Abstract
Alterations in mitochondrial oxidative phosphorylation have long been documented in tumors. Other types of mitochondrial dysfunction, including altered reactive oxygen species (ROS) production and apoptosis, also can contribute to tumorigenesis and cancer phenotypes. Furthermore, mutation and altered amounts of mitochondrial DNA (mtDNA) have been observed in cancer cells. However, how mtDNA instability per se contributes to cancer remains largely undetermined. Mitochondrial transcription factor A (TFAM) is required for expression and maintenance of mtDNA. Tfam heterozygous knock-out (Tfam+/−) mice show mild mtDNA depletion, but have no overt phenotypes. We show that Tfam+/− mouse cells and tissues not only possess less mtDNA but also increased oxidative mtDNA damage. Crossing Tfam+/− mice to the adenomatous polyposis coli multiple intestinal neoplasia (APCMin/+) mouse cancer model revealed that mtDNA instability increases tumor number and growth in the small intestine. This was not a result of enhancement of Wnt/β-catenin signaling, but rather appears to involve a propensity for increased mitochondrial ROS production. Direct involvement of mitochondrial ROS in intestinal tumorigenesis was shown by crossing APCMin/+ mice to those that have catalase targeted to mitochondria, which resulted in a significant reduction in tumorigenesis in the colon. Thus, mitochondrial genome instability and ROS enhance intestinal tumorigenesis and Tfam+/− mice are a relevant model to address the role of mtDNA instability in disease states in which mitochondrial dysfunction is implicated, such as cancer, neurodegeneration, and aging.
Mitochondria are complex and essential organelles involved in many important cellular processes including metabolism, apoptosis, oxygen sensing, and signaling.1–5 Thus, how mitochondrial dysfunction contributes to human disease is multifaceted. For example, in addition to deficits in energy metabolism and ATP production, mitochondrial pathology can involve oxidative stress and aberrant cell death responses.6–8 Mammalian mitochondria contain multiple copies of the double-stranded, circular mitochondrial DNA (mtDNA) molecule that encode 13 protein components of the mitochondrial oxidative phosphorylation (OXPHOS) complexes required for electron transport and ATP synthesis.9 In addition, mtDNA harbors 2 ribosomal RNA genes and 22 transfer RNA genes required for translation of the mtDNA-encoded OXPHOS subunits in the matrix. The remainder of the approximately 1500 proteins in mitochondria is encoded in the nuclear genome and imported into the organelles. In addition to structural components and metabolic enzymes (including ∼80 OXPHOS complex subunits), these include all of the factors required for expression, replication, and maintenance of mtDNA.10,11 One important protein in this latter category is mitochondrial transcription factor A (TFAM), a mitochondrial transcriptional activator of the high-mobility-group box family12,13 involved in mtDNA packaging,14,15 copy number regulation,16,17 and base-excision repair.18
The essential nature of mammalian mtDNA was shown by knock-out of the Tfam gene in mice, resulting in early embryonic lethality caused by loss of mtDNA and respiratory insufficiency.16 Many homozygous, tissue-specific knock-outs of Tfam subsequently have been examined, providing informative yet coarse models of mitochondrial dysfunction and disease.19,20 In most human disease states involving mitochondria, including those caused by maternally inherited mtDNA mutations and cancer, mtDNA is not absent like it is in the Tfam−/− condition.21,22 Thus, additional mouse models are needed to assess the role of mtDNA instability per se in human disease. In this study, we have evaluated and tested the Tfam+/− genetic background as one such model. These animals are viable and have no reported phenotypes despite having approximately 30% to 50% mtDNA depletion in tissues. They also have essentially normal amounts of mtDNA transcripts,16 thus any phenotypes observed in these animals can be interpreted to be largely independent of effects on mitochondrial transcriptional output. Finally, lack of the yeast ortholog of TFAM, Abf2p, results in mtDNA depletion as well as increased mtDNA mutagenesis,23,24 which led us to hypothesize that the Tfam+/− condition in mice likewise would result in increased mtDNA damage (in addition to mtDNA depletion) and hence represent a sensitized genetic background in mice with regard to mtDNA instability.
