The translocase of the outer mitochondrial membrane (TOM) is essential for the import of proteins into mitochondria. Cytosolic protein kinase A phosphorylates the precursor of the channel-forming protein Tom40 and inhibits its import into mitochondria, thus regulating the biogenesis of the protein entry gate of mitochondria.
Abstract
The preprotein translocase of the outer mitochondrial membrane (TOM) functions as the main entry gate for the import of nuclear-encoded proteins into mitochondria. The major subunits of the TOM complex are the three receptors Tom20, Tom22, and Tom70 and the central channel-forming protein Tom40. Cytosolic kinases have been shown to regulate the biogenesis and activity of the Tom receptors. Casein kinase 2 stimulates the biogenesis of Tom22 and Tom20, whereas protein kinase A (PKA) impairs the receptor function of Tom70. Here we report that PKA exerts an inhibitory effect on the biogenesis of the β-barrel protein Tom40. Tom40 is synthesized as precursor on cytosolic ribosomes and subsequently imported into mitochondria. We show that PKA phosphorylates the precursor of Tom40. The phosphorylated Tom40 precursor is impaired in import into mitochondria, whereas the nonphosphorylated precursor is efficiently imported. We conclude that PKA plays a dual role in the regulation of the TOM complex. Phosphorylation by PKA not only impairs the receptor activity of Tom70, but it also inhibits the biogenesis of the channel protein Tom40.
INTRODUCTION
Most mitochondrial proteins are imported from the cytosol. The proteins are synthesized as precursors on cytosolic ribosomes. Targeting signals contained in the precursor proteins direct them to receptors on the mitochondrial surface (Hoogenraad et al., 2002; Dolezal et al., 2006; Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo et al., 2011). The translocase of the outer mitochondrial membrane (TOM complex) functions as the main entry gate for mitochondrial precursor proteins. After passing through the TOM channel, the precursor proteins follow different import routes to the four mitochondrial subcompartments—outer membrane, intermembrane space, inner membrane, and matrix.
The TOM complex consists of seven subunits: three receptors, a central channel-forming protein, and three small subunits (Meisinger et al., 2001; Walther and Rapaport, 2009; Endo et al., 2011). The receptors Tom20 and Tom22 preferentially recognize precursor proteins that carry amino-terminal targeting signals (presequences), whereas the third receptor, Tom70, mainly binds hydrophobic precursor proteins with internal targeting signals (metabolite carriers; Brix et al., 1999; Abe et al., 2000; Young et al., 2003; Li et al., 2009). Each of the receptors is anchored in the outer membrane via a single α-helical transmembrane segment and exposes the preprotein-binding domain to the cytosol. The β-barrel protein Tom40 is the essential core of the TOM complex. Tom40 forms a hydrophilic channel, through which the vast majority of mitochondrial proteins are imported (Hill et al., 1998; Ahting et al., 2001; Suzuki et al., 2004; Becker et al., 2005; Harsman et al., 2010). The small subunits Tom5, Tom6, and Tom7 are involved in the assembly and stability of the TOM complex (Schmitt et al., 2005; Sherman et al., 2005; Meisinger et al., 2006b; Kato and Mihara, 2008; Becker et al., 2010, 2011; Yamano et al., 2010). Each subunit of the TOM complex is encoded by a nuclear gene and thus is synthesized as precursor in the cytosol and imported into mitochondria (Model et al., 2001; Walther and Rapaport, 2009).
For a long time, little has been known about the regulation of the mitochondrial preprotein translocases. Recently, however, mass spectrometric analyses revealed that the subunits of the TOM complex are phosphorylated at multiple sites (Chi et al., 2007; Li et al., 2007; Albuquerque et al., 2008; Gnad et al., 2009; Schmidt et al., 2011). On the basis of prediction programs for kinase target sites and in vitro assays with recombinant Tom proteins and purified kinases, a number of candidate kinases for TOM phosphorylation were identified (Rao et al., 2011; Schmidt et al., 2011). So far, a functional role of TOM phosphorylation has been shown for the Tom receptors in vivo and in organello. 1) Cytosolic casein kinase 2 (CK2) phosphorylates the precursor of Tom22 in the cytosol and thereby stimulates its import into the outer membrane. In addition, CK2 also phosphorylates the mitochondrial import protein Mim1, an outer membrane protein that mediates membrane insertion of the precursors of Tom20 and Tom70 (Becker et al., 2008; Hulett et al., 2008; Popov-Celeketic et al., 2008), and thus promotes the import of these two receptors. The assembly of Tom20 with mature Tom22 in the TOM complex is enhanced when Tom22 is in the phosphorylated state. Taken together, CK2 stimulates the biogenesis of the three Tom receptors (Rao et al., 2011; Schmidt et al., 2011). 2) In contrast, cytosolic protein kinase A (PKA) was found to inhibit the receptor activity of Tom70. Phosphorylation by PKA does not affect the biogenesis of the Tom70 precursor but exerts an inhibitory effect on the mature Tom70 receptor and thus impairs the import of metabolite carriers into mitochondria (Schmidt et al., 2011).
We analyzed a possible effect of kinases on the channel protein Tom40. We show that PKA impairs import of the Tom40 precursor into mitochondria. The inhibitory effect is selectively caused by phosphorylation of a specific serine residue of the Tom40 precursor in the cytosol, whereas mature, imported Tom40 is not accessible to phosphorylation by PKA. We conclude that cytosolic kinases not only regulate the biogenesis and activity of Tom receptors, but they also exert an inhibitory effect on the biogenesis of the channel protein Tom40.
RESULTS
Phosphorylation of Tom40 by yeast PKA
The in vitro screen for TOM phosphorylation by Schmidt et al. (2011) led to the prediction of Ser-54 of Tom40 as PKA target site and the demonstration that purified mouse PKA phosphorylates recombinant Saccharomyces cerevisiae Tom40 at this site. It has not been determined whether the phosphorylation takes place in yeast and whether it is of functional relevance. PKA consists of two catalytic subunits and two regulatory (inhibitory) subunits. In yeast the catalytic subunits are encoded by the genes TPK1, TPK2, and TPK3 and the regulatory subunit by BCY1 (Cannon and Tatchell, 1987; Toda et al., 1987a, 1987b; Thevelein, 1994; Zaman et al., 2008; Smets et al., 2010). PKA is activated on fermentable growth conditions that lead to increased intracellular cAMP levels (Broach, 1991; Thevelein, 1994). cAMP binds to Bcy1, leading to a release of the active catalytic subunits (Taylor et al., 1990; Thevelein, 1994; Tamaki, 2007; Zaman et al., 2008). BCY1-deficient yeast cells lack cAMP-dependent regulation of PKA activity and are unable to grow under nonfermentable conditions (Matsumoto et al., 1983; Toda et al., 1987a; Cameron et al., 1988).
