tubbs R.R., Hussein M., Pettay J., Hsi E.D. Automated colorimetric in situ hybridization (CISH) detection of immunoglobulin (Ig) light chain mRNA expression in plasma cell (PC) dyscrasias and non-Hodgkin lymphoma. Diagn Mol Pathol. 2003;12:14–20. doi: 10.1097/00019606-200303000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Harris L., Fritsche H., Mennel R., Norton L., Ravdin P., Taube S., Somerfield M.R., Hayes D.F., Bast R.C., Jr, American Society of Clinical Oncology American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 20.Annecke K., Schmitt M., Euler U., Zerm M., Paepke D., Paepke S., von Minckwitz G., Thomssen C., Harbeck N. uPA and PAI-1 in breast cancer: review of their clinical utility and current validation in the prospective NNBC-3 trial. Adv Clin Chem. 2008;45:31–45. doi: 10.1016/s0065-2423(07)00002-9. [DOI] [PubMed] [Google Scholar]
- 21.Castelló R., Landete J.M., España F., Vázquez C., Fuster C., Almenar S.M., Ramón L.A., Radtke K.P., Estellés A. Expression of plasminogen activator inhibitors type 1 and type 3 and urokinase plasminogen activator protein and mRNA in breast cancer. Thromb Res. 2007;120:753–762. doi: 10.1016/j.thromres.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen B.S., Sehested M., Duun S., Rank F., Timshel S., Rygaard J., Johnsen M., Danø K. Urokinase plasminogen activator is localized in stromal cells in ductal breast cancer. Lab Invest. 2001;81:1485–1501. doi: 10.1038/labinvest.3780363. [DOI] [PubMed] [Google Scholar]
- 23.Qian X., Lloyd R.V. Recent developments in signal amplification methods for in situ hybridization. Diagn Mol Pathol. 2003;12:1–13. doi: 10.1097/00019606-200303000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Femino A.M. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 26.Nuovo G.J. In situ PCR: protocols and applications. PCR Methods Applications. 1995;4:S151–S167. doi: 10.1101/gr.4.4.s151. [DOI] [PubMed] [Google Scholar]
- 27.Speel E.J., Saremaslani P., Roth J., Hopman A.H., Komminoth P. Improved mRNA in situ hybridization on formaldehyde-fixed and paraffin-embedded tissue using signal amplification with different haptenized tyramides. Histochem Cell Biol. 1998;110:571–577. doi: 10.1007/s004180050319. [DOI] [PubMed] [Google Scholar]
- 28.Ukpo O.C., Flanagan J.J., Ma X.J., Luo Y., Thorstad W.L., Lewis J.S., Jr High-risk human papillomavirus E6/E7 mRNA detection by a novel in-situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2011;35:1343–1350. doi: 10.1097/PAS.0b013e318220e59d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of probe pairs required for generating visible signal in chromogenic RNAscope assay. HeLa cell FFPE pellet was used as a model system for testing the number of probe pairs needed to detect POLR2A mRNAs in FFPE tissue specimens. A: Chromogenic staining was performed with the full PolR2A probe set consisting of 20 target probe pairs or with dapB probe set as negative control. Nuclei were counterstained with hematoxylin. B: The PolR2A probe set was divided into 20 individual target probe pairs, numbered 1 to 20 based on their location along the target (5′ to 3′). Chromogenic staining was then performed using one target pair (TP 10; left) or a combination of two target pairs (TP 10 and TP 11; middle) or a combination of three consecutive target pairs located at the middle of the full probe set (TP 9, TP 10, and TP 11; right). Insets: Higher magnification views. Similar experiments were also performed using one, two, or three target pairs located at the 5′ or 3′ end of the probe set; similar results were observed.
Detection of HPV E6/E7 mRNA. Cells from three cervical cancer cell lines known to contain different HPV subtypes [ie, SiHa (HPV-16), HeLa (HPV-18), and MS751 (HPV-45)] were stained by RNAscope with probe sets targeting HPV-16, HPV-18, and HPV-45. Positive staining was detected with an alkaline phosphatase label probe followed by reaction with Fast Red substrate (red), which was visualized under a fluorescence microscope. Cells were counterstained with DAPI (blue).
Single-copy HER2 mRNA detection in HeLa cells. A: The same fluorescently labeled HER2 probes and signal amplification system were used to detect HER2 genomic DNA (see Figure 3A) and HER2 mRNA (green) in HeLa and SK-BR-3 cells. B: Relative signal intensities of individual fluorescent dots in HeLa cells were quantified using CellProfiler image analysis software for identifying and quantifying cell phenotypes (Broad Institute, Cambridge, MA; http://www.cellprofiler.org) and were plotted as histograms for RNA and DNA.