Defects in mitochondrial OXPHOS in cancer cells originally were documented by Warburg,25 who proposed that a switch in metabolism away from respiration toward glycolysis (ie, aerobic glycolysis) is beneficial for tumorigenesis. In addition to metabolic alterations, mitochondria contribute to tumorigenesis and other aspects of cancer development through their direct involvement in apoptosis and the production of reactive oxygen species (ROS).2,6,26 The latter can promote oxidative stress, increase nuclear genome instability, and affect signaling pathways involved in cellular proliferation, differentiation, and adaptation to hypoxia.27–29 Alterations in mtDNA also have been reported in a variety of human cancers.30,31 For example, the majority of human colon cancer cells harbor specific mtDNA point mutations32,33 and mtDNA mutations associated with increased ROS production enhance the metastatic potential of tumor cells.34 Finally, changes in mtDNA abundance also have been associated with tumorigenesis, with certain cancer cells having a higher or lower mtDNA copy number.35–37 Notably, complete loss of mtDNA (via homozygous knock-out of Tfam) inhibits anchorage-dependent growth of cells in vitro and Kras-mediated lung tumorigenesis in mice.26 However, in most tumor cells, mtDNA is not absent, suggesting that, if mtDNA is involved in tumorigenesis, mtDNA instability (ie, increased damage, mutation load, and/or altered copy number) and its downstream consequences likely are more relevant to cancer than complete loss of mtDNA. In this study, using a sensitized background for mtDNA instability (Tfam+/−), we have addressed the contribution of mtDNA instability to tumorigenesis in the well-characterized APCMin/+ model of intestinal cancer.38,39
Materials and Methods
Cell Culturing and mtDNA Damage Analysis
Mouse embryonic fibroblasts (MEFs) were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum and 5% penicillin/streptomycin at 37°C in 5% CO2 humidified incubators. Kinetics of mtDNA repair was followed using gene-specific quantitative PCR measuring restoration of amplification of the target DNA after removal of H2O2 as described.40 Briefly, MEFs were seeded in 100-mm Petri dishes 15 to 18 hours before the experiment, and immediately before treatment cells were washed once with DMEM without any supplements. A 30-mmol/L stock of H2O2 (Sigma-Aldrich) was prepared in PBS (Invitrogen, Carlsbad, CA) and used to generate the 200-μmol/L solution (in DMEM alone) with which the cells were challenged.40 Cells were treated with H2O2 for 1 hour and were either harvested immediately (time 0) or were allowed to recover in conditioned medium for 6 or 24 hours. Total genomic DNA then was isolated and the integrity of the mtDNA was measured with quantitative PCR using two sets of primers to the mtDNA (10 kb and 117 bp). Total genomic DNA was isolated using the Qiagen (Valencia, CA) genomic tip kit, and mtDNA integrity and copy number were determined using a quantitative PCR method as described.41 Specific primers were used to amplify a 10-kb fragment of mouse mtDNA to determine mtDNA integrity and a small 117-bp fragment to monitor mtDNA copy number for normalization of the data obtained with the 10-kb fragment.41 Relative amplifications were calculated, comparing each group with the average of wild-type controls, and used to assess the damage frequency, assuming a Poisson distribution of damages on the template. The same genomic DNA isolation kit and PCR-based assay was used to assess basal mtDNA damage in snap-frozen small intestine tissues.
SDS-PAGE, Immunoblot, and mtDNA Copy Number Analyses
Lysates from MEFs or small intestine tissues (15 μg) were resolved on 10% to 12% SDS-PAGE gels. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA), immunoblotted with primary antibodies [anti-TFAM, a gift from Dr. David Clayton; anti–voltage-dependent anion-selective channel (anti-VDAC), Abcam (Cambridge, MA) #ab15895; or anti-actin, Sigma-Aldrich #A5060], and detected with peroxidase-linked antibodies and a Western Lightning chemiluminescence detection kit (PerkinElmer, Waltham, MA). For mtDNA copy number analysis, total cellular DNA was extracted from MEFs or snap-frozen tissues. A quantitative, real-time PCR method was used to determine the relative abundance of mtDNA versus nuclear 18S ribosomal RNA using mtDNA and nuclear primer sets in two parallel PCR reactions as described previously.42 Relative mtDNA copy number was calculated as the ratio of the amount of amplification obtained with mtDNA versus nuclear 18S rDNA primer sets for each sample and plotted normalized to the control group.
Flow Cytometry Analysis of Mitochondrial Mass and Membrane Potential
MEF cells were cultured in DMEM 10% fetal bovine serum and stained with 100 nmol/L of Mitotracker Green FM (Invitrogen) or Mitotracker Red (Invitrogen) dissolved in DMEM (incubated at 37°C for 20 minutes). Cells then were washed three times with PBS containing 1% fetal bovine serum and resuspended in 200 μL of 1× PBS containing 1% fetal bovine serum. Stained cells (∼10,000 cells per group) then were analyzed by flow cytometry using a FACSCalibur (BD Biosciences, San Jose, CA). Flow cytometry data were analyzed using FlowJo software version 8.7.3 (Treestar, Ashland, OR) and all experiments were performed in triplicate.
Mouse Strains and Tumorigenesis Analysis
All procedures were approved by the Yale University Animal Care and Use Committee. To produce Tfam+/− mice, we crossed Tfam double-floxed mice with those containing a β-actin promoter-driven Cre recombinase (obtained from Jackson Laboratory, Bar Harbor, ME). The resulting global Tfam+/− mice were back-crossed 10 times to wild-type C57BL/6J (B6) mice to remove the Cre transgene and purify the genetic background. The genotype of Tfam+/− mice was determined by a multiplex PCR assay (see Supplemental Figure S1 at http://ajp.amjpathol.org). To generate APCMin/+ Tfam+/− we crossed male APCMin/+ (obtained from Jackson Laboratory) and female Tfam+/− mice. To produce APCMin/+ MCAT, male APCMin/+ mice were crossed to female MCAT (obtained from Dr. Peter S. Rabinovitch). To score polyposis male APCMin/+, APCMin/+ Tfam+/−, and APCMin/+ MCAT mice were sacrificed, and the small and large intestines were removed immediately. The small intestine was cut into thirds (proximal, middle, and distal), and each segment was flushed gently with PBS to remove fecal material, cut longitudinally, and splayed flat. To visualize polyps more clearly, we applied Indigo amine dye to the mucosal surface of the opened intestine and measured macroadenoma numbers and diameters. For histologic examination, 4% formaldehyde/PBS-fixed intestines were prepared and embedded in paraffin, and 5-μm sections were prepared for H&E staining and immunohistochemistry (anti–β-catenin antibody from BD #610154).
