Acetyltransferases induce transcription by enhancing the activity of transcriptional activators and opening chromatin domains. A third avenue is described by which gene activation is accomplished by acetylation through the targeted destruction of the Ume6p repressor.
Abstract
Ume6p represses early meiotic gene transcription in Saccharomyces cerevisiae by recruiting the Rpd3p histone deacetylase and chromatin-remodeling proteins. Ume6p repression is relieved in a two-step destruction process mediated by the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase. The first step induces partial Ume6p degradation when vegetative cells shift from glucose- to acetate-based medium. Complete proteolysis happens only upon meiotic entry. Here we demonstrate that the first step in Ume6p destruction is controlled by its acetylation and deacetylation by the Gcn5p acetyltransferase and Rpd3p, respectively. Ume6p acetylation occurs in medium lacking dextrose and results in a partial destruction of the repressor. Preventing acetylation delays Ume6p meiotic destruction and retards both the transient transcription program and execution of the meiotic nuclear divisions. Conversely, mimicking acetylation induces partial destruction of Ume6p in dextrose medium and accelerates meiotic degradation by the APC/C. These studies reveal a new mechanism by which acetyltransferase activity induces gene expression through targeted destruction of a transcriptional repressor. These findings also demonstrate an important role for nonhistone acetylation in the transition between mitotic and meiotic cell division.
INTRODUCTION
The acetylation and deacetylation of histones play an important role in both transcriptional repression and activation. For example, the acetylation of nucleosomes, primarily histones H3 and H4, promotes transcription initiation (Berger, 2007). In budding yeast, the SAGA complex contains the Gcn5p histone acetyltransferase (HAT; Rodriguez-Navarro, 2009), which induces the transcription of many gene sets, especially those regulated by environmental stimuli (Huisinga and Pugh, 2004). Conversely, transcriptional repression is mediated by histone deacetylases (HDACs), which are recruited to promoters by sequence-specific DNA-binding proteins (Thiagalingam et al., 2003). The opposing activities of these enzymes represent a major regulatory mechanism controlling transcription initiation.
In addition to histones, there is a rapidly growing list of transcription factors that are also modified by acetylation (for review see Spange et al., 2009). For example, acetylation of p53 and GATA-1 by p300 increases their respective DNA-binding activities (Gu et al., 1993; Boyes et al., 1998). Acetylation of TAL1 is reported to enhance DNA-binding activity, as well as prevent association of the corepressor mSin3 (Huang et al., 2000). Acetylation increases the stability of myc (Patel et al., 2004) and promotes protein:protein interactions for α-importin (Bannister et al., 2000). Therefore, similar to histone modifications, the acetylation of transcription factors is also a mechanism to elevate gene expression.
In Saccharomyces cerevisiae, many of the genes required for meiosis and spore formation are repressed during mitotic cell division but transiently expressed at defined stages during development (Chu et al., 1998; Primig et al., 2000). Vegetative repression of the early meiotic gene (EMG) class requires the sequence-specific DNA-binding protein Ume6p (Strich et al., 1994). Ume6p represses transcription by recruiting both the HDAC Rpd3p (Kadosh and Struhl, 1997) and the chromatin-remodeling factor Isw2p (Goldmark et al., 2000) to EMG promoters. To relieve this repression in meiotic cells, Ume6p is destroyed in a two-step process by the Cdc20p-activated, anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase (Mallory et al., 2007). The first step partially degrades Ume6p and occurs when cultures are shifted to non–dextrose-containing growth medium. Shifting to sporulation medium, which lacks nitrogen and dextrose, induces the expression of Ime1p, a meiosis-specific factor whose association induces final Ume6p destruction and full EMG transcription. In this study, we report that acetylation of Ume6p by the Gcn5p-dependent SAGA complex provides the signal for the first step in Ume6p destruction during vegetative growth. In addition, Ume6p acetylation promotes step 2 destruction, thus stimulating the transition between mitotic cell division and meiotic development.
RESULTS
Gcn5p acetylates Ume6p in vitro
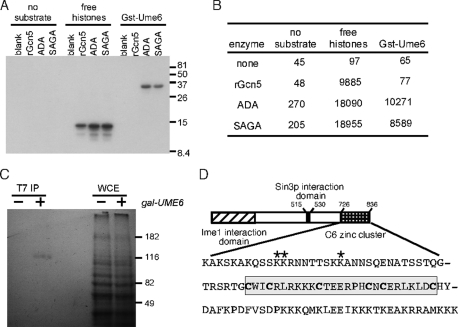
A previous study found that Gcn5p is required for meiotic gene induction (Burgess et al., 1999). We obtained a similar result in cells that failed to destroy Ume6p (Mallory et al., 2007), suggesting a mechanistic link between the two factors. Because Ume6p is a basic protein (pI 10.4) with a high concentration of lysines in the carboxy terminus of the protein (amino acids 726–836), we tested whether a glutathione S-transferase (GST) fusion protein (GST-Ume6726-836) was an in vitro substrate of Gcn5p. As expected, two Gcn5p-containing HAT complexes, SAGA and ADA, as well as recombinant Gcn5 (rGcn5), acetylated free histone H3 or H4 (Figure 1A; quantitated in Figure 1B). GST-Ume6726-836 was also a substrate of both the SAGA and ADA complexes, but not rGcn5. Previous studies also found that only SAGA, and not rGcn5, is able to acetylate nucleosomes (Yang et al., 1996; Grant et al., 1997). Therefore the finding that rGcn5 is not active suggested that Ume6p acetylation by SAGA may be physiological. To verify that Ume6p acetylation occurs in vivo, cultures expressing either a single-epitope tagged allele of UME6 (T7-UME6) under the control of the GAL1 promoter or the vector control were grown for 2 h in galactose medium containing 14C-acetyl-CoA. Extracts prepared from these cultures were incubated with a T7 monoclonal antibody (mAb) and the immunoprecipitates subjected to PAGE and fluorography. A signal migrating at the expected size of Ume6p was observed in the GAL-T7-UME6 extract but not in the control immunoprecipitate (Figure 1C). These results indicate that Ume6p is acetylated in vivo.