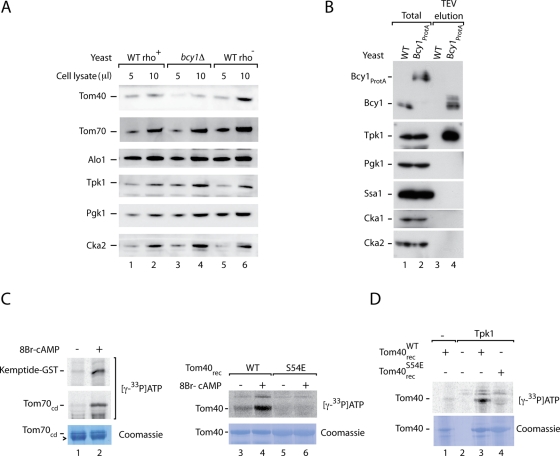
To test for a possible effect of PKA on Tom40 in vivo, we grew yeast cells on fermentable medium containing sucrose as carbon source (Lemaire et al., 2004; Van de Velde and Thevelein, 2008). We compared bcy1Δ yeast to rho+ wild-type yeast as well as to rho− wild-type yeast that lack mitochondrial DNA and are thus also unable to grow on nonfermentable medium. The steady-state protein levels of Tom40 were reduced in bcy1Δ yeast cells in comparison to rho+ as well as rho− wild-type yeast (Figure 1A). The levels of several control proteins, including cytosolic kinases, were not affected. Moreover, the levels of Tom70 were not changed, in agreement with the findings of Schmidt et al. (2011) that PKA affects neither the biogenesis nor the level of Tom70 but inhibits the receptor activity of the mature, imported Tom70 (in the study by Schmidt et al., 2011, glucose was used as carbon source). These results suggest that a constitutive activation of PKA exerts an inhibitory influence on the level of Tom40.
FIGURE 1:
Phosphorylation of Tom40 by yeast PKA. (A) Yeast cells from rho+ wild-type (WT), rho− wild type, and bcy1Δ were grown on sucrose-containing medium at 30°C. Protein extracts were prepared by postalkaline lysis and analyzed by SDS–PAGE and Western blotting. Alo1, d-arabinono-1,4-lactone oxidase; Pgk1, 3-phosphoglycerate kinase. (B) Yeast PKA (Bcy1-Tpk) was purified by affinity chromatography using a Bcy1ProtA yeast strain as described in Materials and Methods. Total (yeast lysate) and TEV eluate were analyzed by SDS–PAGE and Western blotting. Total, 10%; eluate, 100%. Cka1, Cka2, catalytic subunits of CK2; Ssa1, cytosolic member of heat shock protein 70 family. (C) Kemptide, the cytosolic domain of Tom70, and recombinant Tom40 (WT and S54E mutant form) were incubated with purified yeast PKA (Bcy1-Tpk) and [γ-33P]ATP in the presence of 8Br-cAMP as indicated. The samples were analyzed by SDS–PAGE, digital autoradiography, and staining with Coomassie brilliant blue R-250. Arrowhead, bovine serum albumin. (D) Recombinant Tom40WT and Tom40S54E were incubated with affinity-purified yeast Tpk1 as indicated and [γ-33P]ATP. The samples were analyzed as described for C.
To analyze whether Tom40 is a substrate of PKA in a homologous system, we generated a yeast strain expressing protein A–tagged Bcy1. Purification of Bcy1 by affinity chromatography led to the efficient copurification of PKA catalytic subunits, shown here with antibodies directed against Tpk1 (Figure 1B; the protein A tag was removed by cleavage with tobacco etch virus [TEV] protease). On stimulation with 8Br-cAMP, the purified yeast PKA was active and phosphorylated the PKA model substrate Kemptide and the receptor domain of Tom70 in the presence of [γ-33P]ATP (Figure 1C, lane 2; Schmidt et al., 2011). Purified recombinant yeast Tom40 was phosphorylated by yeast PKA in the presence of 8Br-cAMP (Figure 1C, lane 4). The phosphorylation was specific for the predicted PKA target residue Ser-54 since replacement of this serine by glutamic acid blocked the phosphorylation of Tom40 (Figure 1C, lane 6). As an independent assay, we used a yeast strain containing tagged Tpk1 and purified the enzyme. Purified wild-type Tom40, but not the mutant form Tom40S54E, was phosphorylated by Tpk1 (Figure 1D, lanes 3 and 4). We conclude that Tom40 is a substrate of yeast PKA.
PKA phosphorylates the precursor of Tom40
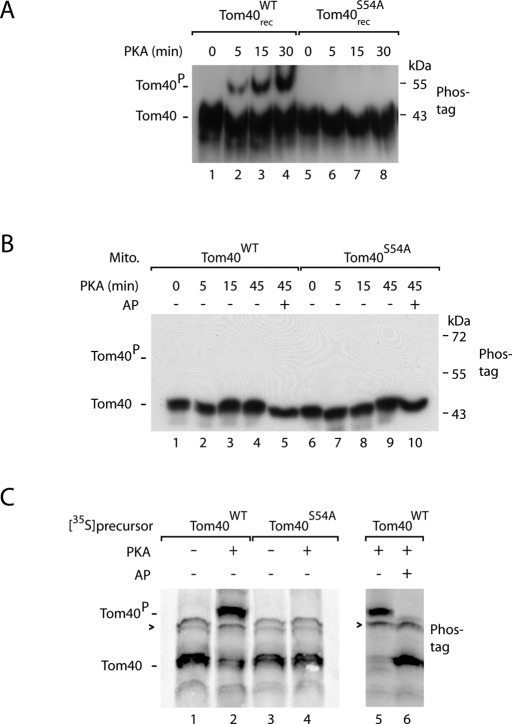
Phosphate-affinity (Phos-tag) SDS–PAGE leads to a lower gel mobility of many phosphorylated proteins compared with the nonphosphorylated forms (Kinoshita et al., 2006). We asked whether the phosphorylation of Tom40 at Ser-54 by PKA could be directly monitored by Phos-tag SDS–PAGE. We incubated purified Tom40 with PKA and indeed observed a slower-migrating form that was generated in a PKA- and time-dependent manner (Figure 2A, lanes 2–4). When Ser-54 was replaced by alanine, no phosphorylated form of Tom40 was detected (Figure 2A, lanes 6–8).