Multiplex PCR for Tfam+/− Mouse Genotyping
Standard protocols were used for PCR. Genotyping was performed by multiplex PCR for 35 cycles with primers A, B, and C (see Supplemental Figure S1 at http://ajp.amjpathol.org). The primer sequences were as follows: primer A: 5′-CTCTAGCCCGGGTCCTATCT-3′, primer B: 5′-GTAACAGCAGACAACTTGTG-3′, and primer C: 5′-CAGTGGTGTGGTGGTTGAAG-3′.
Real-Time Quantitative PCR
Total RNA was isolated from polyps and neighboring normal intestine tissues using TRIzol (Invitrogen), and 1 μg RNA converted to cDNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems) following the manufacturer's instructions. One tenth of the cDNA was subjected to a 25 μL PCR performed in an iCycler thermal cycler (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) with PCR primers [c-MYC (alias c-myc): forward: 5′-TGAGGAAACGACGAGAACAGTTG-3′, and reverse: 5′-CAAGGTTGTGAGGTTAGGCTTTGAG-3′; cyclin D1: forward: 5′-CATCAAGTGTGACCCGGACTG-3′, and reverse: 5′-CCTCCTCCTCAGTGGCCTTG-3′ and β-actin primer sets (forward: 5′-GGTCATCACTATTGGCAACG-3′, and reverse: 5′-CCTCACCAAGCTAAGGATGC-3′). Expression quantities were normalized to the β-actin transcript and the ΔΔCt method was used to calculate the relative levels of expression.
Mitochondrial ROS Production
Mitochondria were isolated from small intestine tissues from APCMin/+ and APCMin/+ Tfam+/− mice by differential centrifugation as described.43 Mitochondrial protein concentration was determined by the Bradford method. Mitochondrial H2O2 production was measured using Amplex Red (Invitrogen) in the presence of horseradish-peroxidase, respiration substrates (glutamate and malate), and, when indicated, a respiratory inhibitor (antimycin A; Sigma-Aldrich) according to the manufacturer's instructions. Data are expressed as the change in arbitrary fluorescence units produced from equal amounts of mitochondria (based on total protein input) as a function of time. H2O2 production in mitochondria isolated from Tfam+/−, APCMin/+, and APCMin/+ MCAT mice were performed as described earlier. Immunoblot analysis for catalase in MCAT mice was performed using anti-catalase antibody (Sigma-Aldrich #C0979).
Histologic Analysis and Immunohistochemistry
Mouse small intestine sections containing polyps from APCMin/+ and APCMin/+ Tfam+/− (n = 5 per group) were subjected to H&E staining and β-catenin immunohistochemistry. Stained sections were analyzed for tumor histology and Wnt/β-catenin signaling by a gastrointestinal pathologist without their knowledge of the identity of the samples. Anti–β-catenin antibody (BD Biosciences #610154) was used for immunohistochemistry.
Statistical Analysis
Error bars in all Figures represent the mean ± SEM. The Student's two-tailed t-test was used for the determination of statistical relevance between groups, and a P value of <0.05 was considered significant.
Results
Mice Heterozygous for Tfam (Tfam+/−) Have Increased Oxidative mtDNA Damage Susceptibility in Addition to mtDNA Depletion
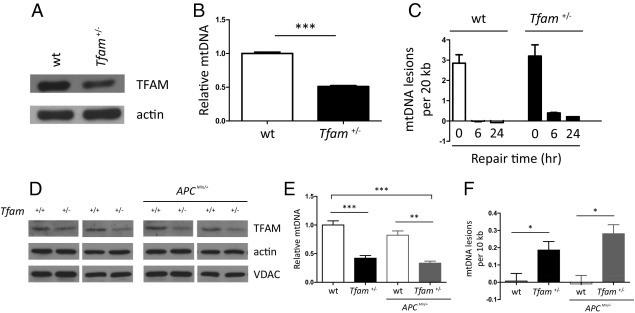
We sought to determine whether mtDNA instability contributes to cancer development using the APCMin/+ mouse model of intestinal tumorigenesis.39 We reasoned that the Tfam+/− background would be a salient for cancer studies of this type because mtDNA would still be present, but unstable, similar to the situation in tumors. Accordingly, we found that MEFs isolated from Tfam+/− mice not only had reduced amounts of TFAM (Figure 1A) and mtDNA (Figure 1B) as expected, but displayed a reduced ability to repair oxidative mtDNA damage (Figure 1C) compared with those isolated from the wild-type (Tfam+/+) background. A small decrease in mitochondrial mass and membrane potential also was observed in Tfam+/− MEFs (see Supplemental Figure S2 at http://ajp.amjpathol.org), however, no decrease in VDAC (used as a mitochondrial housekeeping marker; compared with actin) were observed in intestine tissue (Figure 1D). Importantly, reduced amounts of TFAM and mtDNA also were observed in intestinal tissue from Tfam+/− mice (Figure 1, D and E) and, consistent with an increased susceptibility to endogenous oxidative insults, higher basal oxidative mtDNA damage was observed in this tissue (Figure 1F). These phenotypes were maintained in the APCMin/+ Tfam+/− genetic background (Figure 1, D–F) and there was an obvious trend toward even greater mtDNA damage, although this was not statistically significant (Figure 1F). Altogether, these results led us to conclude that the Tfam+/− condition is indeed sensitized for oxidative mtDNA damage, which, in combination with the mtDNA depletion, makes it an appropriate background in which to assess the contribution of mtDNA instability to tumorigenesis.
