Some components of the fission yeast septation initiation network (SIN) localize asymmetrically to spindle pole bodies during anaphase. Symmetric localization of these proteins correlates with cytokinesis defects. It is shown that the SIN-kinase Sid2 mediates SIN asymmetry, in part via the scaffold Cdc11, revealing a previously unknown feedback loop operating to generate SIN asymmetry.
Abstract
The Schizosaccharomyces pombe septation initiation network (SIN) is an Spg1-GTPase–mediated protein kinase cascade that triggers actomyosin ring constriction, septation, and cell division. The SIN is assembled at the spindle pole body (SPB) on the scaffold proteins Cdc11 and Sid4, with Cdc11 binding directly to SIN signaling components. Proficient SIN activity requires the asymmetric distribution of its signaling components to one of the two SPBs during anaphase, and Cdc11 hyperphosphorylation correlates with proficient SIN activity. In this paper, we show that the last protein kinase in the signaling cascade, Sid2, feeds back to phosphorylate Cdc11 during mitosis. The characterization of Cdc11 phosphomutants provides evidence that Sid2-mediated Cdc11 phosphorylation promotes the association of the SIN kinase, Cdc7, with the SPB and maximum SIN signaling during anaphase. We also show that Sid2 is crucial for the establishment of SIN asymmetry, indicating a positive-feedback loop is an important element of the SIN.
INTRODUCTION
To ensure proper segregation of genetic material to daughter cells during cell division, the onset of cytokinesis must be coordinated with the completion of mitosis. In the yeast Schizosaccharomyces pombe, a GTPase-driven signaling pathway termed the septation initiation network (SIN) participates in this control (reviewed in Krapp et al., 2004b; Wolfe and Gould, 2005). The SIN controls the final stages of cell division, including actomyosin ring contraction and formation of the division septum. Loss-of-function mutations in sin genes result in elongated multinucleate cells, due to multiple rounds of nuclear division and cell growth in the absence of cell division. Reciprocally, prolonged SIN activity (caused by mutations in SIN inhibitors) leads to the formation of multiple actomyosin contractile rings and septa in the absence of nuclear division.
SIN signaling is initiated by activation of the GTPase, Spg1 (Schmidt et al., 1997; Sohrmann et al., 1998). Spg1 is first activated in a symmetrical fashion at both spindle pole bodies (SPBs) during metaphase and recruits its effector protein kinase Cdc7. As anaphase ensues, SIN symmetry is broken, with Spg1 and Cdc7 becoming inactivated at the old SPB (Schmidt et al., 1997; Grallert et al., 2004) and Cdc7 increasing in abundance at the new SPB, which retains active Spg1 (Sohrmann et al., 1998; Garcia-Cortes and McCollum, 2009). The Sid1 protein kinase complex is then recruited to the new SPB, which has active Spg1-Cdc7 (Guertin et al., 2000). The persistence of symmetrical localization of Cdc7 during anaphase, or precocious localization of Cdc7 to SPBs during interphase, correlates with elevated SIN activity (Sohrmann et al., 1998). However, the signaling mechanisms that underlie the retention or loss of Cdc7 at SPBs are not completely understood.
The final SIN protein kinase, Sid2, also localizes to SPBs but in an Spg1-independent manner (Sparks et al., 1999). Unlike the aforementioned SIN components, Sid2, with its binding partner Mob1, localizes to the medial ring when Sid1 binds the SPBs; Sid2 medial localization depends on all other upstream SIN components (Sparks et al., 1999; Hou et al., 2000; Salimova et al., 2000). It is therefore speculated that Sid2-mediated phosphorylation events provide the final output of the SIN, but substrates other than the Cdc14 family phosphatase, Clp1 (Chen et al., 2008), have yet to be identified.
SIN inactivation at the end of division involves removal of a positive regulator, Etd1, from the daughter cell that inherited active Spg1-Cdc7 (Garcia-Cortes and McCollum, 2009) and increases in Spg1 GAP activity at both SPBs (Li et al., 2000). Byr4 and Cdc16 comprise the Spg1 GAP (Furge et al., 1998). These components localize to the SPB during interphase to prevent Spg1 activation, are transiently lost from SPBs during early mitosis when Cdc7 localizes to both SPBs, and then localize to the old SPB during anaphase (Li et al., 2000).
