The evolution of life has been closely tied to the evolution of light-harvesting systems. Even the most primitive ability to use the sun's energy would have given the earliest photoautotrophs tremendously expanded opportunities to proliferate and diversify. As early as 2.5–3.3 billion years ago (1, 2), the cyanobacteria had evolved a sophisticated linear electron transport chain with two photosystems that could pull electrons from water and give rise to molecular oxygen. One of these cyanobacteria became the ancestor of all modern chloroplasts (Fig. 1). Although there is now an enormous diversity in photosynthetic eukaryotes, the molecular structures of the two photosynthetic reaction centers and their core light-harvesting antennas have been remarkably conserved (3), along with their almost exclusive use of one type of chlorophyll (Chl), Chl a.
Figure 1.
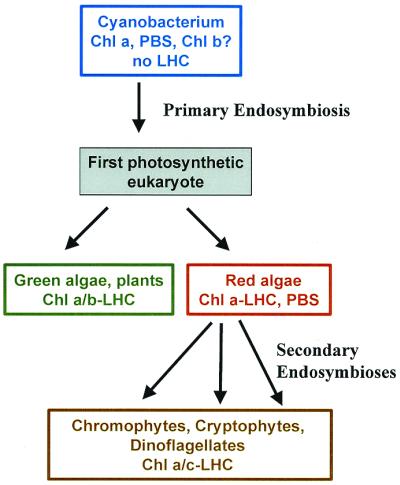
Evolution of the three major types of photosynthetic eukaryotes, based on a consensus of ultrastructure, pigment, and molecular phylogenetic data. LHC, membrane-intrinsic LHC; PBS, phycobilisome.
Where photosynthetic eukaryotes differ is in their peripheral light-harvesting antennas and accessory pigments. The red algal chloroplasts, like their cyanobacterial ancestors, use phycobilisomes, eleborate multisubunit structures with open-chain tetrapyrrole chromophores associated with the cytoplasmic surface of the thylakoid membrane. The green algal and plant chloroplasts have membrane-intrinsic antenna proteins binding Chl b as well as Chl a, whereas the many groups of brown and yellow algae (dinoflagellates, cryptophytes, and chromophytes sensu lato) have related antenna proteins binding Chl c and Chl a. The antenna proteins of these two groups are members of a very large protein family, the light-harvesting complex (LHC) superfamily, which also has members involved in photoprotection rather than light harvesting (4).
This simple story took an unexpected turn a few years ago when Elizabeth Gantt and her colleagues (5) reported that red algae had at least five proteins related immunologically to the Chl a/b proteins of higher plants and the fucoxanthin Chl a/c proteins of chromophytes. Sequencing showed that they were indeed members of the LHC superfamily, even though they bound only Chl a and were associated only with photosystem I (6–8). This showed that phycobilisomes (associated primarily with PSII) and membrane-intrinsic antennas of the LHC superfamily could coexist in the same chloroplast, and strongly supported a common evolutionary origin for all chloroplasts (8). In addition, it suggested that ancestral members of the LHC family may have had a considerable degree of flexibility in pigment binding, so they were able to adapt to binding a variety of different Chls and carotenoids.
It turns out that this adaptibility was not restricted to the past. In this issue of PNAS, Grabowski et al. (9) demonstrate quite clearly that a red algal LHC apoprotein can be reconstituted with the pigments from either a higher plant or a chromophyte alga, and that the reconstituted pigment-protein can perform the critical function of transferring energy absorbed by the accessory Chl (Chl b or Chl c) to Chl a. Furthermore, approximately eight Chls are bound per polypeptide chain, the same number bound when the protein is reconstituted with its own pigments, and close to the number found in the native complex (5). It therefore appears that the Chl-binding sites on the LHC protein may be conserved, but they are not particularly specific for which Chl they bind, and in fact, can bind Chls that their ancestors had never encountered.
Does this promiscuity result from Chl structures being very similar to each other? This could be the case for Chls a and b, because they differ only by a methyl side chain in Chl a versus a formyl side chain in Chl b (Fig. 2A). Reconstitution and mutagenesis experiments on Chl a/b proteins suggest that at least some of the eight Chl-binding sites common to all Chl a/b proteins (10, 11) can ligate either Chl a or b (11–13). One of these proteins can even be reconstituted almost exclusively with Chl b, although with somewhat altered energy transfer characteristics (13). The several variants of Chl c are similar in structure to divinyl protochlorophyllide a (Fig. 2B), the common precursor of all Chls, which also can act as a minor light-harvesting pigment (14–16). They have a completely unsaturated ring system and lack the hydrophobic phytyl tail. These features might be expected to impede the binding of Chl c to a Chl a site and even diminish protein folding, but this was not found in the experiments of Grabowski et al. (9). All of these experiments suggest that the differences in sequence among the LHC proteins (8) may have more to do with optimizing energy transfer, binding carotenoids, and forming protein–protein interactions than with binding a specific Chl.