Figure 1.
Tfam+/− MEFs and intestinal tissues exhibit mtDNA instability. A: Western blot of TFAM protein in wild-type (wt) and Tfam+/− MEFs, with actin probed as a loading control. B: Relative mtDNA copy number in the same MEFs as in A. C: MEFs in A were exposed to H2O2 for 60 minutes and harvested immediately (0 hours recovery point) or allowed to recover for 6 or 24 hours (6-hour and 24-hour recovery points). Repair of H2O2-induced oxidative mtDNA damage at the indicated time points was measured by a quantitative PCR method and plotted as the number of mtDNA lesions/20 kb. D: Western blot of TFAM protein in mouse intestinal tissues from wt (Tfam+/+) and Tfam+/− with the APC+/+ (left two panels) and APCMin/+ (right two panels) backgrounds. Actin was probed as a loading control and VDAC as an indicator of mitochondrial abundance. E: Relative mtDNA copy number (E) and oxidative mtDNA damage levels (F) in the same samples as in D. All cell culture experiments were performed at least in triplicate. E and F: Five mice were analyzed of each genotype and the mean ± SD is plotted. *P < 0.05, **P < 0.01, and ***P < 0.001.
Intestinal Tumorigenesis in APCMin/+ Mice Is Increased in the Tfam+/− mtDNA-Instability Background
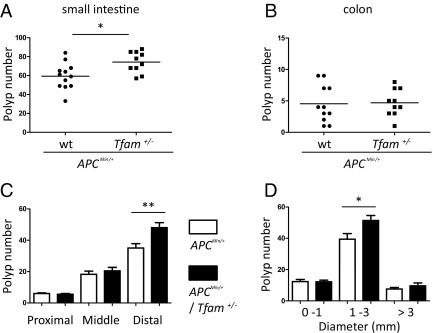
To test directly the contribution of enhanced mtDNA instability to tumorigenesis, we examined intestinal polyp formation in APCMin/+ Tfam+/− mice (ie, tumor-prone mice in an mtDNA-instability background) compared with APCMin/+ mice with wild-type levels of Tfam (ie, APCMin/+ Tfam+/+). Compared with sex- and age-matched APCMin/+ Tfam+/+ cohorts, we found a significant increase in the number of macroadenomas in the small intestine of APCMin/+ Tfam+/− mice (Figure 2A), whereas polyposis in colon was comparable (Figure 2B). The polyps formed in APCMin/+ Tfam+/− mice are histopathologically similar to those in APCMin/+ Tfam+/+ mice, suggesting they are benign adenomas (see Supplemental Figure S3A at http://ajp.amjpathol.org). In the distal part of the small intestine, there was an increase in polyp number in APCMin/+ Tfam+/− mice (Figure 2C) and there was an increase in medium-sized (1 to 3 mm in diameter) polyps throughout the small intestine (Figure 2D). These results suggest that tumor growth as well as initiation is enhanced by loss of mtDNA stability in APCMin/+ Tfam+/− mice.
Figure 2.
Mitochondrial genomic instability (Tfam+/−) increases intestinal polyposis in APCMin/+ mice. Total polyp number in the small intestine (A) and colon (B) were scored in 18- to 20-week-old male mice (APCMin/+ n = 12, and APCMin/+ Tfam+/− n = 10). Each point in the graph indicates the total polyp number from one mouse. C: Polyp number in the proximal, middle, and distal small intestine. D: Small intestinal polyp number as a function of size. C and D: White bars indicate APCMin/+ and black bars indicate APCMin/+ Tfam+/− genetic backgrounds and the mean ± 1 SD are shown. *P < 0.05 and **P < 0.01.