Assembly of SIN signaling components and their regulators occurs at the SPB on a platform comprised of the SIN scaffolding components Cdc11 and Sid4 (Chang and Gould, 2000; Krapp et al., 2001, 2004a; Tomlin et al., 2002; Morrell et al., 2004). Cdc11 binds directly to Sid2, Cdc16, and Spg1 through its N-terminal domain, while its C-terminal, leucine-rich repeats tether it to Sid4 (Krapp et al., 2001; Morrell et al., 2004). The Cdc11 N-terminus also interacts with Cdk1-Cdc13/cyclin B, and multiple sites of Cdk1 phosphorylation are predicted (Morrell et al., 2004). Indeed, Cdc11 is hyperphosphorylated during mitosis, although this modification has been correlated with SIN activation and depends upon both Cdc7 and its upstream regulator, the Plo1 kinase (Krapp et al., 2003, 2004a). However, the specific kinase involved in Cdc11 phosphorylation and the functional consequence of Cdc11 phosphorylation remained undefined.
In this study, we investigated the regulation of Cdc11 phosphorylation and its involvement in SIN function. In addition to our previous finding that Cdk1 phosphorylates Cdc11 during mitosis (Morrell et al., 2004), we found that Sid2 also phosphorylates Cdc11 on multiple sites. Sid2 phosphorylation of Cdc11 affects the dynamics of Cdc7 and Sid2 and promotes the robustness of SIN signaling. Unexpectedly, our study also revealed that Sid2 is required for the development of Cdc7 asymmetry, uncovering a positive-feedback autoamplification loop in the SIN.
RESULTS
Sid2 phosphorylates Cdc11
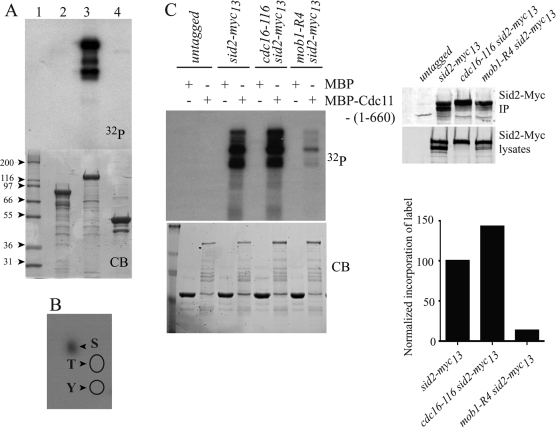
We had previously shown that Cdc11 binds Sid2 directly (Krapp et al., 2004a; Morrell et al., 2004), and we also found that Cdc11 copurified in a Sid2 tandem affinity purification (TAP; Supplemental Figure S1). Therefore, we investigated whether Sid2 phosphorylated Cdc11. While neither maltose binding protein (MBP) nor the C-terminal half of Cdc11 was phosphorylated by Sid2-Myc13 immunoprecipitates prepared from cdc16-116 cells (in which Sid2 is active [ Sparks et al., 1999]), the N-terminal 660 amino acids of Cdc11 were phosphorylated (Figure 1A). Phosphorylation of Cdc11 occurred exclusively on serine residues (Figure 1B). Cdc11 phosphorylation was due to Sid2-Myc13 and not a coprecipitating kinase, because Sid2-Myc13 from mob1-R4 mutant cells phosphorylated MBP-Cdc11-(1-660) very inefficiently (Figure 1C). Mob1 is an essential subunit of the Sid2 kinase (Hou et al., 2000; Salimova et al., 2000). Furthermore, Sid2-Myc13 from cdc16-116 mutant cells phosphorylated MBP-Cdc11-(1-660) better (Figure 1C).
FIGURE 1:
Sid2-Mob1 phosphorylates Cdc11. (A) Sid2-Myc13 was immunoprecipitated from cdc16-116 cells that had been shifted to 36ºC for 2.5 h, and the immunoprecipitate was divided into three portions. MBP-Cdc11-(632-1045) (lane 2), MBP-Cdc11-(1-660) (lane 3), or MBP (lane 4) was added to each portion in the presence of labeled ATP. Following 30 min at 30ºC, the reactions along with molecular mass standards (lane 1) were resolved by SDS–PAGE. The gel was stained with Coomassie (CB; bottom panel), dried, and then exposed to film [32P] (top panel). (B) Phosphoamino acid analysis of Cdc11(1-660) phosphorylated by Sid2-Mob1. The positions of the phosphothreonine and phosphotyrosine standards are indicated by the circles. (C) Anti-Myc immunoprecipitates from the indicated strains shifted to 36ºC for 4 h were incubated with MBP or MBP-Cdc11-(1-660) in the presence of labeled ATP. Following 30 min at 30ºC, the reactions were stopped and resolved by SDS–PAGE. The gel was stained with Coomassie blue (bottom, left panel), dried, and then exposed to film (top, left panel). The levels of Sid2-Myc13 in total lysates and in the immunoprecipitates used for the kinase reactions were determined by immunoblotting (top, right panels). The relative activity of Sid2-Myc13 was normalized in the various strains (bottom, right panel) and the relative activity in wild-type cells was set at 100%.