FIGURE 1:
Ume6p is a substrate of acetyltransferase complexes. (A) Affinity-purified acetyltransferase fractions or recombinant Gcn5p were incubated with GST-Ume6721-831 or the four histones. The reactions were split, with one-half separated by PAGE and fluorographed or quantitated (B) by liquid scintillation spectroscopy (dpm). (C) Wild-type strain RSY10 harboring GAL-T7-UME6 expression plasmid (+) or vector control (–) was grown in the presence of 14C-acetyl-CoA. Extracts derived from these cultures were immunoprecipitated with T7 mAb, and the immunoprecipitates were separated by PAGE and then visualized by fluorography. Whole-cell extracts (WCEs) controlled for equal protein loading and labeling. (D) Acetylation occurs in a region adjacent to the C6 zinc-cluster DNA-binding domain of Ume6p. The sequence of the last 110 amino acids of Ume6p (726–836) is presented. The acetylated residues are indicated by the asterisks. The cysteines making up the C6 zinc cluster are in boldface and boxed. The Ime1p and Sin3p interaction domains are indicated.
Three lysines are modified by Gcn5p in vitro and in vivo
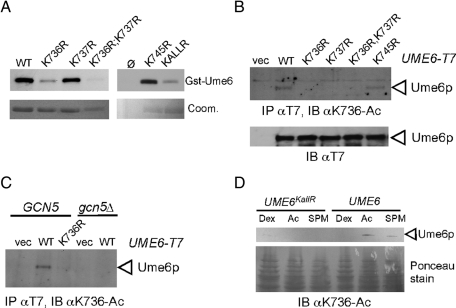
The in vitro data indicated that the last 110 amino acids of Ume6p are acetylated by a Gcn5p-containing HAT complex. To identify which residues are modified, we performed mass spectrometry on the GST-Ume6 following an in vitro acetylation assay. These spectra revealed three clustered acetylated lysine residues (for representative spectra, see Supplemental Figure S1A). The three residues K736, K737, and K745 were modified either individually or in tandem for K736 and K737. These residues (identified by asterisks, Figure 1D) lie outside of the Zn2Cys6 zinc-cluster DNA-binding domain (cysteines in boldface). To verify the mass spectroscopy results, the ability of SAGA to acetylate Gst-Ume6p mutants containing K → R substitutions was tested. We observed a significant reduction in activity on the GST-Ume6K736R peptide or the K736R;K737R double mutant. Only a modest reduction was observed with the K737R or K745R single-mutant derivatives (Figure 2A). Acetylation was reduced, but was still present, on the triple mutant (KallR) peptide, indicating that additional modified residues exist. These findings indicate that K736, K737, and K745 are acetylated by SAGA in vitro and that K736 is the preferred residue for modification.
FIGURE 2:
In vivo Ume6p acetylation requires Gcn5p. (A) In vitro acetylation assays described in Figure 1 were repeated with the indicated Gst-Ume6 substitution mutants as substrate. Gels were Coomassie stained (Coom.) before fluorography and used to control for substrate addition. (B) Extracts prepared from log-phase RSY10 cultures expressing the indicated GAL-T7-UME6 alleles (or the vector control) were blotted and probed with α-K736-Ac antibodies. (C) The experiment described in B was repeated in the wild-type (RSY10) and gcn5∆ mutant (RSY622) strains. (D) Endogenous Ume6p (RSY1079) or Ume6pKallR (RSY1149) was detected in extracts derived from mid-log cultures growing in dextrose, acetate, or after the switch from acetate to SPM. K736 acetylation was monitored by Western blot analysis probing with α-K736-Ac. Ponceau staining of the gel is shown as loading control.
To determine whether these lysine residues were modified in vivo, antibodies were raised against modified peptides acetylated at K736 and K745 (Supplemental Figure S1B). Antibodies directed against acetylated K737 were not sufficiently specific for further study. To determine whether Ume6p is acetylated on these residues in vivo, wild-type T7-Ume6p or the lysine-to-arginine substitution mutant proteins just described were expressed under the control of the GAL1 promoter. Extracts prepared from mid-log cultures were immunoprecipitated with T7 mAb and then probed with the αK736-Ac antibody. A signal was detected for Ume6p and Ume6pK745R but not with the K736R, K737R, or double K736R;K737R substitution mutant proteins (Figure 2B). These findings confirm that Ume6p is modified on K736 in vivo and suggest that K736 and K737 modifications may be linked, whereas K745 acetylation is independent. Next we determined whether in vivo K736 acetylation was dependent on SAGA. For this experiment, GCN5 was deleted in the strain expressing GAL1-T7-UME6. Extracts prepared from mid-log-phase galactose cultures were immunoprecipitated with T7 mAb and the immunoprecipitates subjected to Western blot analysis probing for acetylated K736. This study revealed that K736 acetylation was dependent on Gcn5p (Figure 2C), indicating that K736 is an in vitro and in vivo target of SAGA.