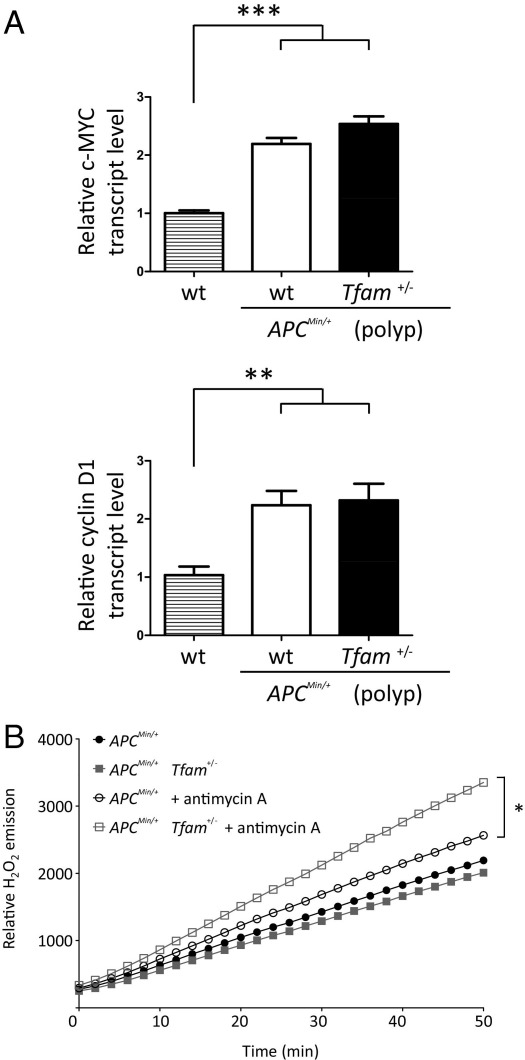
A key event that initiates polyposis in APCMin/+ mice is activation of the Wnt signaling pathway, resulting in β-catenin–mediated target gene expression.44 Thus, we examined the possibility that the increased tumorigenesis observed in APCMin/+ Tfam+/− mice was caused by further activation of this pathway. Although we observed increased expression of two well-characterized Wnt target genes, cyclin D1 and c-MYC, in polyps from APCMin/+ mice, their expression was not enhanced in the APCMin/+ Tfam+/− background (Figure 3A). We also observed no exacerbation of β-catenin nuclear localization in the APCMin/+ Tfam+/− background (see Supplemental Figure S3B at http://ajp.amjpathol.org). Altogether, these results indicate that the increased intestinal tumorigenesis in APCMin/+ Tfam+/− mice is not attributable to additional activation of Wnt/β-catenin signaling, but rather through a separate pathway.
Figure 3.
Wnt/β-catenin signaling activation is comparable in polyps from APCMin/+ and APCMin/+ Tfam+/− mice, but mitochondrial ROS production capacity is higher in the APCMin/+ Tfam+/− background. A: Amount of c-MYC (top) and cyclin D1 (center) transcripts from size-matched polyps in APCMin/+ and APCMin/+ Tfam+/− mice are shown normalized to that in wild-type (wt). Normal tissues adjacent to polyps were used as wild-type controls. Four mice of each genotype were analyzed and the mean ± 1 SD is plotted. B: Rates of H2O2 production (Amplex Red assay; Invitrogen) in mitochondria purified from intestine of the indicated mice are shown. A representative plot from three trials is shown. *P < 0.05, **P < 0.01, and ***P < 0.001.
Tfam+/− mice are viable and show no obvious phenotypes on their own, suggesting that basal mitochondrial function in tissues is not compromised to a significant extent.16 In addition, our own results show that Tfam+/− MEFs have only minor changes in mitochondrial mass and membrane potential (see Supplemental Figure S2 at http://ajp.amjpathol.org) and that mitochondrial biogenesis is not affected markedly in Tfam+/− tissues (Figure 1D). However, a common manner in which mitochondria affect normal and disease states is through the production of ROS.27,45 In fact, it has been suggested that mitochondrial ROS can act as signaling molecules (eg, to promote cell division).3,28 To test the hypothesis that changes in mitochondrial ROS production may underlie some of the enhanced tumorigenesis in the APCMin/+ Tfam+/− mice, we measured H2O2 production from isolated mitochondria from mouse intestine. Although basal mitochondrial ROS generation was comparable between APCMin/+ Tfam+/− and APCMin/+ Tfam+/+ mice, mitochondrial ROS production in response to antimycin A (an inhibitor that affects complex III, a known source of ROS) was significantly higher in mitochondria from APCMin/+ Tfam+/− mice (Figure 3B). There was no difference in basal or induced ROS production from mitochondria isolated from Tfam+/+ and Tfam+/− mouse intestine (ie, in the absence of APCMin/+; see Supplemental Figure S4A at http://ajp.amjpathol.org). These data indicate that, in the tumor-prone APCMin/+ background, mtDNA instability caused by the Tfam+/− circumstance could result in increased mitochondrial ROS production that contributes to tumorigenesis.
Reducing Mitochondrial ROS via Targeted Expression of Catalase Decreases Tumorigenesis in APCMin/+ Mice
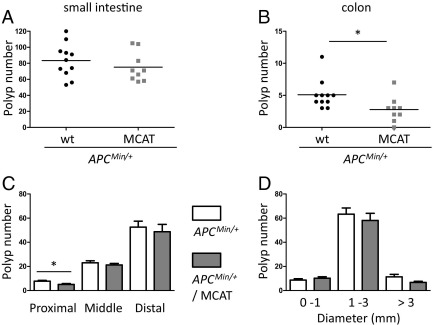
To test the hypothesis that mitochondrial ROS production is involved in intestinal tumorigenesis, we crossed APCMin/+ mice to transgenic mice that target catalase to mitochondria (MCAT), which have reduced mitochondrial ROS and oxidative stress in a variety of tissues and circumstances.46–48 Compared with control APCMin/+ mice, APCMin/+ MCAT mice showed fewer polyps in the colon (Figure 4B) and in the proximal region of the small intestine (Figure 4C). In the small intestine, there was a trend toward fewer larger polyps (>3 mm) (Figure 4D), perhaps indicating a late effect on tumor growth. We confirmed that catalase was overexpressed in the intestine of the MCAT mice used in this study (see Supplemental Figure S4C at http://ajp.amjpathol.org), and determined that the basal rate of hydrogen peroxide production in mitochondria isolated from the intestine of APCMin/+ MCAT mice was reduced significantly relative to APCMin/+ without MCAT (see Supplemental Figure S4B at http://ajp.amjpathol.org).