FIGURE 2:
PKA phosphorylates the precursor of Tom40 at serine 54. (A) Recombinant Tom40WT or Tom40S54A was incubated with purified PKA (New England BioLabs). The samples were lysed in Laemmli buffer and analyzed by Phos-tag SDS–PAGE and Western blotting using antiserum directed against Tom40. (B) Mitochondria were isolated from WT and Tom40S54A yeast strains and incubated with purified yeast PKA (Bcy1-Tpk) and 8Br-cAMP for the indicated periods. Where indicated, the samples were subsequently incubated with alkaline phosphatase (AP). The samples were analyzed as described for A. Similarly, mammalian PKA did not phosphorylate Ser-54 of Tom40 in intact mitochondria. (C) 35S-Labeled Tom40WT and Tom40S54A were synthesized in reticulocyte lysate in the presence or absence of PKA as indicated. Sample 6 was subsequently treated with AP. The samples were analyzed by Phos-tag SDS–PAGE and digital autoradiography. Arrowhead, nonspecific band; Tom40P, phosphorylated form of Tom40.
When isolated yeast mitochondria were incubated with PKA, however, a phosphorylated form of Tom40 was not detected (Figure 2B; neither mammalian PKA nor yeast PKA in the presence of 8Br-cAMP led to a detectable phosphorylation of Ser-54). Thus mature Tom40 that is stably integrated into the mitochondrial outer membrane was not accessible to PKA, whereas purified Tom40 in detergent was phosphorylated by PKA (Figures 1, C and D, and 2A). We therefore asked whether Tom40 can be phosphorylated in the cytosol. We synthesized the precursor of Tom40 in reticulocyte lysate in the presence of [35S]methionine. When the synthesis was performed in the presence of PKA, a slower-migrating form of Tom40 was observed by Phos-tag SDS–PAGE (Figure 2C, lanes 2 and 5). This form was sensitive to treatment with alkaline phosphatase (Figure 2C, lane 6). When Tom40S54A was synthesized in reticulocyte lysate, it was not affected by PKA (Figure 2C, lane 4). Taken together, these results indicate that the precursor of Tom40 is phosphorylated by PKA in the cytosol, whereas mature imported Tom40 is not a substrate of PKA.
Phosphorylation impairs the biogenesis of Tom40
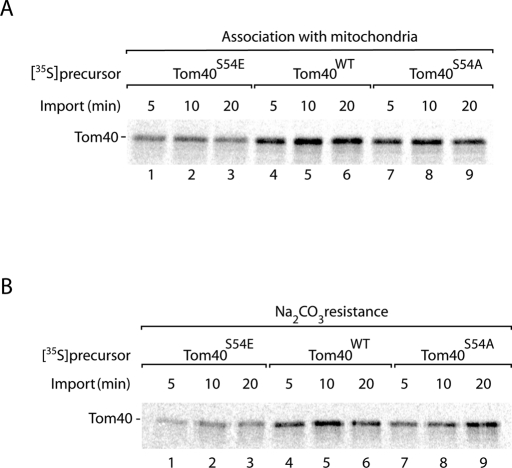
To analyze the biogenesis of Tom40, we incubated the 35S-labeled precursor with isolated mitochondria. We used the wild-type precursor of Tom40 and two mutant forms of Ser-54. When serine was replaced by alanine, the binding of Tom40 to mitochondria was only mildly affected compared with the wild-type precursor (Figure 3A, lanes 4–9). When serine was replaced by the phosphomimetic residue glutamate, however, binding of Tom40 to mitochondria was reduced (Figure 3A, lanes 1–3). Treatment of mitochondria with sodium carbonate at alkaline pH leads to the extraction of soluble and peripheral membrane proteins, whereas integral membrane proteins remain in the membrane sheets (Fujiki et al., 1982; Stojanovski et al., 2007). Mitochondria were incubated with [35S]Tom40 and then treated at alkaline pH, demonstrating that the replacement of Ser-54 by glutamate considerably reduced the membrane integration of Tom40 (Figure 3B, lanes 1–3).
FIGURE 3:
Replacement of serine 54 by glutamate impairs import of Tom40 into mitochondria. 35S-Labeled Tom40S54E, Tom40WT, and Tom40S54A precursors were incubated with isolated yeast wild-type mitochondria at 25°C for the indicated periods. The samples were split in half, and the mitochondria were reisolated. (A) One half was lysed in Laemmli buffer and analyzed by SDS–PAGE and digital autoradiography. (B) The other half was resuspended in Na2CO3, pH 11.5, and incubated for 30 min on ice. Membrane sheets were pelleted by centrifugation at 100,000 × g and analyzed by SDS–PAGE and digital autoradiography.
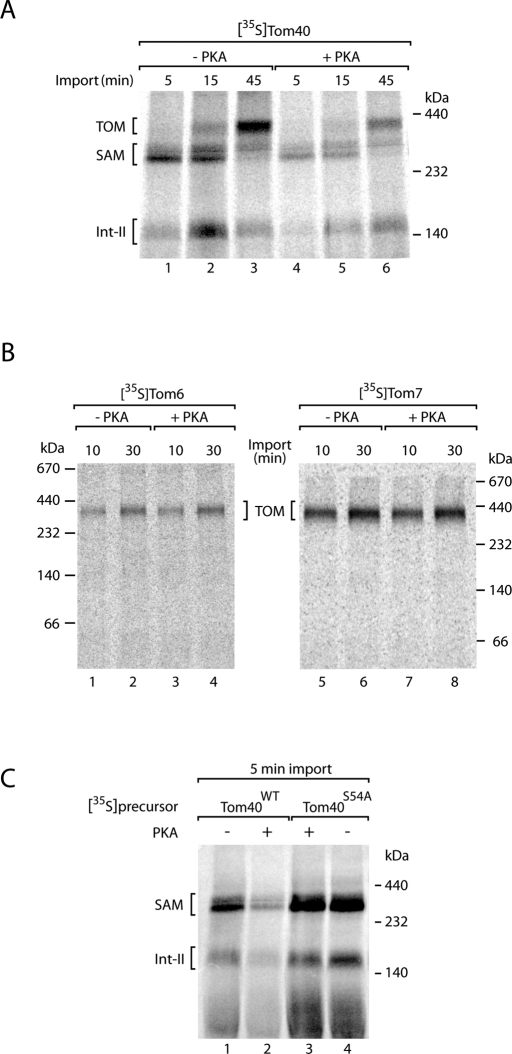
The assembly pathway of Tom40 involves several steps. On initial import of the precursor by the TOM complex to the intermembrane space side, intermembrane space chaperone complexes transfer Tom40 to the sorting and assembly machinery (SAM complex) of the outer membrane (Model et al., 2001; Kozjak et al., 2003; Paschen et al., 2003; Wiedemann et al., 2003, 2004; Gentle et al., 2004; Hoppins and Nargang, 2004). The interaction of the Tom40 precursor with the SAM complex can be directly monitored by blue native electrophoresis after lysis of the mitochondria with the nonionic detergent digitonin (Ryan et al., 2001; Wittig et al., 2006; Stojanovski et al., 2007). The SAM intermediate of ∼250 kDa is followed by a second intermediate (Int-II) of lower molecular mass and finally the assembly of imported Tom40 into the mature TOM complex of ∼400 kDa (Figure 4A, lanes 1–3; Wiedemann et al., 2003; Kutik et al., 2008). When the precursor of Tom40 was phosphorylated by PKA, formation of the assembly steps was considerably impaired. The inhibition occurred already at an early stage, since the generation of the SAM intermediate was impaired (Figure 4A, lanes 4–6). To exclude that PKA inhibited the assembly of the TOM complex in general, we imported two small Tom precursors—Tom6 and Tom7—whose assembly can be efficiently monitored by blue native electrophoresis (Dembowski et al., 2001; Model et al., 2001). Both proteins were assembled into the TOM complex independent of the presence or absence of PKA (Figure 4B).