Dextrose prevents Ume6p acetylation
Previous studies found that Gcn5p was recruited to promoters under low-dextrose culture conditions (van Oevelen et al., 2005). To determine whether Ume6p acetylation was regulated by carbon source, wild-type T7-UME6 or T7-UME6KallR alleles were integrated into the genome, placing these constructions under the control of the natural UME6 promoter. Western blot analysis was performed on extracts prepared from cultures growing in medium using dextrose or acetate as a carbon source. Probing the blot with the αK736-Ac–specific antibody revealed a signal in the acetate culture but not in the culture growing on dextrose (Figure 2D). In addition, a K736-Ac signal was also observed in extracts obtained from a culture incubated in sporulation medium (SPM) that contains acetate as the carbon source but lacks nitrogen. This signal was reduced most likely due to the destruction of Ume6p associated with meiotic entry. These findings indicate that Ume6p acetylation occurs in the absence of dextrose. To expand these results, we conducted a time-course study to determine whether Ume6p acetylation status changed upon meiotic entry. For these studies, endogenous T7-Ume6p was immunoprecipitated from extracts prepared from a culture before and after transfer to SPM. Overall levels of Ume6p were reduced as expected upon meiotic entry (Supplemental Figure S1C). Probing these extracts with αK736-Ac or αK745-Ac antibodies revealed a loss of signal consistent with the observed reduction in Ume6p levels. These results suggest that the acetylation status of these two residues did not significantly change upon meiotic induction.
Acetylation mediates Ume6p destruction
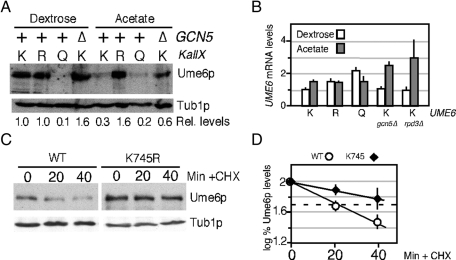
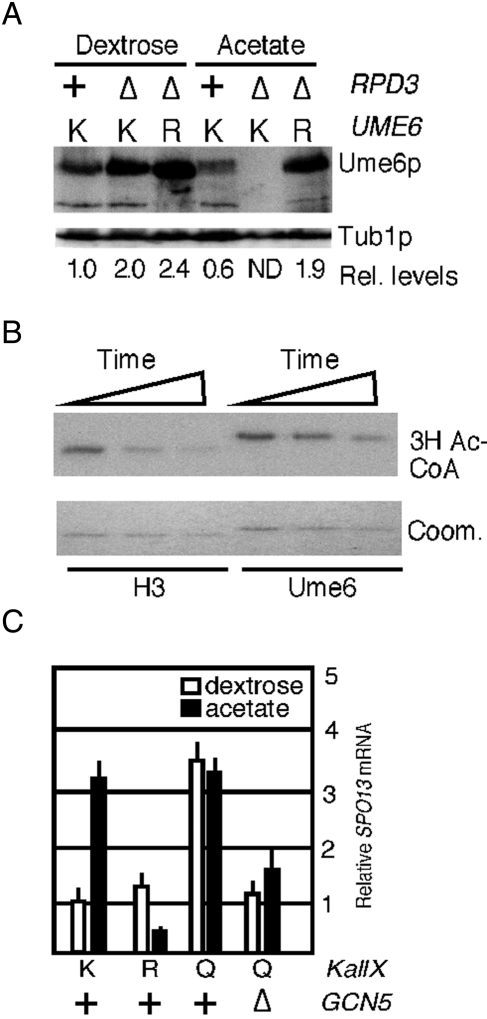
Our previous studies found that switching cultures from dextrose- to acetate-based medium reduces Ume6p steady state levels ∼50% (Mallory et al., 2007). To determine whether acetylation regulated Ume6p stability, integrated mutant alleles of UME6 that substituted all three K736, K737, and K745 residues to arginine (KallR) or glutamine (KallQ) were integrated into the UME6 locus. The arginine substitutions prevent acetylation while preserving the basic charge, and the introduction of glutamine residues can mimic Gcn5p-dependent acetylation (Zhang et al., 1998). As observed previously, wild-type Ume6p levels were reduced >50% in acetate cultures compared with dextrose-grown cells (Figure 3A). However, levels of the acetylation-deficient Ume6pKallR remained unchanged in either medium, suggesting that modification of these residues is required for acetate-induced degradation. Consistent with this model, the KallQ mutant exhibited reduced steady-state levels in either dextrose- or acetate-containing medium. If Gcn5p-dependent acetylation reduced Ume6p levels in acetate medium, then eliminating Gcn5p should prevent this destruction. Therefore wild-type Ume6p levels were monitored in gcn5∆ culture growing in either dextrose or acetate medium. As predicted, Ume6p levels remained elevated in acetate gcn5∆ culture. These results indicate that Gcn5p-dependent acetylation is required for Ume6p degradation in acetate medium.
FIGURE 3:
Acetylation promotes Ume6p destruction. (A) Steady-state levels of endogenous Ume6p (RSY1079), Ume6pKALLR (RSY1149), Ume6pKALLQ (RSY1226), or Ume6p in a gcn5∆ mutant (∆, RSY1091) were determined by Western blot analysis of extracts derived from log-phase dextrose or acetate cultures as indicated. The blots were stripped and reprobed for tubulin to calculate relative levels of the Ume6p derivatives. UME6 alleles: K, wild type; R, KallR; Q, KallQ. (B) Quantitative PCR was used to monitor UME6 mRNA in strains containing the chromosomal wild-type (K), KallR (R), and KallQ (Q) alleles grown to mid-log phase in dextrose or acetate medium as indicated. Experiments conducted in gcn5∆ or rpd3∆ mutants with wild-type UME6 are indicated. All values were normalized to internal ENO1 mRNA concentrations with UME6 levels in dextrose medium set at 1. Error bars indicate SEM from triplicate replicates from two independent cultures. (C) Half-life determinations of Ume6p and Ume6pK745R. Extracts prepared from G2-arrested (0 min) and released cultures were probed for Ume6p or Ume6pK745R. See Materials and Methods for details. Tub1p levels served as a loading control. (D) Quantification of the results indicated in C are shown (see Materials and Methods for details). Half-lives were determined by linear regression analysis (linear correlation, >0.9) for two independent experiments. Dashed line indicates 50% initial protein levels.