Figure 4.
Targeted overexpression of catalase to mitochondria decreases intestinal polyposis in APCMin/+ mice. Total polyp numbers in small intestine (A) and colon (B) from 20-week-old male mice (APCMin/+ n = 11, and APCMin/+ MCAT n = 9). Each point in the graph indicates the total polyp number from one mouse. C: Polyp number in the proximal, middle, and distal small intestine is shown. D: Polyp number in the small intestine as a function of size is shown. C and D: White bars indicate APCMin/+ and gray bars indicate APCMin/+ MCAT. The mean ± SD are plotted. *P < 0.05.
Discussion
Nuclear genome instability is involved in cancer at many levels.49 However, despite alterations in mtDNA being associated with a variety of tumors,30,31,35 the contribution of mtDNA instability to the tumorigenesis process has not been assessed directly. Here, we addressed this important issue by crossing a Tfam+/− mouse strain, which shows mtDNA depletion and increased susceptibility to oxidative mtDNA damage (Figures 1 and 2), to the APCMin/+ mouse model of intestinal cancer. The main conclusion we draw from our results is that mtDNA instability can contribute to tumorigenesis in the APCMin/+ mouse model by a mechanism that is independent of canonical Wnt/β-catenin signaling and likely involves increased oxidative mtDNA damage and mitochondrial ROS production (Figures 2 and 3).
Tfam homozygous knock-outs (Tfam−/−) have been performed in a variety of tissues to address the requirement of mtDNA in tissue function and disease in the mouse. Although these models have been useful to a significant degree, they result in fast and complete loss of mtDNA and mitochondrial OXPHOS, limiting their use in analyzing the effects of persistent mtDNA damage. Here, we have used Tfam+/− mice as a genetic background that is sensitized to mtDNA instability owing to mtDNA depletion and increased oxidative damage susceptibility (Figure 1). Our logic for using this model is that, under many pathogenic circumstances, including cancer, mtDNA is not lost, but rather present in a depleted, damaged, and/or mutated state. Our results show that this premise is reasonable and provide the first direct in vivo demonstration that mtDNA instability per se can contribute to tumorigenesis. This new mouse model may be generally useful for assessing the role of mtDNA instability in other disease states, especially those that involve oxidative stress. It also may be instrumental in determining the role of oxidative mtDNA damage in aging.
Even though common in human colon cancers, specific mtDNA mutations do not result in major perturbations of mitochondrial oxygen consumption or respiratory chain enzymatic activities as one might predict.32 This suggests that these are passenger mutations either do not contribute to the tumorigenesis process or they work in concert with other polymorphic variations in mtDNA or nuclear DNA.32 Our results show clearly that mtDNA instability can enhance tumorigenesis in vivo (Figure 2) and are consistent with the idea that in certain nuclear genetic backgrounds mitochondrial function is more prone to the effects of mtDNA damage and mutation. In particular, we find that mtDNA instability leads to an enhanced capacity to generate mitochondrial ROS when superimposed on the already tumor-prone APCMin/+ nuclear genetic background (Figure 3B). These results highlight that nuclear and mitochondrial genome instability likely cross-talk and cooperate in the tumorigenesis process. Similar complex interactions between mtDNA and nuclear genetic background potentially hold significance under other cancer scenarios in which mtDNA mutagenesis has been implicated and in other human disease states.7 Finally, that increased ROS production in purified mitochondria from APCMin/+ Tfam+/− intestine is observed only when complex III is inhibited (Figure 3B), suggests that the effects of mitochondrial ROS in vivo might only be manifest after tumorigenesis has begun and mitochondrial respiration is down-regulated (eg, after initiation of the “Warburg Effect”25).
With regard to the role of mitochondrial ROS per se in tumorigenesis, our results show that increasing mitochondrial hydrogen peroxide detoxification via mitochondria-targeted expression of catalase has a positive effect in the APCMin/+ cancer model (Figure 4). At this point we do not know the precise role of mitochondrial ROS in the process, but promoting mitochondrial or cellular oxidative stress and genome instability are likely possibilities. For example, the increased ROS could promote nuclear genome instability that leads to increased rates of APC loss of heterozygosity or activation/inactivation of oncogenes/tumor suppressors. Alternatively, mitochondrial ROS may be acting as signaling molecules3,28 that enhance tumor initiation and/or growth by affecting proliferation or differentiation of intestinal stem cells involved in polyp formation.50 Our results show that ROS derived from inhibition of mitochondrial complex III are increased in mitochondria from the APCMin/+ Tfam+/− mouse intestine. That complex III–derived ROS are implicated in oxygen sensing51 and in activating the mitogen-activated protein kinases/extracellular-signal-regulated kinases (MAPK/ERK) pathway to support K-ras-induced anchorage-dependent cell growth26 is consistent with mitochondrial ROS signaling being one component of the effects we observe on tumorigenesis in APCMin/+ mice.