FIGURE 4:
Phosphorylation of Tom40 by PKA impairs formation of the SAM import intermediate. (A) 35S-Labeled precursor of Tom40 was synthesized in the presence or absence of PKA (New England BioLabs) as indicated and incubated with isolated yeast wild-type mitochondria for the indicated periods at 25°C. The mitochondria were solubilized in digitonin-containing buffer and analyzed by blue native electrophoresis and digital autoradiography. (B) 35S-Labeled precursors of Tom6 and Tom7 were imported into isolated mitochondria in the presence or absence of PKA. The mitochondria were analyzed as described for A. (C) [35S]Tom40WT and [35S]Tom40S54A precursors were synthesized in the presence or absence of PKA and incubated with isolated mitochondria for 5 min at 25°C. The mitochondria were analyzed as described for A.
To analyze whether the inhibitory effect of PKA was specific for the phosphorylation of Ser-54 of Tom40, we compared formation of the SAM intermediate of the wild-type Tom40 precursor to the mutant precursor Tom40S54A. In the absence of PKA, wild-type precursor and mutant precursor accumulated at the SAM after a short-term import reaction (Figure 4C, lanes 1 and 4). PKA only inhibited the formation of the SAM intermediate of the wild-type precursor and not of the mutant precursor (Figure 4C, lanes 2 and 3), demonstrating that PKA inhibits the biogenesis of Tom40 selectively via phosphorylation of Ser-54. Taking the data together, we conclude that phosphorylation by PKA impairs the biogenesis pathway of Tom40 at an early that is, at or before formation of the SAM intermediate.
Mitochondria import the nonphosphorylated form of Tom40
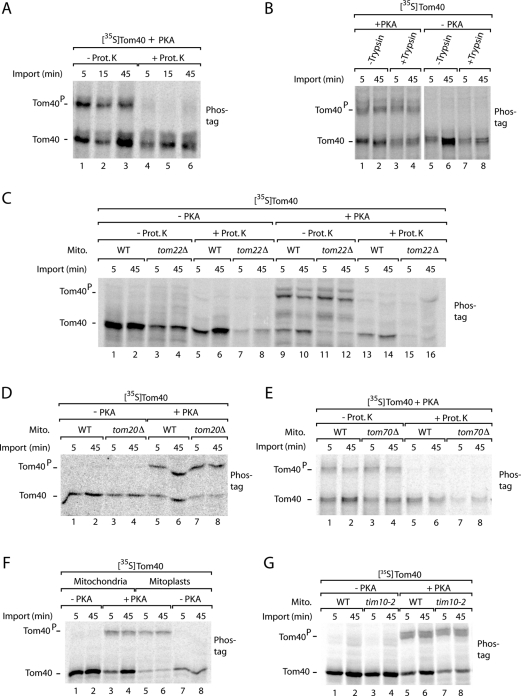
The initial stage of Tom40 import, that is, translocation via the TOM complex to the intermembrane space side, does not involve a blue native–stable intermediate and thus cannot be directly monitored by native gel analysis (Wiedemann et al., 2004). Therefore we used the accessibility to externally added protease to determine a possible role of PKA phosphorylation in this initial import step. To distinguish between phosphorylated and nonphosphorylated Tom40 precursors, we analyzed the mitochondria by Phos-tag SDS–PAGE. The nonphosphorylated Tom40 was protected against added proteinase K, indicating that it was imported into mitochondria (Figure 5A; Model et al., 2001; Wiedemann et al., 2003). The phosphorylated Tom40 precursor, however, was almost completely degraded by proteinase K (Figure 5A), demonstrating that the precursor was still located on the mitochondrial surface and not imported.
FIGURE 5:
Mitochondria import the nonphosphorylated precursor of Tom40, whereas phosphorylated Tom40 remains on the mitochondrial surface. (A) [35S]Tom40 was synthesized in the presence of PKA (New England BioLabs) and incubated with isolated wild-type mitochondria for the indicated periods at 25°C. The mitochondria were treated with proteinase K (Prot. K; Stojanovski et al., 2007) where indicated and analyzed by Phos-tag SDS–PAGE and digital autoradiography. (B) [35S]Tom40 was synthesized in the presence or absence of PKA and imported into mitochondria that had been pretreated with trypsin (Ryan et al., 2001) as indicated. The mitochondria were analyzed by Phos-tag SDS–PAGE. (C) [35S]Tom40 was imported into mitochondria, which were isolated from tom22Δ yeast or the corresponding wild-type (WT) strain, in the presence or absence of PKA. The mitochondria were treated with proteinase K where indicated and analyzed by Phos-tag SDS–PAGE. (D) [35S]Tom40 was imported into mitochondria, which were isolated from tom20Δ and wild-type yeast, in the presence or absence of PKA. The mitochondria were analyzed by Phos-tag SDS–PAGE. (E) [35S]Tom40 was imported into mitochondria, which were isolated from tom70Δ and wild-type yeast, in the presence of PKA. The mitochondria were treated with proteinase K where indicated and analyzed by Phos-tag SDS–PAGE. (F) Mitochondria were preincubated in isotonic or hypotonic (swelling) buffer for 30 min on ice (Stojanovski et al., 2007). The mitochondria/mitoplasts were reisolated and incubated with [35S]Tom40 in the presence or absence of PKA and analyzed by Phos-tag SDS–PAGE. (G) [35S]Tom40 was imported into mitochondria, which were isolated from tim10-2 yeast or the corresponding wild-type strain, in the presence or absence of PKA. The mitochondria were analyzed by Phos-tag SDS–PAGE.