To test whether the differences in Ume6p levels observed in Figure 3A were due to changes in transcription, we determined mRNA levels of UME6 UME6KallR and UME6KallQ by quantitative PCR (qPCR) in mid-log-phase dextrose or acetate cultures. Despite the low levels of Ume6pKallQ in both glucose and acetate medium, no significant differences in mRNA levels were observed between UME6 and UME6KallQ. These finding indicate that the reduction of Ume6pKallQ in dextrose or in acetate cultures cannot be explained by changes in mRNA levels. Next we observed an approximate twofold elevation in UME6 mRNA levels in the gcn5∆ strain growing in acetate (Figure 3B). However, Ume6p concentrations were slightly reduced in the gcn5∆ acetate culture compared with the wild-type dextrose control. Taken together, these studies did not find a correlation between mRNA and protein levels, suggesting that the differences in concentration observed between Ume6p and the two derivatives were due to a posttranscriptional event.
To determine whether the differences in steady-state Ume6p levels reflected changes in stability, we calculated Ume6p and Ume6pK745R half-lives in G2 cells (see Materials and Methods for details). The K745R mutation was used in this study, as it appears to provide a similar level of stabilization as the KallR allele (see later discussion of Figure 5; also, unpublished data). It was important to enrich the cultures in G2, as APC/CCdc20 is only active from anaphase through early G1 (Kramer et al., 2000). Wild-type cultures harboring plasmids containing either UME6 or UME6K745R under the control of the galactose-inducible promoter GAL1 were grown to mid-log phase in acetate medium. Acetate medium permits Ume6p acetylation and does not induce or repress the GAL1 promoter. GAL1-UME6 and GAL1-UME6K745R were induced by the addition of galactose, and the cultures were arrested in G2 by the addition of nocodazole. Under these conditions APC/CCdc20 is not functional, due to spindle checkpoint activation (Hwang et al., 1998), allowing Ume6p accumulation. After 2.5 h, the G2-arrested cultures (determined by 4′,6-diamidino-2-phenylindole [DAPI] staining) were released by washing out the drug, allowing APC/C reactivation. Glucose and cycloheximide were then added to the medium to inhibit GAL1-UME6 and GAL1-UME6K745R mRNA transcription and translation, respectively. Western blot analysis from samples taken of arrested cultures and following release revealed that Ume6pK745R was approximate twofold more stable than the wild-type protein (Figure 3C; quantified in Figure 3D). Taken together with the mRNA analysis, these results indicate that Gcn5p-dependent acetylation increases Ume6p turnover.
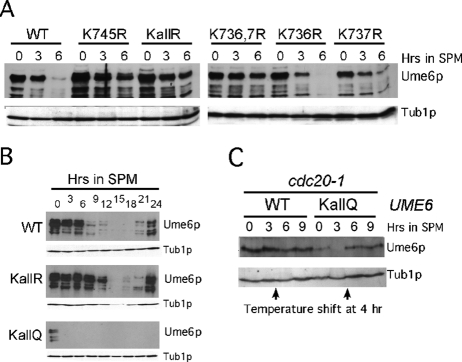
FIGURE 5:
Acetylation is required for meiotic Ume6p destruction. (A) Steady-state levels of endogenous T7-Ume6p (RSY1079), T7-Ume6pK745R (RSY1148), T7-Ume6pKallR (RSY1149), T7-Ume6pK736,737R (RSY1147), T7-Ume6pK736R (RSY1145), and T7-Ume6pK737R (RSY1146) were determined by Western blot analysis of extracts prepared from cultures in acetate medium (0 h) or after transfer to SPM (at indicated hours). Tub1p served as a loading control. (B) The experiments described in A were repeated with strains RSY1079 (WT), RSY1149 (T7-Ume6pKallR), and RSY1226 (T7-Ume6pKallQ) during an extended meiotic time course. (C) A cdc20-1 mutant strain expressing T7-tagged, wild-type UME6 (RSY1101) or UME6KALLQ (RSY1623) were grown at the permissive temperature (23°C) and then transferred to SPM. After 4 h, the cultures were shifted to the restrictive temperature (34.5°C) and the time course continued. Levels of Ume6p and Ume6pKallQ were determined by Western blot analysis. Tub1p levels were monitored as a loading control.
Ume6p is a substrate for HDAC1
Ume6p tethers the HDAC Rpd3p to early meiotic gene promoters. We next examined whether Rpd3p antagonizes Gcn5p activity to maintain high Ume6p levels in dextrose cultures. As described, Ume6p steady-state levels are reduced ∼50% when the culture is shifted from dextrose to acetate medium (Figure 4A). Surprisingly, Ume6p and Ume6pKallR levels increased approximately twofold in the dextrose rpd3∆ mutant cultures (Figure 4A). These results suggest that Ume6p levels are modulated by acetylation even in dextrose medium. In acetate cultures, the loss of Rpd3p activity exacerbated the negative affect of this medium on Ume6p stability. This result was not due to changes in UME6 mRNA concentrations in the rpd3∆ strain (Figure 3B). Consistent with our model, preventing acetylation (KallR) protected Ume6p from destruction in rpd3∆ mutants growing in acetate medium. These results are consistent with a model in which Rpd3p promotes Ume6p stability through deacetylation of these lysines.