Although dysregulation of Wnt/β-catenin signaling in intestinal epithelial cells is a well-characterized event initiating intestinal polyposis,50 tumorigenesis in APCMin/+ mice is complex. In addition to the earlier-mentioned potential effects of increased mitochondrial ROS production on the tumor or initiating cells, oxidative stress can influence innate immune responses and inflammation that are involved in intestinal tumorigenesis in APCMin/+ mice.52,53 Thus, it is a formal possibility that some of the effects we observe with regard to mtDNA instability and ROS on tumorigenesis are occurring via effects on immune system–related cells acting extrinsically on tumor or initiating cells.
Although our results with the MCAT model clearly implicate mitochondrial ROS in intestinal tumorigenesis, we emphasize that we have not unequivocally shown the direct involvement of mitochondrial ROS in the increased tumorigenesis observed in the APCMin/+ Tfam+/− mouse model. Given that Tfam is a multifunctional protein it is expected that the effects of reduced Tfam levels on intestinal tumorigenesis in APCMin/+ may include those other than, or in addition to, increased ROS production and oxidative mtDNA damage. Nonetheless, the results of this study open new avenues of investigation into the precise role of mitochondrial dysfunction and ROS in cancer development and provide a new experimental paradigm to investigate the role of mtDNA instability in other physiological and disease states in Tfam+/− mice.
Acknowledgments
We thank Drs. Eddie Fox and Lawrence Loeb for sharing data and insights related to this study that were ultimately not included; Drs. Namiko Hoshi, Hui-Young Lee, and Zimei Zhang for technical assistance; and Dr. Peter Rabinovitch for providing the MCAT mouse strain.
Footnotes
Supported by Program Project Grant ES-011163 from the National Institutes of Health (G.S.S.), and Department of Defense grant 56027LS from the Army Research Office (J.H.S.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi:10.1016/j.ajpath.2011.10.003.
Supplementary data
Tfam+/− mouse genotyping strategy. Shown are PCR products resolved in a 2% agarose gel. A Tfam wild-type allele produces a 396-bp PCR product from PCR primers A and B, whereas A Tfam deleted allele produces a 438-bp PCR product from PCR primers A and C.
Mitochondrial mass and potential in cultured Tfam+/− MEFs. Mitochondrial mass (A) and potential (B) were assessed by flow cytometry using Mitotracker Green FM (Invitrogen) and Mitotracker Red (Invitrogen) dyes, respectively. Data are expressed as arbitrary fluorescence units.
Analysis of APCMin/+ and APCMin/+ Tfam+/− H&E staining and β-catenin immunostaining. Representative H&E staining (A) for polyp histology and β-catenin immunostaining (B) for Wnt/β-catenin signaling are shown. Mouse genotypes are indicated below each image.
Mitochondrial ROS production and catalase expression in mouse intestine. A and B: Mitochondria were isolated from small intestine of wild-type, Tfam+/−, and MCAT mice by differential centrifugation and mitochondrial ROS were measured as described in Materials and Methods and plotted in Figure 3B. C: Steady-state levels of catalase in mouse intestine tissues from wild-type and MCAT mice examined by Western immunoblot analysis. Actin was probed as a loading control.
References
- 1.Jones R.G., Thompson C.B. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gogvadze V., Orrenius S., Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 2008;18:165–173. doi: 10.1016/j.tcb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Green D.R., Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 6.Orrenius S., Gogvadze V., Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 7.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kujoth G.C., Leeuwenburgh C., Prolla T.A. Mitochondrial DNA mutations and apoptosis in mammalian aging. Cancer Res. 2006;66:7386–7389. doi: 10.1158/0008-5472.CAN-05-4670. [DOI] [PubMed] [Google Scholar]
- 9.Shadel G.S., Clayton D.A. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 10.Shutt T.E., Shadel G.S. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonawitz N.D., Clayton D.A., Shadel G.S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Parisi M.A., Xu B., Clayton D.A. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shutt T.E., Lodeiro M.F., Cotney J., Cameron C.E., Shadel G.S. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci U S A. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanki T., Ohgaki K., Gaspari M., Gustafsson C.M., Fukuoh A., Sasaki N., Hamasaki N., Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 17.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 18.Canugovi C., Maynard S., Bayne A.C., Sykora P., Tian J., de Souza-Pinto N.C., Croteau D.L., Bohr V.A. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trifunovic A., Larsson N.G. Tissue-specific knockout model for study of mitochondrial DNA mutation disorders. Methods Enzymol. 2002;353:409–421. doi: 10.1016/s0076-6879(02)53065-2. [DOI] [PubMed] [Google Scholar]
- 20.Vempati U.D., Torraco A., Moraes C.T. Mouse models of oxidative phosphorylation dysfunction and disease. Methods. 2008;46:241–247. doi: 10.1016/j.ymeth.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H.C., Chang C.M., Chi C.W. Somatic mutations of mitochondrial DNA in aging and cancer progression. Ageing Res Rev. 2010;9(Suppl 1):S47–S58. doi: 10.1016/j.arr.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Schon E.A., Bonilla E., DiMauro S. Mitochondrial DNA mutations and pathogenesis. J Bioenerg Biomembr. 1997;29:131–149. doi: 10.1023/a:1022685929755. [DOI] [PubMed] [Google Scholar]