Tom20, Tom22, and Tom70 function as receptors for import of nuclear-encoded precursor proteins into mitochondria (Kiebler et al., 1993; Brix et al., 1997, 1999; van Wilpe et al., 1999; Yamano et al., 2008; Rimmer et al., 2011; Shiota et al., 2011). Pretreatment of mitochondria with trypsin removes the receptor domains and inhibits preprotein import into mitochondria (Ryan et al., 2001). Trypsin pretreatment considerably impaired the interaction of the nonphosphorylated precursor of Tom40 with mitochondria but not the phosphorylated precursor (Figure 5B). We thus studied mutant mitochondria deficient in Tom receptors. The import of radiolabeled Tom40 precursor was strongly inhibited in mitochondria lacking the central receptor Tom22 (Figure 5C, lanes 7 and 8; Model et al., 2001). Analysis by Phos-tag SDS–PAGE revealed that interaction of the nonphosphorylated precursor with mitochondria was strongly inhibited when Tom22 was lacking, whereas binding of the phosphorylated form to mitochondria was not affected (Figure 5C, lanes 11 and 12). As observed with wild-type mitochondria, the phosphorylated Tom40 interacting with tom22Δ mitochondria was digested by externally added protease (Figure 5C, lanes 13–16), demonstrating that phosphorylated Tom40 was not imported into mitochondria. Similarly, mitochondria lacking Tom20 or Tom70 were impaired in the interaction with nonphosphorylated Tom40, whereas the binding of phosphorylated Tom40 to the isolated mitochondria was not affected by lack of the receptors (Figure 5, D and E).
Swelling of mitochondria leads to a release of intermembrane space chaperones and thus inhibits the biogenesis of Tom40 (Wiedemann et al., 2004). Swollen mitochondria (mitoplasts) were inhibited in the interaction with nonphosphorylated Tom40 but not phosphorylated Tom40 (Figure 5F). tim10-2 mutant mitochondria are impaired in the activity of the Tim9–Tim10 intermembrane space chaperone and thus in the import of Tom40 (Truscott et al., 2002; Wiedemann et al., 2004). Only nonphosphorylated Tom40, and not the phosphorylated form of Tom40, was affected by the tim10-2 mutant (Figure 5G).
Taking the data together indicates that the nonphosphorylated precursor of Tom40 shows the characteristics of specific import into mitochondria, including dependence on Tom receptors and intermembrane space chaperones. In contrast, phosphorylated Tom40 remains on the mitochondrial surface in a receptor-independent manner and is not imported into mitochondria, indicating that the binding observed with mitochondria is nonproductive. We conclude that mitochondria specifically import the nonphosphorylated form of Tom40.
PKA inhibits Tom40 import independently of Tom70 phosphorylation
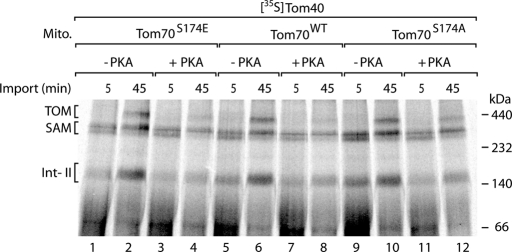
Phosphorylation of the receptor Tom70 by PKA impairs the interaction of the cytosolic chaperone Hsp70 with Tom70 (Schmidt et al., 2011). Hsp70 delivers hydrophobic precursor proteins such as the inner membrane metabolite carriers to Tom70 (Young et al., 2003; Li et al., 2009; Zara et al., 2009). PKA selectively phosphorylates Ser-174 of Tom70, which is located close to the chaperone-binding site of Tom70, and thus disturbs the Hsp70–Tom70 interaction (Schmidt et al., 2011). We asked whether the phosphorylation of Tom70 influenced the import of Tom40. In yeast mitochondria in which Ser-174 of Tom70 was replaced by alanine, the import of carrier precursors was enhanced, whereas a replacement of Ser-174 by glutamate inhibited carrier import (Schmidt et al., 2011). In the case of Tom40, however, neither replacement of Ser-174 by alanine nor replacement by glutamate affected the import of the precursor in comparison to wild-type mitochondria (Figure 6, lanes 1, 2, 5, 6, 9, and 10). Phosphorylation of the Tom40 precursor by PKA inhibited its import into Tom70S174A and Tom70S174E mitochondria like that into wild-type mitochondria (Figure 6, lanes 3, 4, 7, 8, 11. and 12). Given that Ser-174 is the only PKA target site of Tom70 (Schmidt et al., 2011), these results demonstrate that the inhibitory effect of PKA on the import of Tom40 occurs independently of the phosphorylation of Tom70.
FIGURE 6:
Inhibition of Tom40 import by PKA is not connected to the phosphorylation of Tom70 by PKA. 35S-Labeled precursor of Tom40 was imported into mitochondria, which were isolated from Tom70S174E, Tom70WT, or Tom70S174A yeast strains, at 25°C in the presence or absence of PKA (New England BioLabs) as indicated. The mitochondria were solubilized in digitonin-containing buffer and analyzed by blue native electrophoresis and digital autoradiography.
DISCUSSION
We report a new mechanism of how cytosolic kinases regulate the preprotein translocase of the outer mitochondrial membrane. PKA phosphorylates the precursor of Tom40, the channel-forming core component of the TOM complex, and thus inhibits the import of Tom40 into mitochondria. So far, cytosolic kinases had been shown either to stimulate the biogenesis of Tom subunits (phosphorylation of Tom22 and Mim1 by CK2) or to inhibit the receptor activity of a Tom receptor without affecting its biogenesis (phosphorylation of Tom70 by PKA; Schmidt et al., 2011). We found that the PKA effect on Tom40 biogenesis was independent of the phosphorylation status of Tom70, demonstrating that PKA exerts two different inhibitory effects on the TOM complex: inhibition of receptor activity (Tom70) and inhibition of precursor import (Tom40). In agreement with these observations, in a yeast strain lacking the inhibitory PKA subunit Bcy1, the steady-state level of Tom40, but not that of Tom70, was reduced.