FIGURE 4:
Ume6p is a substrate for the Rpd3p HDAC. (A) Ume6p levels were monitored in the wild-type (RSY1079) or rpd3∆ (RSY1632) strains and Ume6pKallR in the rpd3∆ strain (RSY1622) in acetate or dextrose medium as indicated. The relative levels of the Ume6p signal were normalized to Tub1p and then standardized to the wild-type protein in dextrose medium. ND, not detectable above background. (B) In vitro 3H-acetyl-CoA–modified Ume6p or histone H3 generated as described in Figure 2A was incubated with HDAC1 for increasing time (0, 2, 4 h). Reactions were separated by PAGE and then subjected to Coomassie staining and fluorography. (C) SPO13 mRNA levels were quantitated by qPCR in wild-type (+, RSY1079), gcn5∆ (RSY1634), or rpd3∆ (RSY1636) mutants expressing the indicated UME6 alleles as described in Figure 3A. Cultures were grown to mid-log phase in either dextrose-based (open bars) or acetate-based (closed bars) medium. SPO13 mRNA levels were determined by qPCR and presented following standardization to ENO1 transcript concentrations for each sample. SPO13 mRNA level in the dextrose-grown UME6 culture was set at 1. Error bars were calculated from three biological replicates.
We next determined whether Rpd3p directly regulates the acetylation status of Ume6p. First, His6-Ume6p726-836 was acetylated by SAGA in the presence of 3H-acetyl-CoA as described in Figure 2. Histone H3 was also modified to serve as a positive control. The acetylation reactions were stopped and then incubated with HDAC1, the human orthologue of Rpd3p (Taunton et al., 1996). Time points were taken and the products separated by SDS–PAGE and subjected to fluorography. These experiments revealed that Ume6p acetylation was reduced in a time-dependent manner (Figure 4B), indicating that it is indeed a substrate for HDAC1. Taken together, these results indicate that Ume6p acetylation, and ultimately its stability, is a dynamic process controlled by the opposing activities of Gcn5p and Rpd3p. Shifting the balance of these activities may represent an important switch in the decision between cellular proliferation and meiotic differentiation.
Acetylation-induced destruction partially relieves Ume6p repression
To determine whether changes in Ume6p levels affected its ability to function as a transcriptional repressor, we measured mRNA levels of the EMG SPO13 in vegetative cultures growing in dextrose or acetate medium. An approximate threefold increase in SPO13 mRNA levels was observed in wild-type UME6-expressing culture upon its transfer to acetate medium (Figure 4C). This acetate-induced derepression was lost in cells expressing the KallR mutant, indicating that maintaining Ume6p at dextrose levels decreased SPO13 mRNA levels in this medium. Consistent with this model, the introduction of the glutamine residues derepressed SPO13 in dextrose medium to levels observed in acetate-grown cultures. No additive effect in SPO13 derepression was observed when the UME6KallQ-expressing culture was grown in acetate medium. This result argues that the derepression observed in the KallQ mutation and in acetate medium function through the same pathway. These results indicate that the EMG derepression observed in acetate medium is controlled by acetylation-induced reduction in Ume6p levels. Next we determined whether the KallQ mutation relieved the requirement for Gcn5p in EMG derepression. To address this question, we examined SPO13 mRNA levels in a gcn5∆ mutant expressing Ume6pKallQ. In both dextrose and acetate medium, Gcn5p was required for SPO13 depression in the KallQ-expressing strain, indicating that additional targets (e.g., nucleosomes) are modified to allow SPO13 mRNA induction. Taken together, our findings indicate that the deacetylation and acetylation of Ume6p control both its stability and its ability to fully repress SPO13 transcription.
Acetylation accelerates Ume6p meiotic destruction
Early meiotic gene induction requires complete Ume6p destruction as cells transit from mitotic to meiotic cell division. To determine whether the acetylation status of Ume6p affects its meiotic destruction kinetics, we performed a series of time-course experiments. Plasmids expressing the various K → R substitution mutant proteins were introduced into a wild-type strain and their destruction followed by Western blot analysis. The K736R and K737R single-mutant proteins displayed destruction kinetics similar to the wild type (Figure 5A). However, the K745R single mutant and the K736R;K737R double mutant were protected from rapid degradation upon meiotic induction. To more fully investigate this observation, an extended time course was conducted with strains expressing either the wild-type or the integrated UME6KallR allele. Western blot analysis revealed that Ume6pKallR destruction was delayed approximately 6 h compared with wild type (Figure 5B). These results suggest that acetylation of the lysine cluster is required for the rapid meiotic destruction of Ume6p but is not essential for the process to eventually occur. Consistent with this model, Ume6pKallQ displayed accelerated destruction kinetics, with its levels reduced below the limits of detection within 3 h after the shift to SPM (Figure 5B, bottom). Of interest, unlike wild type or Ume6pKallR, Ume6pKallQ was not detected late in meiosis during spore wall assembly (at 24 h). These results suggest that maintaining newly synthesized Ume6p in the deacetylated state is important to allow reaccumulation of Ume6p after spore morphogenesis.
The Cdc20p-directed APC/C is required for Ume6p destruction as cells enter meiosis (Mallory et al., 2007). Therefore we next determined whether the rapid destruction of Ume6pKallQ was dependent on APC/CCdc20 or was due to nonspecific protein instability. Because Cdc20p is required for cell growth, we used a temperature-sensitive allele of CDC20 (cdc20-1). Meiotic time-course experiments were repeated with a cdc20-1 stain expressing either Ume6p or Ume6pKallQ. Cultures were grown at the permissive temperature (23°C) and then transferred to SPM at the same temperature. After 4 h, the cultures were shifted to the restrictive temperature (34.5°C). Western blot analysis before and after the temperature shift revealed that although Ume6pKallQ levels dropped below the limits of detection by 3 h, they rebounded to detectable levels after inactivation of Cdc20-1p (Figure 5C). These findings indicate that Ume6pKallQ instability is dependent on APC/CCdc20.