- 23.O'Rourke T.W., Doudican N.A., Mackereth M.D., Doetsch P.W., Shadel G.S. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebedeva M.A., Shadel G.S. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA inheritance without compromising mitochondrial gene expression. Cell Cycle. 2007;6:2048–2057. doi: 10.4161/cc.6.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerutti P.A. Prooxidant states and tumor promotion. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 28.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 29.Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D., Simon M.C., Hammerling U., Schumacker P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Penta J.S., Johnson F.M., Wachsman J.T., Copeland W.C. Mitochondrial DNA in human malignancy. Mutat Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- 31.Brandon M., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 32.Polyak K., Li Y., Zhu H., Lengauer C., Willson J.K., Markowitz S.D., Trush M.A., Kinzler K.W., Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 33.He Y., Wu J., Dressman D.C., Iacobuzio-Donahue C., Markowitz S.D., Velculescu V.E., Diaz L.A., Jr, Kinzler K.W., Vogelstein B., Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 35.Lee H.C., Yin P.H., Lin J.C., Wu C.C., Chen C.Y., Wu C.W., Chi C.W., Tam T.N., Wei Y.H. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 36.Chandra D., Singh K.K. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2011;1807:620–625. doi: 10.1016/j.bbabio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan Q., Lim U., Liu C.S., Weinstein S.J., Chanock S., Bonner M.R., Virtamo J., Albanes D., Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112:4247–4249. doi: 10.1182/blood-2008-05-157974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser A.R., Pitot H.C., Dove W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 39.Moser A.R., Luongo C., Gould K.A., McNeley M.K., Shoemaker A.R., Dove W.F. ApcMin: a mouse model for intestinal and mammary tumorigenesis. Eur J Cancer. 1995;31A:1061–1064. doi: 10.1016/0959-8049(95)00181-h. [DOI] [PubMed] [Google Scholar]
- 40.Santos J.H., Hunakova L., Chen Y., Bortner C., Van Houten B. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. J Biol Chem. 2003;278:1728–1734. doi: 10.1074/jbc.M208752200. [DOI] [PubMed] [Google Scholar]
- 41.Santos J.H., Mandavilli B.S., Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol Biol. 2002;197:159–176. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- 42.D'Souza A.D., Parikh N., Kaech S.M., Shadel G.S. Convergence of multiple signaling pathways is required to coordinately up-regulate mtDNA and mitochondrial biogenesis during T cell activation. Mitochondrion. 2007;7:374–385. doi: 10.1016/j.mito.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 44.Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 45.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 46.Schriner S.E., Linford N.J., Martin G.M., Treuting P., Ogburn C.E., Emond M., Coskun P.E., Ladiges W., Wolf N., Van Remmen H., Wallace D.C., Rabinovitch P.S. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 47.Dai D.F., Santana L.F., Vermulst M., Tomazela D.M., Emond M.J., MacCoss M.J., Gollahon K., Martin G.M., Loeb L.A., Ladiges W.C., Rabinovitch P.S. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H.Y., Choi C.S., Birkenfeld A.L., Alves T.C., Jornayvaz F.R., Jurczak M.J., Zhang D., Woo D.K., Shadel G.S., Ladiges W., Rabinovitch P.S., Santos J.H., Petersen K.F., Samuel V.T., Shulman G.I. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 2010;12:668–674. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 50.Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 51.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rakoff-Nahoum S., Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 53.Chae W.J., Gibson T.F., Zelterman D., Hao L., Henegariu O., Bothwell A.L. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:5540–5544. doi: 10.1073/pnas.0912675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tfam+/− mouse genotyping strategy. Shown are PCR products resolved in a 2% agarose gel. A Tfam wild-type allele produces a 396-bp PCR product from PCR primers A and B, whereas A Tfam deleted allele produces a 438-bp PCR product from PCR primers A and C.
Mitochondrial mass and potential in cultured Tfam+/− MEFs. Mitochondrial mass (A) and potential (B) were assessed by flow cytometry using Mitotracker Green FM (Invitrogen) and Mitotracker Red (Invitrogen) dyes, respectively. Data are expressed as arbitrary fluorescence units.
Analysis of APCMin/+ and APCMin/+ Tfam+/− H&E staining and β-catenin immunostaining. Representative H&E staining (A) for polyp histology and β-catenin immunostaining (B) for Wnt/β-catenin signaling are shown. Mouse genotypes are indicated below each image.
Mitochondrial ROS production and catalase expression in mouse intestine. A and B: Mitochondria were isolated from small intestine of wild-type, Tfam+/−, and MCAT mice by differential centrifugation and mitochondrial ROS were measured as described in Materials and Methods and plotted in Figure 3B. C: Steady-state levels of catalase in mouse intestine tissues from wild-type and MCAT mice examined by Western immunoblot analysis. Actin was probed as a loading control.