PKA is rapidly activated by addition of fermentable carbon sources such as glucose and sucrose that lead to increased intracellular cAMP levels in yeast cells (Broach, 1991; Thevelein, 1994; Santangelo, 2006; Tamaki, 2007; Zaman et al., 2008; Smets et al., 2010). PKA was shown to affect numerous cellular processes, including morphology of the mitochondrial membranes, programmed cell death, and oxidative metabolism. In addition, a few precursor proteins were found to be phosphorylated in a cAMP-dependent manner, affecting their interaction with molecular chaperones and translocation into mitochondria (Anandatheerthavarada et al., 1999; Cho et al., 2001; Robin et al., 2002, 2003; Pagliarini and Dixon, 2006; Chang and Blackstone, 2007; Carlucci et al., 2008; De Rasmo et al., 2008; Santel and Frank, 2008; Soubannier and McBride, 2009). The inhibitory effects of PKA on TOM biogenesis (this study) and function (Schmidt et al., 2011) provide a direct means to control the mitochondrial preprotein entry gate upon shift to fermentable growth conditions, under which a lower mitochondrial activity is required. Under nonfermentable, respiratory conditions the cAMP levels and PKA activity are low (Russell et al., 1993; Thevelein, 1994; Zaman et al., 2008; Smets et al., 2010), and thus Tom40 is efficiently imported.
The biogenesis pathway of Tom40 can be dissected into several steps, involving transport by the TOM complex, intermembrane space chaperones, and the SAM complex (Model et al., 2001; Wiedemann et al., 2003; Waizenegger et al., 2004; Habib et al., 2005; Kutik et al., 2008). Phosphorylation of the Tom40 precursor by PKA inhibits the initial stage of precursor translocation through the TOM complex to a protease-protected location. Whereas nonphosphorylated Tom40 precursor is efficiently imported, the phosphorylated form remains nonproductively bound to the mitochondrial surface and is not translocated. Numerous mutational studies have been performed with Tom40 (Rapaport and Neupert, 1999; Rapaport et al., 2001; Taylor et al., 2003, Humphries et al., 2005; Sherman et al., 2006; Kutik et al., 2008). A linear sorting signal for binding of Tom40 to the SAM complex has been identified in the carboxy-terminal region of the precursor (Kutik et al., 2008); however, a targeting signal that is responsible for the initial recognition of Tom40 by the TOM complex has not been found. It is conceivable that several elements of the precursor may contribute to the targeting process. Serine 54 is not essential for the targeting of Tom40, since this residue can be deleted (Rapaport and Neupert, 1999; Rapaport et al., 2001; Taylor et al., 2003; Humphries et al., 2005; Sherman et al., 2006; Kutik et al., 2008) or replaced by alanine (this study) without blocking Tom40 targeting. Harsman et al. (2010) reconstituted purified Tom40WT and Tom40S54E into planar lipid bilayers and observed a similar gating behavior of the Tom40 channel of wild type and mutant, indicating that the replacement of Ser-54 by the phosphomimetic residue glutamate did not disturb the overall folding of Tom40. Of interest, the association rate of positively charged presequence peptides with Tom40 was altered when Ser-54 was replaced by glutamate (Harsman et al., 2010), suggesting that modification of this residue can affect the interaction properties of Tom40 in vitro. We analyzed the biogenesis of Tom40S54E in organello and observed an impairment of import into mitochondria. Using PKA, we showed that phosphorylation of Ser-54 inhibits the translocation of Tom40 via the TOM complex, demonstrating that phospho–Ser-54 interferes with the targeting process. Although the exact nature of the Tom40 targeting signal remains elusive, the strong inhibitory effect of a single phosphorylation event provides an efficient mechanism to control the import of Tom40.
Taking our results together with the findings by Schmidt et al., (2011), we conclude that cytosolic kinases regulate biogenesis and function of the TOM complex at multiple levels. The main protein entry gate of mitochondria is not functioning autonomously but is tightly integrated into a network of regulatory kinases.
MATERIALS AND METHODS
Yeast strains and cloning
Most S. cerevisiae strains used in this study are derived from the strain YPH499 (MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801; Sikorski and Hieter, 1989). YPH499-tom40Δ+pFL39-TOM40 was made by transforming the shuffling strain tom40Δ+Yep-TOM40 (Kutik et al., 2008) with the plasmid pFL39-TOM40 as described (Schmidt et al., 2011). The mutations Tom40S54A and Tom40S54E were generated by site-directed mutagenesis. Clones were verified by sequencing. The Tom70S174E and Tom70S174A yeast strains, tom20Δ strain, tom22Δ strain, tom70Δ strain, tim10-2 strain, and bcy1Δ strain have been described previously (Moczko et al., 1994; van Wilpe et al., 1999; Truscott et al., 2002; Stojanovski et al., 2007; Schmidt et al., 2011). The Bcy1ProtA strain was constructed by amplifying the HIS3MX6-pNOP-ProtA-TEV plasmid (Meisinger et al., 2007) with primers A1 (5′ ATTACAACAAGC AGATTATTTTCAAAAGACAACAGTAAGAATAAACGGGAATACGAATTCGAGCTC 3′) and A2 (5′ GTTCTGGAACAGTTGCAATTCGGCTTGCGATTCCTTGGGCAAAGA AGATACCACGTCACTCATACCCTGA 3′) and integrating into the BCY1 open reading frame by homologous recombination. The Tom70WT/pET19 and Kemptide-GST/pETGEXct constructs were reported previously (Brix et al., 1997; Schmidt et al., 2011).
Isolation of mitochondria and preparation of yeast protein extracts
Yeast strains were typically grown on nonfermentable YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] bactopeptone, 3% [wt/vol] glycerol, pH 5.0) at 23°C until an OD600 of 0.5–1.0 was reached. Mitochondria were isolated by differential centrifugation (Meisinger et al., 2006a). Mitochondria were resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, and 10 mM 3-(N-morpholino)propanesulfonic acid–KOH, pH 7.2), adjusted to a protein concentration of 10 mg/ml, frozen in liquid nitrogen, and stored at −80°C. For preparation of yeast protein extracts, yeast cells were grown on fermentable YPS medium (1% [wt/vol] yeast extract, 2% [wt/vol] bactopeptone, 2% [wt/vol] sucrose), and extracts corresponding to the same OD600 were prepared by postalkaline lysis.