Ume6p acetylation is required for normal meiotic gene induction
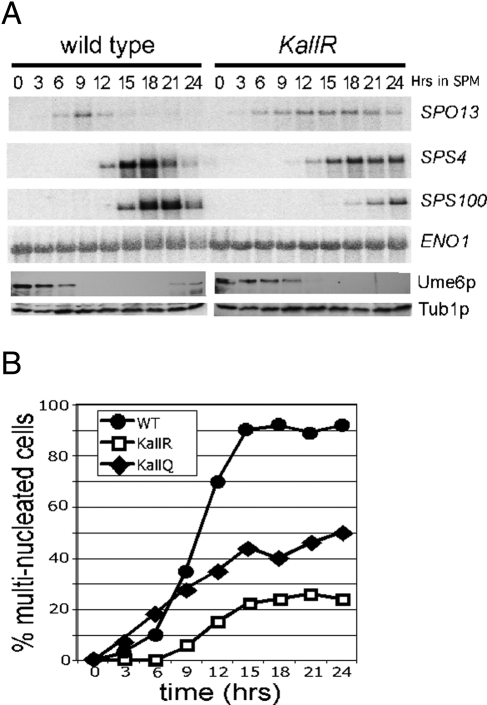
The persistence of Ume6pKallR after meiotic entry suggested the possibility that EMG transcription was delayed, as observed when Ume6p was protected from destruction by mutating its destruction box (Mallory et al., 2007). To test this possibility, we evaluated the mRNA expression profiles of genes from the early (SPO13), middle (SPS4), or late (SPS100) expression classes by Northern blot analysis of samples collected from an extended time-course experiment. This study revealed a complex transcription profile for these loci. Both SPO13 and SPS4 mRNA were observed at the same time point (6 and 12 h, respectively) in both strains (Figure 6A). However, their induction kinetics were altered, in that peak expression intensity was reduced in the Ume6pKallR strain, and the genes failed to reestablish repression. A caveat to this conclusion is that the synchrony of meiotic progression may be perturbed in the Ume6pKallR strain, thus broadening the expression peaks (see later discussion). SPS100 mRNA accumulation was delayed compared with wild type and did not peak within the time frame of this experiment. Taken together, these results indicate that efficient acetylation-dependent destruction of Ume6p is required to maintain the normal meiotic transcription program.
FIGURE 6:
Ume6p acetylation is required for normal meiotic gene expression and progression. (A) Northern blot analysis of early (SPO13), middle (SPS4), and late (SPS100) meiotic mRNAs in either UME6 (RSY1079)-expressing or UME6KallR (RSY1149)-expressing stains after transfer to SPM as indicated. Western blot analysis was used to monitor Ume6p and Ume6pKallR levels (bottom) at the same time points. (B) Wild-type (RSY1079, open circles) and Ume6pKallR (RSY1149)-expressing (open squares) and Ume6pKallQ (RSY1226)-expressing (closed diamonds) strains were induced to enter meiosis and time points taken as indicated. The percentages of cells containing multiple nuclei indicating the execution of at least one meiotic nuclear division were determined by DAPI staining.
Genes composing the middle and late expression classes are required for execution of the meiotic nuclear divisions and spore wall assembly, respectively. Therefore we next examined the affect of the altered transcription of middle and late genes on these processes. DAPI staining of meiotic nuclei was used to quantitate the fraction of the population undergoing meiosis I (binucleates) or meiosis I and II (tetranucleates). These studies revealed that the appearance of binucleated or tetranucleated cells was delayed in the Ume6pKallR-expressing strain compared with wild type (Figure 6B). By 24 h, 27% of the Ume6pKallR-expressing strain had undergone both divisions compared with 92% for the wild type. Although the initial appearance of multinucleated cells in Ume6pKallQ-expressing strains occurred with similar kinetics as wild type, the final percentage of the population undergoing any meiotic divisions was reduced. These results indicate that the correct timing of Ume6p destruction is important for normal execution of the meiotic nuclear divisions (see Discussion). Finally, we analyzed the viability of spores derived from these diploids. No differences were observed in spore appearance, viability, or sensitivity to enzymatic digestion (unpublished data). These results indicate that although the overall meiotic division completion efficiency was reduced, the spores that were produced appeared normal.
DISCUSSION
Ume6p destruction is required for induction of EMGs and execution of meiosis and spore formation. This proteolysis is accomplished by a two-step process that is controlled by both endogenous and exogenous signals (Mallory et al., 2007; for review see Cooper and Strich, 2011). The first step, which results in ∼50% reduction in Ume6p levels and partial derepression of EMG, occurs when vegetative cultures are switched from dextrose to a nonfermentable carbon-based medium. The second step, resulting in the complete elimination of Ume6p and full EMG induction, requires the withdrawal of a nitrogen source and the direct association of the meiotic protein Ime1p. In the present study, we find that the initial step in this degradation program is regulated by Ume6p acetylation and deacetylation by the Gcn5p-dependent acetyltransferase complex SAGA and the HDAC Rpd3p, respectively. Specifically, acetylation at a cluster of three lysines is required for step 1 Ume6p destruction and partial EMG derepression. In addition, substitution mutations that either prevent or mimic acetylation retard or accelerate step 2 meiotic Ume6p destruction, respectively. These results suggest that these two steps are connected. Of interest, either hastening or delaying Ume6p destruction reduced the efficiency of meiosis I and II execution. These findings indicate that the fine-tuning of Ume6p destruction by acetylation is important for normal meiotic progression.