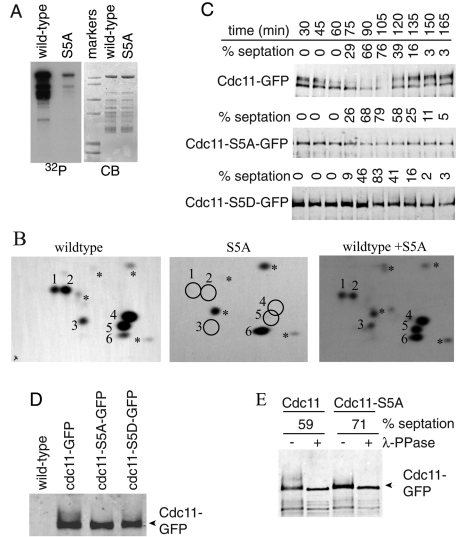
To narrow down the region of Cdc11 that was phosphorylated, we tested additional Cdc11 fragments fused to MBP as Sid2-Myc13 substrates. MBP-Cdc11-(488-660) was not phosphorylated by Sid2-Myc13 (Figure S2A), but Cdc11 residues 1–282, 110–160, and 301–493 were (Figure S2B and unpublished data), indicating that multiple Sid2-Myc13 phosphorylation sites reside in the first 488 amino acids of Cdc11. Accordingly, phosphotryptic peptide mapping of MBP-Cdc11-(1-660) generated five major (1–5), one minor (6), and a few very minor phosphopeptides (Figures 2B, left panel, and S2B, top panel). All of these were accounted for by phosphopeptides derived from various N-terminal Cdc11 fragments (Figure S2B and unpublished data).
FIGURE 2:
Identification of Sid2-mediated Cdc11 phosphorylation sites. (A) Sid2-Myc13 was immunoprecipitated from cdc16-116 cells that had been shifted to 36ºC for 2.5 h and incubated with MBP-Cdc11-(1-660) (wild-type) or MBP-Cdc11-S5A. (B) Phosphotryptic peptides from the indicated Cdc11 proteins were separated in two dimensions with the anode on the left. The positions of major phosphopeptides are numbered. The asterisks indicate minor phosphopeptides. Equal counts of wild-type MBP-Cdc11-(1-660) or the S5A mutant were analyzed individually (left two panels) or mixed together (right panel) to prove the correct assignment of phosphorylation sites. (C) Sid2 phosphorylation sites contribute to Cdc11 mitotic phosphorylation. cdc11-GFP, cdc11-S5A-GFP, and cdc11-S5D-GFP strains were synchronized in G2 phase using temperature shift of cdc25-22 to 36ºC for 4 h, and samples were taken every 15 min after return to 25ºC. Cdc11 proteins were isolated from cell pellets by immunoprecipitation and detected by immunoblotting with anti-GFP antibody. Cell cycle progression was monitored by formation of septa. (D) Samples from the 60-min time point in (C) were run on the same gel to demonstrate comigration of the most rapidly migrating Cdc11 band. (E) An anti-GFP immunoprecipitate of wild-type Cdc11 or Cdc11-S5A isolated from a similar cdc25 block-and-release experiment at the indicated septation indices was treated (+) or not (−) with λ-phosphatase before immunoblotting.
Sid2 substrate specificity was defined previously as RXXS (Chen et al., 2008). Cdc11 contains nine RXXS motifs within its first 488 amino acids. To determine which of these was phosphorylated in vivo, we purified Cdc11 using both C-terminal and N-terminal TAP fusions and subjected it to two-dimensional liquid chromatography tandem mass spectrometry (2D-LC MS/MS) analysis. Phosphorylation of residues 121, 122, 264, 345, and 418, in addition to eight Cdk1 consensus (S/T)P sites, which we previously showed were not essential to Cdc11 function (Morrell et al., 2004), were detected multiple times (Table 1 and Supplemental Table S1). Mutating serines 121, 122, 264, 345, and 418 to alanines (S5A) greatly diminished the ability of Sid2-Mob1 to phosphorylate MBP-Cdc11-(1-660) in vitro (Figure 2A), and tryptic phosphopeptides 1–5 were eliminated (Figures 2B, middle panel, and S3A). From these data, we conclude that residues 121, 122, 264, 345, and 418 are the major Sid2 targets within Cdc11. Like Sid2-mediated phosphorylation of the Clp1 phosphatase (Chen et al., 2008), phosphorylation of Cdc11 by Sid2 created recognition sites for 14-3-3 protein binding in vitro (Figure S3B).