Purification of yeast PKA
The yeast strain expressing Bcy1ProtA and the corresponding wild-type strain (YPH499) were grown in YPG medium at 30°C to an OD600 of 2.0, harvested, and lysed as described for the preparation of mitochondria (Meisinger et al., 2006a) but using homogenization buffer with 2 mg/ml bovine serum albumin, 2 mM phenylmethylsulfonyl fluoride (PMSF), and complete EDTA-free protease inhibitors (Roche, Indianapolis, IN). The lysate was cleared by centrifugation (30,000 × g) and was added to human immunoglobulin G–coupled Sepharose beads. Unbound material was removed by washing with excess homogenization buffer. The Bcy1-Tpk complexes were eluted by cleavage with TEV protease in homogenization buffer. After addition of 10–20% (vol/vol) glycerol and 2 mM dithiothreitol (DTT), the eluates were stored at −80°C. Alternatively, a yeast strain expressing Tpk1TAP was grown on fermentable YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] bactopeptone, 2% [wt/vol] glucose) until an OD600 of 1.0, and extracts were purified as described (Mah et al., 2005). The kinase was eluted by cleavage with TEV protease in kinase buffer (100 mM Tris, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 20% glycerol, 1 mM PMSF, 1× PhosStop [Roche]). Eluates were stored at −80°C.
In vitro phosphorylation assays
Tom40 was purified from inclusion bodies as described (Hill et al., 1998). The inclusion bodies were solubilized in urea buffer (8 M urea, 50 mM Tris/HCl pH 8.0, 1 mM EDTA, and 100 mM DTT). Kemptide-GST and Tom70cd were prepared as described (Schmidt et al., 2011).
In vitro phosphorylation reactions with recombinant mouse PKA (New England BioLabs, Ipswich, MA) and [γ-33P]ATP were performed as described (Schmidt et al., 2011). For analysis by Phos-tag SDS–PAGE and immunoblotting, 5–10 mM ATP and 50 U/μl PKA (New England BioLabs) were used. Tom40 in vitro phosphorylation was performed in a buffer with 25 mM tricine, 7.5 mM BisTris, and 0.5% (vol/vol) heptyl-thio-glucopyranoside, pH 7.0.
Bcy1-Tpk complexes were supplemented with 1× PKA assay buffer (New England BioLabs), 1× PhosStop (Roche), 2 mM PMSF, 0.2 mM ATP, 5 mCi of [γ-33P]ATP (PerkinElmer, Waltham, MA), 50–100 μM 8Br-cAMP (Sigma-Aldrich, St. Louis, MO; or Biaffin, Kassel, Germany) where indicated, Kemptide-GST, or Tom70cdWT in a total volume of 18 μl and incubated for 30 min at 25°C under constant shaking. The reactions were stopped by addition of Laemmli buffer and heating to 95°C.
For Tom40, the Bcy1-Tpk complexes or the Tpk1 eluate were incubated with 1 μg of Tom40WT, Tom40S54E, or Tom40S54A, 50–100 μM 8Br-cAMP (where indicated), 0.2 mM ATP, and 5 mCi [γ-33P]ATP (PerkinElmer) in a total volume of 20 μl for 90 min at 25°C under constant shaking. The reactions were stopped by addition of Laemmli buffer and heating to 95°C.
Isolated mitochondria were supplemented with 1× PKA assay buffer, 1× PhosStop, 5–10 mM ATP, and either 50 U/μl recombinant PKA in SEM buffer or Bcy1-Tpk complexes with 50–100 μM 8Br-cAMP and incubated at 25°C. For Phos-tag and immunoblot analysis, 5–10 mM ATP and recombinant Tom40WT or Tom40S54A were used in a total volume of 15 μl.
For Phos-tag SDS–PAGE, standard discontinuous 8–12.5% polyacrylamide gels were prepared with the modification that 50 μM Phos-tag reagent (Kinoshita et al., 2006) and 100 μM MnCl2 were added to the separation gel mix prior to polymerization.
Protein import assays
35S-Labeled precursor proteins were generated by in vitro translation and incubated with isolated mitochondria in import buffer as described (Ryan et al., 2001; Stojanovski et al., 2007). Where indicated, the reactions were supplemented with 50 U/μl recombinant PKA (New England BioLabs) during translation or import. The import efficiency of different precursor forms was directly compared by adjusting the levels of the radiolabeled proteins added to the import reactions.
The import reactions were stopped on ice, and mitochondria were reisolated, washed in SEM buffer, and either lysed directly in sample buffer or treated with 100 mM Na2CO3 (pH 11.5) and centrifuged at 100,000 × g for 1 h before lysis in sample buffer to test for membrane integration (Fujiki et al., 1982; Stojanovski et al., 2007). SDS–PAGE or Phos-tag SDS–PAGE was followed by digital autoradiography (GE Healthcare, Piscataway, NJ). To analyze protein complex formation, mitochondria were lysed in 1% digitonin-containing buffer and analyzed by blue native electrophoresis (Ryan et al., 2001; Wittig et al., 2006; Stojanovski et al., 2007) and digital autoradiography.
Acknowledgments
We thank C. Gerbeth and B. Kulawiak for discussion. This work was supported by the Deutsche Forschungsgemeinschaft, Excellence Initiative of the German Federal and State Governments (GSC-4 Spemann Graduate School; EXC 294 BIOSS), Trinationales Graduiertenkolleg GRK 1478, Sonderforschungsbereich 746, Bundesministerium für Bildung und Forschung (Dynamo), Landesforschungspreis Baden-Württemberg, and Gottfried Wilhelm Leibniz Program.
Abbreviations used:
- CK2
casein kinase 2
- Mim1
mitochondrial import protein 1 of the outer membrane
- PKA
protein kinase A
- TEV
tobacco etch virus
- TOM
translocase of the outer mitochondrial membrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0933) on March 14, 2012.