Acetyltransferases induce transcription by enhancing the activity of transcriptional activators and opening chromatin domains. The present study defines a third avenue by which gene activation is accomplished by acetylation, through the targeted destruction of the Ume6p repressor. Lysine acetylation is usually associated with protecting proteins from proteolysis by serving as a block to ubiquitylation (Sadoul et al., 2008). A potential exception to this rule was the report that acetylation of the hypoxic transcription factor HIF1-α by the N-terminal acetyltransferase ARD1 stimulated its proteolysis (Jeong et al., 2002). This result was surprising, given the mostly cytoplasmic localization of ARD1 and its function as an N-terminal α-acetyltransferase rather than as targeting the ε-amino group of lysine like ubiquitin ligases or Gcn5p (reviewed in Kuo and Hung, 2010). Moreover, two subsequent reports found no connection between ARD1 function and HIF1-α stability (Arnesen et al., 2005; Bilton et al., 2005), bringing the initial result into question. The combination of in vitro and in vivo studies presented in the present study provides solid evidence for the regulated acetylation of a transcriptional repressor and the role of this modification in promoting APC/C-dependent destruction.
Gcn5p-dependent acetyltransferase activity stimulates partial Ume6p degradation when the cells are switched from dextrose to acetate media. In many differentiation pathways, transcriptional programs provide a series of gates that allows flexibility in cell fate decisions. For example, in hematopoietic stem cell differentiation, changes in the levels of the transcription factor PU.1 help to refine the gene expression profile to push the cell toward macrophage differentiation (Laslo et al., 2006). A similar scenario may be occurring when yeast cells switch from a fermentable to a nonfermentable carbon source. Yeast will ferment various sugars, producing ethanol as a byproduct. When the sugar is exhausted, yeast will use the ethanol as a carbon source through mitochondrial-dependent respiration. Partial reduction in Ume6p levels induced by acetylation results in low-level derepression of EMG and rapid execution of meiosis upon further stimulation. A similar fate is observed for the yeast C-type cyclin. Along with its kinase Cdk8p, cyclin C also represses EMG transcription (Strich et al., 1989) through its interaction with the mediator complex (Cooper and Strich, 1999). Similar to Ume6p, cyclin C levels are reduced to approximately half when cultures are shifted from dextrose to ethanol- or acetate-based medium (Cooper et al., 1999). Final destruction of cyclin C, which is also required for full EMG induction, occurs upon meiotic entry (Cooper et al., 1997). These findings argue that a general down-regulation of EMG transcriptional repressors in acetate medium represents a “potentiated state” of the cell that allows rapid transition from mitosis to meiosis when nitrogen sources are exhausted.
The transition from mitotic to meiotic cell division requires Ime1p, a protein necessary for the complete Ume6p destruction (Mallory et al., 2007). Although still a possibility, it seems unlikely that acetylation recruits Ime1p, as the Ume6p-Ime1p association domain (Rubin-Bejerano et al., 1996) and acetylated lysines are on opposite ends of the protein (Figure 1D). In addition, in vitro studies revealed no difference between the association of Ime1p to Ume6p or to Ume6pKallQ (unpublished results). A previous study suggested that one potential function of Ime1p is to disrupt the association of Ume6p to the Sin3p-Ume1p-Rpd3p HDAC complex (Pnueli et al., 2004). The removal of Rpd3p from EMG promoters could serve two purposes. First, it would allow further acetylation of Ume6p, which would stimulate its destruction. Second, the surrounding histones would also become more likely acetylated due to the loss of Rpd3p from the promoter. Under step 1 destruction conditions (acetate-grown vegetative cells), Ime1p is not expressed, thus retaining Rpd3p at the EMG promoter and setting up a competition between this HDAC and Gcn5p activity to determine Ume6p acetylation status. This model is supported by our analysis of cis and trans mutations that throw the acetylation balance in one direction or the other. For example, mimicking acetylation (KallQ) promotes Ume6p step 1 destruction in the absence of Gcn5p. Similarly, deleting RPD3 enhances Ume6p destruction in acetate cultures. This constant interplay between these two activities is illustrated by our finding that although Ume6p is acetylated in acetate cultures, it is only partially destroyed compared with the KallQ mutant, which the cell may recognize as always acetylated. Taken together, these findings suggest that the interplay between Rpd3p and Gcn5p activities controls the acetylation status and stability of Ume6p, which in turn regulates meiotic entry and progression.
MATERIALS AND METHODS
Plasmids and strains
All strains used in this study are listed in Supplemental Table S1 and derived from a cross between a rapid sporulating (SK1) background and a strain (W303) that does not precociously enter meiosis. All UME6 derivatives were constructed by oligonucleotide-directed mutagenesis and then introduced into its normal chromosomal location using the pop-in, pop-out method (Rothstein, 1991) and were verified by sequence analysis of the chromosomal allele. The various derivatives were introduced into this vector by replacing the SphI-HindIII fragment of the wild-type and mutant constructs. Conditional alleles were generated by placing the UME6 or UME6K745R coding region under the control of the GAL1 promoter in pGALSET985 (Enomoto et al., 1998), forming pMM225 and pMM482, respectively. GAL1pro-UME6 induction was accomplished in acetate cultures by addition of galactose (0.5%). GST-Ume6p721-831 (pMM421) was constructed by insertion of the PCR-amplified DNA into pGEX1λT. Site-directed mutagenesis on these plasmids was accomplished as described (Kunkel, 1985) on M13-derived, single-stranded DNA.
Acetylation/deacetylation assays
For in vitro assays, SAGA and ADA complexes were obtained using affinity purification (His6-ADA2) and further separated by ion exchange chromatography as described previously (Henry et al., 2003). Recombinant His6-Gcn5p (rGcn5) was purified from Escherichia coli as described (Candau et al., 1996). Approximately 10 μg of GST-Ume6p substrate purified from E. coli or unacetylated histones was used in each reaction. Acetylation activity of SAGA and ADA HAT complexes with Ume6p and histones was quantitated by spotting the reactions on P81 paper, washing the filters extensively to remove unbound 3H-acetyl-CoA, and counting the remaining radioactivity by liquid scintillation spectroscopy. In vivo labeling of acetylated proteins was achieved by incubating mid-log cultures harboring the GAL1pro-T7-UME6 expression construct (pMM225) or the vector control with 14C-acetyl-CoA (500 μCi) for 2 h. Deacetylation assays were conducted using the in vitro Ume6p acetylation reaction just described, followed by heat inactivation of SAGA and addition of 0.4 μg of HDAC1 (Cell Sciences, Canton, MA). Reactions were inactivated by boiling after 0, 2, and 4 h and separated by PAGE, and the resulting gel was incubated in Amplify (GE Healthcare, Piscataway, NJ) before fluorography.