TABLE 1:
Phosphosites identified by MS analysis of Cdc11 purified from cells.
| Consensus kinase | Residue(s) | Number of phosphopeptides |
|---|---|---|
| 1. Cdk1: (S/T)P | S98 | 43 |
| S103 | 3 | |
| S136 | 5 | |
| S199 | 6 | |
| S208 | 7 | |
| S360 | 12 | |
| S393 | 3 | |
| S558 | 61 | |
| 2. Sid2: RXXS | S121,122 | 3 |
| S264 | 8 | |
| S301 | 3 | |
| S345 | 3 | |
| S418 | 5 |
The results are collated from six separate experiments. The phosphorylated residue(s) number is given, followed by the number of phosphopeptides identified containing the phosphorylated residue.
We next tested whether phosphorylation of Cdc11 by Sid2 affected its function. Plasmids expressing cdc11-S5A or the corresponding aspartic acid mutations (cdc11-S5D) rescued cdc11-123, cdc11-136, and cdc11::ura4+ cells (Figure S4, A and B, and unpublished data), indicating Sid2 phosphorylation is not essential for Cdc11 function. Each mutant was then integrated into the genome at the cdc11 locus by replacing cdc11::ura4+. In the following experiments, we used untagged and epitope-tagged cdc11 alleles so the phosphorylation state, localization, and function of the phosphomutants could be evaluated.
Cdc11 phosphorylation was detected previously as gel shifts that increased during mitosis (Krapp et al., 2003). The SDS–PAGE mobilities of Cdc11-GFP, Cdc11-S5A-GFP, and Cdc11-S5D-GFP were therefore examined during a synchronous cell cycle generated by arrest–release of the cdc25-22 mutant. As described previously (Krapp et al., 2003), Cdc11 migrated in at least two bands during interphase and became extensively modified during mitosis (Figure 2C). This is in accord with the > 13 phosphorylation sites on mitotic Cdc11 identified by 2D-LC MS/MS (Tables 1 and S1). In contrast with wild-type, Cdc11-S5A migrated throughout the cell cycle as a more discrete band in SDS–PAGE (Figure 2C) that comigrated with the faster-migrating bands of Cdc11 (Figure 2D), indicating that phosphorylation of Sid2 sites generates a significant fraction of modified Cdc11 during mitosis. Cdc11-S5D also did not display significant gel shifts compared with wild-type Cdc11 (Figure 2, C and D). Phosphatase treatment of Cdc11 immunoprecipitates confirmed that the slowly migrating forms of Cdc11 are due to phosphorylation (Figure 2E). Phosphatase treatment of Cdc11-S5A immunoprecipitates showed that phosphorylation still occurred on this mutant protein (Figure 2E), consistent with the presence of Cdk1 consensus phosphorylation sites identified by 2D-LC MS/MS (Tables 1 and S1).
Sid2-mediated Cdc11 phosphorylation affects Cdc7 localization and SIN signaling
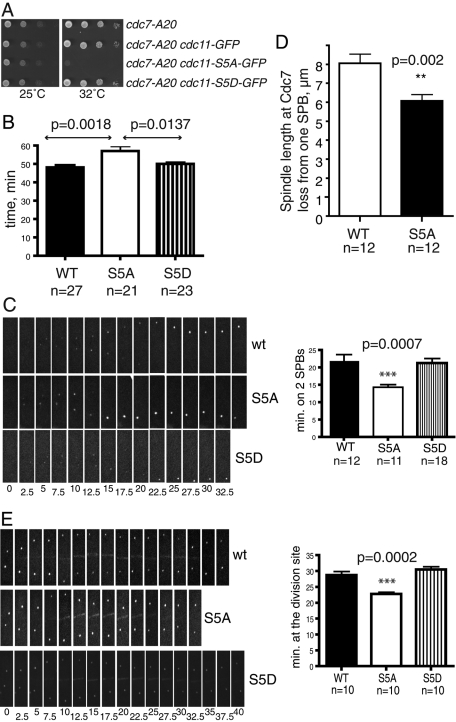
To assay whether Cdc11 phosphorylation at Sid2 sites affects cytokinesis, we first took a genetic approach. Although we did not detect synthetic genetic interactions with mutations in sid4, spg1, sid1, or sid2 (unpublished data), cdc11-S5A and cdc11-S5A-GFP were synthetically lethal with cdc7-24 and synthetically sick with cdc7-A20, whereas the cdc11-S5D mutation had no effect on the growth of these strains (Figures 3A and S4C, and unpublished data). These results indicate that loss of Cdc11 phosphorylation at Sid2 sites compromises Cdc11 function, and this is particularly deleterious if Cdc7 function is also reduced.