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Biswas G, Mullick J, Sepuri NB, Otvos L, Pain D, Avadhani NG. Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at ser128. EMBO J. 1999;18:5494–5504. doi: 10.1093/emboj/18.20.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Bannwarth M, Meisinger C, Hill K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, Wagner R. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol. 2005;353:1011–1020. doi: 10.1016/j.jmb.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Becker T, Guiard B, Thornton N, Zufall N, Stroud DA, Wiedemann N, Pfanner N. Assembly of the mitochondrial protein import channel: role of Tom5 in two-stage interaction of Tom40 with the SAM complex. Mol Biol Cell. 2010;21:3106–3113. doi: 10.1091/mbc.E10-06-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Thornton N, Stroud D, Meisinger C, Wiedemann N, Pfanner N. Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J Mol Biol. 2011;405:113–124. doi: 10.1016/j.jmb.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- Brix J, Rüdiger S, Bukau B, Schneider-Mergener J, Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- Broach JR. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Cannon JF, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;8:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci A, Lignitto L, Feliciello A. Control of mitochondria dynamics and oxidative metabolism by cAMP, AKAPs and the proteasome. Trends Cell Biol. 2008;18:604–613. doi: 10.1016/j.tcb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Lee YK, Chae CB. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim Biophys Acta. 2001;1522:175–186. doi: 10.1016/s0167-4781(01)00333-5. [DOI] [PubMed] [Google Scholar]
- De Rasmo D, Panelli D, Sardanelli AM, Papa S. cAMP-dependent protein kinase regulates the mitochondrial import of the nuclear encoded NDUFS4 subunit of complex I. Cell Signal. 2008;20:989–997. doi: 10.1016/j.cellsig.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Dembowski M, Künkele KP, Nargang FE, Neupert W, Rapaport D. Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa. J Biol Chem. 2001;276:17679–17685. doi: 10.1074/jbc.M009653200. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta. 2011;1808:955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, de Godoy LM, Cox J, Neuhauser N, Ren S, Olsen JV, Mann M. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 2009;9:4642–4652. doi: 10.1002/pmic.200900144. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. J Biol Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. [DOI] [PubMed] [Google Scholar]
- Harsman A, Krüger V, Bartsch P, Honigmann A, Schmidt O, Rao S, Meisinger C, Wagner R. Protein conducting nanopores. J Phys Condens Matter. 2010;22:454102. doi: 10.1088/0953-8984/22/45/454102. [DOI] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta. 2002;1592:97–105. doi: 10.1016/s0167-4889(02)00268-9. [DOI] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likic VA, Gooley PR, Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Humphries AD, Streimann IC, Stojanovski D, Johnston AJ, Yano M, Hoogenraad NJ, Ryan MT. Dissection of the mitochondrial import and assembly pathway of human Tom40. J Biol Chem. 2005;280:11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- Kato H, Mihara K. Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem Biophys Res Commun. 2008;369:958–963. doi: 10.1016/j.bbrc.2008.02.150. [DOI] [PubMed] [Google Scholar]
- Kiebler M, Keil P, Schneider H, van der Klei IJ, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Kutik S, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Li J, Qian X, Hu J, Sha B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem. 2009;284:23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Mah AS, Elia AE, Devgan G, Ptacek J, Schutkowski M, Snyder M, Yaffe MB, Deshaies RJ. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Uno I, Ishikawa T, Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants unable to synthesize it. J Bacteriol. 1983;156:898–900. doi: 10.1128/jb.156.2.898-900.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006a;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schönfisch B, Müller H, Kozjak V, Pfanner N. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem. 2006b;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- Meisinger C, et al. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Ehmann B, Gärtner F, Hönlinger A, Schäfer E, Pfanner N. Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J Biol Chem. 1994;269:9045–9051. [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Popov-Celeketic J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Rao S, Gerbeth C, Harbauer A, Mikropoulou D, Meisinger C, Schmidt O. Signaling at the gate: phosphorylation of the mitochondrial protein import machinery. Cell Cycle. 2011;10:2083–2090. doi: 10.4161/cc.10.13.16054. [DOI] [PubMed] [Google Scholar]
- Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Taylor RD, Käser M, Langer T, Neupert W, Nargang FE. Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria. Mol Biol Cell. 2001;12:1189–1198. doi: 10.1091/mbc.12.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer KA, Foo JH, Ng A, Petrie EJ, Shilling PJ, Perry AJ, Mertens HD, Lithgow T, Mulhern TD, Gooley PR. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J Mol Biol. 2011;405:804–818. doi: 10.1016/j.jmb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Robin MA, Anandatheerthavarada HK, Biswas G, Sepuri NB, Gordon DM, Pain D, Avadhani NG. Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J Biol Chem. 2002;277:40583–40593. doi: 10.1074/jbc.M203292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin MA, Prabu SK, Raza H, Anandatheerthavarada HK, Avadhani NG. Phosphorylation enhances mitochondrial targeting of GSTA4-4 through increased affinity for binding to cytoplasmic Hsp70. J Biol Chem. 2003;278:18960–18970. doi: 10.1074/jbc.M301807200. [DOI] [PubMed] [Google Scholar]
- Russell M, Bradshaw-Rouse J, Markwardt D, Heideman W. Changes in gene expression in the Ras/adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol Biol Cell. 1993;4:757–765. doi: 10.1091/mbc.4.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S. Shaping mitochondria: the complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Schmidt O, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Ahting U, Eichacker L, Granvogl B, Go NE, Nargang FE, Neupert W, Nussberger S. Role of Tom5 in maintaining the structural stability of the TOM complex of mitochondria. J Biol Chem. 2005;280:14499–14506. doi: 10.1074/jbc.M413667200. [DOI] [PubMed] [Google Scholar]
- Sherman EL, Go NE, Nargang FE. Functions of the small proteins in the TOM complex of Neurospora crassa. Mol Biol Cell. 2005;16:4172–4182. doi: 10.1091/mbc.E05-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EL, Taylor RD, Go NE, Nargang FE. Effect of mutations in Tom40 on stability of the translocase of the outer mitochondrial membrane (TOM) complex, assembly of Tom40, and import of mitochondrial preproteins. J Biol Chem. 2006;281:22554–22565. doi: 10.1074/jbc.M601630200. [DOI] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci USA. 2011;108:15179–15183. doi: 10.1073/pnas.1105921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet. 2010;56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- Soubannier V, McBride HM. Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta. 2009;1793:154–170. doi: 10.1016/j.bbamcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kadowaki T, Maeda M, Sasaki H, Nabekura J, Sakaguchi M, Mihara K. Membrane-embedded C-terminal segment of rat mitochondrial TOM40 constitutes protein-conducting pore with enriched beta-structure. J Biol Chem. 2004;279:50619–50629. doi: 10.1074/jbc.M408604200. [DOI] [PubMed] [Google Scholar]
- Tamaki H. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J Biosci Bioeng. 2007;104:245–250. doi: 10.1263/jbb.104.245. [DOI] [PubMed] [Google Scholar]
- Taylor RD, McHale BJ, Nargang FE. Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J Biol Chem. 2003;278:765–775. doi: 10.1074/jbc.M208083200. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Signal transduction in yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, McMullen B, Hurwitz M, Krebs EG, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987a;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987b;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde S, Thevelein JM. Cyclic AMP-protein kinase A and Snf1 signaling mechanisms underlie the superior potency of sucrose for induction of filamentation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:286–293. doi: 10.1128/EC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilpe S, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D. Tob38, a novel essential component in the biogenesis ofβ-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochim Biophys Acta. 2009;1793:42–51. doi: 10.1016/j.bbamcr.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem. 2010;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Yatsukawa Y, Esaki M, Hobbs AE, Jensen RE, Endo T. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem. 2008;283:3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J. 2009;419:369–375. doi: 10.1042/BJ20082270. [DOI] [PMC free article] [PubMed] [Google Scholar]