Western blot analysis
Extracts were prepared from vegetative and meiotic cultures as described previously (Cooper et al., 1997) in the presence of 10 mM Na butyrate and 0.1% trichostatin A to inhibit HDAC activity. Either 1 mg or 100 μg of soluble protein was used for immunoprecipitation and straight Western blot experiments, respectively. Polyclonal antibodies were generated in rabbits against peptides containing acetylated K736, K737, K745, and the double-modified K736,737 (Open Biosystems, Thermo Biosystems, Huntsville, AL). Only α-K736-Ac and α-K745-Ac antibodies were produced with sufficient specificity to be useful. The antibodies were affinity purified and concentrated after preclearing with the unmodified peptide. T7 antibodies were obtained from Novagen (EMD4Biosciences, Gibbstown, NJ). Chemiluminescence signals were quantitated by charge-coupled device camera imaging (Kodak, Rochester, NY).
Gene expression analyses
Northern blot analysis was conducted as previously described (Cooper et al., 2000). Quantitative PCR was conducted using SYBR green protocols (Applied Biosystems, Foster City, CA) with the oligonucleotides described in Supplemental Table 2. Using delta Ct values, we first standardized all mRNA levels to internal ENO1 transcript levels. We then set SPO13 or UME6 mRNA levels in wild-type cultures grown in dextrose medium at 100%. The SEM from three biological repeats is indicated.
Cell culture conditions
Cell growth and meiotic time-course experiments were conducted essentially as described previously (Mallory et al., 2007). Meiotic time-course experiments were conducted by transferring washed, mid-log-phase cells grown in potassium acetate medium to SPM containing 2% potassium acetate. For protein stability assays, UME6 or UME6K745R was placed under the control of the galactose promoter and introduced into a wild-type strain (RSY10). Cultures were grown to mid-log phase in minimal acetate medium to allow Ume6p acetylation by Gcn5p. Galactose (0.5%) and nocodazole (15 μg/ml) were added for 3 h to induce UME6 and UME6K745R expression and arrest the cultures in G2, respectively. Culture arrest was confirmed by DAPI analysis, with >80% of the cells containing a large bud with a single nucleus. After arrest, the cells were washed and placed into glucose medium to repress the GAL1 promoter, and cycloheximide was added (50 μg/ml) to stop protein synthesis. Samples were taken as indicated in the text, and Ume6p and Ume6pK745R levels were determined by Western blot analysis. The corresponding signals were quantified (Kodak Imager), and half-life calculations were made by linear regression after normalization to the Tub1p internal standard. For all lines used in the analysis r = −0.99. The graph represents the average of two independent experiments.
Supplementary Material
Acknowledgments
We thank K. Cooper and M. Primig for helpful discussions during the course of this work. We also thank S. Seeholzer and the Biochemistry and Biotechnology Facility at the Fox Chase Cancer Center for mass spectroscopy analysis. This work was supported by National Institute of Health Grants GM082013 (M.L.) and GM086788 (R.S.).
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- EMG
early meiotic genes
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0536) on March 21, 2012.
REFERENCES
- Arnesen T, Kong X, Evjenth R, Gromyko D, Varhaug JE, Lin Z, Sang N, Caro J, Lillehaug JR. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. FEBS Lett. 2005;579:6428–6432. doi: 10.1016/j.febslet.2005.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Miska EA, Gorlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bilton R, Mazure N, Trottier E, Hattab M, Dery MA, Richard DE, Pouyssegur J, Brahimi-Horn MC. Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1alpha and is not induced by hypoxia or HIF. J Biol Chem. 2005;280:31132–31140. doi: 10.1074/jbc.M504482200. [DOI] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci USA. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cooper KF, Egeland DE, Mallory MJ, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Smith JS, Strich R. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Strich R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires the phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol. 1999;19:3338–3348. doi: 10.1128/mcb.19.5.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Strich R. Functional analysis of the yeast C-type cyclin Ume3p/Srb11p- RNA polymerase II holoenzyme interaction. Gene Exp. 1999;8:43–57. [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Strich R. Meiotic control of the APC/C: similarities and differences from mitosis. Cell Div. 2011;6:16. doi: 10.1186/1747-1028-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Chen G, Berman J. Vectors for expressing T7 epitope- and His6 affinity-tagged fusion proteins in S. cerevisiae. BioTech. 1998;24:782–786. doi: 10.2144/98245st01. [DOI] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Gu W, Cechova K, Tassi V, Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci USA. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Qiu Y, Shi Y, Xu Z, Brandt SJ. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 2000;19:6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HP, Hung MC. Arrest-defective-1 protein (ARD1): tumor suppressor or oncoprotein? Am J Transl Res. 2010;2:56–64. [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Mallory MJ, Cooper KF, Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol Cell. 2007;27:951–961. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Edry I, Cohen M, Kassir Y. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol. 2004;24:5197–5208. doi: 10.1128/MCB.24.12.5197-5208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast; pp. 281–301. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90:306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Strich R, Slater MR, Esposito RE. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann NY Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- van Oevelen CJ, van Teeffelen HA, Timmers HT. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol Cell Biol. 2005;25:4863–4872. doi: 10.1128/MCB.25.12.4863-4872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.