Figure 2.
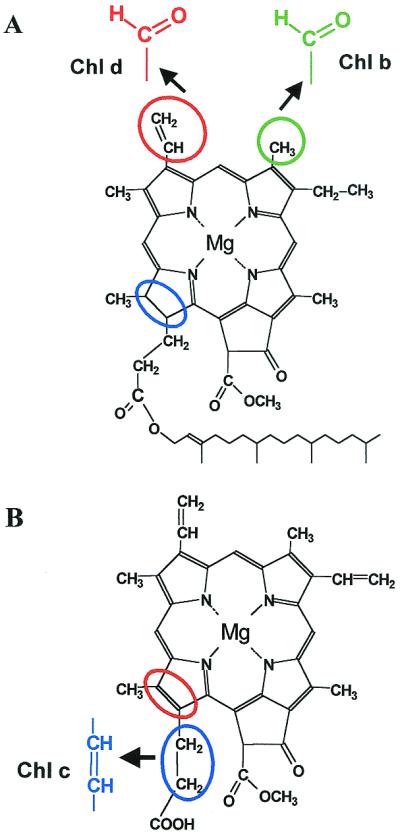
Structures of eukaryotic Chls. (A) Chlorophyll a. Note the reduced ring D (blue circle) and phytyl tail (not drawn to scale). Side-chain modifications resulting in Chl b (green) and Chl d (red) are shown. (B) Mg-divinyl protochlorophyllide a and Chl c2 (blue). Note the double bond in ring D (red circle).
There is another protein family whose members are adapted to bind different types of Chl. Three unrelated genera of cyanobacteria (often referred to collectively as prochlorophytes) have Chl a/b antenna proteins that are completely unrelated to the LHC superfamily. Instead, they belong to the same protein family as the extremely conserved Chl a-binding proteins CP47 and CP43 of the photosystem II core antenna (17, 18). This is a classic example of parallel evolution, and it suggested that the ability to make Chl b evolved separately from the evolution of the proteins that bind it (17, 18). This was confirmed when the gene for Chl a oxygenase (CAO), the enzyme that makes Chl b from Chl a was finally discovered (19), and sequences from two prochlorophytes, two green algae, and several higher plants all were found to be related (20). Unlike the Chl-binding proteins, CAO did have a common ancestor in prokaryotes and chloroplasts. This led Tomitani et al. (20) to propose that all cyanobacteria originally had the ability to make both Chl b and phycobilisomes; the ability to make phycobilisomes was lost in the four lines that led to prochlorophytes and chloroplasts, whereas CAO was lost in all other cyanobacterial lines. This is an interesting suggestion, but it requires multiple independent losses of CAO because the closest relatives of each kind of prochlorophyte do not make Chl b either (21). A more parsimonious possibility is that lateral gene transfer spread the CAO gene among the three prochlorophytes and the ancestor of the chloroplast, which was not closely related to the any of the prochlorophytes (21). It subsequently would have been lost in the line leading to the red and Chl a/c containing chloroplasts; the enzyme for the synthesis of Chl c from protochlorophyllide a would have evolved only after the secondary endosymbios(es) that gave rise to the chromophyte and dinophyte lines (Fig. 1).
The most striking example of Chl substitution is that of the prokaryote Acaryochloris marina (22), in which almost all of the Chl a has been replaced by Chl d (Fig. 2A). All of these observations suggest that the Chl complement of both prokaryotes and eukaryotes depends on which enzymes for Chl synthesis they happened to inherit from their ancestors, and that proteins of both the photosynthetic core and the distal antennas can adapt to using whatever Chl is available. In that case, why have large groups of organisms specialized in binding a particular set of Chls? The usual argument is that the addition of accessory Chls increased the absorption of light in the “green gap” between 470 and 650 nm where Chl a (in vivo) has limited absorption (e.g., ref. 23). However, the addition of Chl b decreases this gap by only a few tens of nm. The double bond in Chl c shifts the red absorption a bit more into the green region, but makes absorption less efficient. Examination of the absorption spectra of Chl a/c proteins (24) suggests that it is the carotenoids of the Chl c algae, with their absorptions extending out to 550 nm, that contribute the most to increased light absorption in the blue-green enriched aquatic environment (25). The red algae with their phycobilisomes are the only group that efficiently harvests light across the complete visible range (400–700 nm). It is therefore unclear how much of an advantage it could have been to replace phycobilisomes with Chl c and some carotenoids in a membrane-intrinsic LHC.
The study of molecular adaptation attempts to determine which amino acid substitutions were essential for a given function, to see whether change was gradual or rapid, and to detect evidence for positive selection against a large background of neutral changes (26). Molecular evolution is now an underlying theme of many papers, even when the word evolution is not in the title. The dramatic increase in high-resolution structures and the explosion of sequence data are providing new insights into the evolution of many proteins, even if understanding the evolution of their owners is more problematic. One of the most exciting new approaches is the fusion of phylogenetic methods and protein engineering to recreate ancestral proteins (27). The next stage in studying molecular evolution should be to follow the evolution of cells with engineered proteins under the relevant physiological conditions to test hypotheses about what factors might actually have favored growth or survival in their ancestors. We are lucky to live in such exciting times!
Footnotes
See companion article on page 2911.
References
- 1.Schopf J W, Packer B M. Science. 1987;237:70–73. doi: 10.1126/science.11539686. [DOI] [PubMed] [Google Scholar]
- 2.Summons R E, Jahnke L L, Hope J M, Logan G A. Nature (London) 1999;400:554–557. doi: 10.1038/23005. [DOI] [PubMed] [Google Scholar]
- 3.Schubert W-D, Klukas O, Saenger W, Witt H T, Fromme P, Krauss N. J Mol Biol. 1998;280:297–314. doi: 10.1006/jmbi.1998.1824. [DOI] [PubMed] [Google Scholar]
- 4.Green B R, Durnford D G. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- 5.Wolfe G R, Cunningham F X, Jr, Durnford D, Green B R, Gantt E. Nature (London) 1994;367:566–568. [Google Scholar]
- 6.Tan S, Cunningham F X, Jr, Gantt E. Plant Mol Biol. 1997;33:157–167. doi: 10.1023/a:1005715528297. [DOI] [PubMed] [Google Scholar]
- 7.Tan S, Ducret A, Aebersold R, Gantt E. Photosynth Res. 1997;53:129–140. [Google Scholar]
- 8.Durnford D G, Deane J A, Tan S, McFadden G I, Gantt E, Green B R. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 9.Grabowski B, Cummingham F X, Gantt E. Proc Natl Acad Sci USA. 2001;98:2911–2916. doi: 10.1073/pnas.031587198. . (First Published February 13, 2001; 10.1073/pnas.031587198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 11.Bassi R, Croce R, Cugini D, Sandoná D. Proc Natl Acad Sci USA. 1999;96:10056–10061. doi: 10.1073/pnas.96.18.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogl H, Kühlbrandt W. Biochemistry. 1999;38:16214–16222. doi: 10.1021/bi990739p. [DOI] [PubMed] [Google Scholar]
- 13.Kleima F J, Hobe S, Calkoen F, Urbanus M L, Peterman E J G, van Grondelle R, Paulsen H, van Amerongen H. Biochemistry. 1999;38:6587–6596. doi: 10.1021/bi982823v. [DOI] [PubMed] [Google Scholar]
- 14.Fawley M W. Biochim Biophys Acta. 1993;1183:85–90. [Google Scholar]
- 15.Helfrich M, Ross A, King G C, Turner A G, Larkum A W D. Biochim Biophys Acta. 1999;1410:262–272. doi: 10.1016/s0005-2728(99)00004-3. [DOI] [PubMed] [Google Scholar]
- 16.Larkum A W D, Scaramuzzi C, Cox G C, Hiller R G, Turner A G. Proc Natl Acad Sci USA. 1994;91:679–683. doi: 10.1073/pnas.91.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Roche J, van der Staay G W M, Partensky F, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W D, Green B R. Proc Natl Acad Sci USA. 1996;93:15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Staay G W M, Yurkova N, Green B R. Plant Mol Biol. 1998;36:709–716. doi: 10.1023/a:1005930210515. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka A, Ito H, Tanaka R, Tanaka N K, Yoshida K, Okada K. Proc Natl Acad Sci USA. 1998;95:12719–12723. doi: 10.1073/pnas.95.21.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomitani A, Okada K, Miyashita H, Matthijs H C P, Ohno T, Tanaka A. Nature (London) 1999;400:159–162. doi: 10.1038/22101. [DOI] [PubMed] [Google Scholar]
- 21.Wilmotte A. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1–25. [Google Scholar]
- 22.Miyashita H, Adachi K, Kurano N, Ikemoto H, Chihara M, Miyachi S. Plant Cell Physiol. 1997;38:274–281. [Google Scholar]
- 23.Anderson J M, Barrett J. In: Photosynthesis III, Photosynthetic Membranes, and Light Harvesting Systems. Staehelin L H, Arntzen C J, editors. Berlin: Springer; 1986. pp. 269–285. [Google Scholar]
- 24.Hiller R G. In: The Photochemistry of Carotenoids. Frank H A, Young A J, Britton G, Cogdell R J, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 81–98. [Google Scholar]
- 25.Kirk J T O. Light and Photosynthesis in Aquatic Ecosystems. 2nd Ed. Cambridge: Cambridge Univ. Press; 1994. [Google Scholar]
- 26.Golding G B, Dean A M. Mol Biol Evol. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- 27.Chang B S W, Donoghue M J. Trends Ecol Evol. 2000;15:109–114. doi: 10.1016/s0169-5347(99)01778-4. [DOI] [PubMed] [Google Scholar]