FIGURE 3:
Sid2 phosphorylation contributes to Cdc11 function. (A) The indicated strains were grown to mid-log phase in YE medium, spotted on YE plates in 10-fold serial dilutions, and incubated at the indicated temperatures. (B) sid4-RFP cdc15-GFP strains containing the indicated cdc11 alleles were analyzed by time-lapse microscopy. The average time for Cdc15 ring formation, constriction, and removal from the division site was measured for the indicated number of cells and analyzed by unpaired two-tailed t test. p value and SE are included. (C) Still images from representative movies of Cdc7-GFP in cdc11, cdc11-S5A, and cdc11-S5D cells. Time in minutes from the beginning of the movies is indicated. The difference in times of Cdc7-GFP proteins detected at 2 SPBs was analyzed by unpaired two-tailed t test. p value and SE are included. (D) The average length of the spindle when Cdc7 broke symmetry was measured for the indicated number of cells for each strain, and analyzed by unpaired two-tailed t test. p value and SE are included. (E) Still images from representative movies of Sid2-GFP in cdc11, cdc11-S5A, and cdc11-S5D cells. The difference in times of Sid2-GFP proteins detected at the cell division site was analyzed as in C. p value and SE are included.
To examine how cytokinesis was altered in cdc11-S5A cells, we used time-lapse microscopy to measure the period from the onset of contractile ring (CR) assembly to the completion of CR constriction in the cdc11 phosphomutant strains. CRs form within 1 min of SPB separation (Wu et al., 2003). Therefore we tracked SPBs with the Sid4–red fluorescent protein (RFP) marker (Chang and Gould, 2000), together with the CR component, Cdc15–green fluorescent protein (GFP) (Fankhauser et al., 1995). Whereas wild-type and cdc11-S5D cells took 48 and 50 min respectively from SPB separation and CR formation to clearance of Cdc15-GFP from the division site, cdc11-S5A cells took significantly longer, 57 min (Figure 3B). Furthermore, every step from CR formation through constriction was slower in each cell (unpublished data).
To explain the delayed and compromised cytokinesis, we examined whether the cdc11-S5A or cdc11-S5D mutations affected the subcellular distribution of SIN components. The Cdc11 phosphosite mutants themselves localized normally to the SPB throughout the cell cycle (Figure S4D). Similarly, there were no changes to the constitutive SPB localization of Sid4 or Spg1 (unpublished data). However, time-lapse imaging of Cdc7-GFP revealed a reproducible and statistically significant change in its distribution in cdc11-S5A cells. Whereas Cdc7-GFP remained at two SPBs for 22.0 ± 1.7 min in wild-type cells, it became asymmetric more rapidly in cdc11-S5A cells (14.4 ± 0.7 min; Figure 3C). Following initial symmetry breaking, Cdc7 became undetectable at one SPB when the spindle reached a length of 8 μm in wild-type cells (tracked using Sid4-RFP), whereas this occurred at a spindle length of 6 μm in cdc11-5A cells (Figure 3D). These results indicated that Cdc11 phosphorylation influenced the retention of Cdc7 at the old SPB. Time-lapse imaging of Sid2-GFP also revealed a difference in the persistence of Sid2-GFP at the division site in the cdc11-S5A mutant. In wild-type cells, Sid2-GFP remained an average of 28.75 ± 1.1 min, whereas Sid2-GFP was detected at the division site for only 22.75 ± 0.6 min in the cdc11-S5A mutant (Figure 3E). In the cdc11-S5D mutant, Sid2-GFP remained an average of 30.50 ± 0.9 min. Thus the phosphostatus of Cdc11 at the SPBs likely affects propagation of the SIN signal to the cell division site.
Cdc7 binds Cdc11 directly
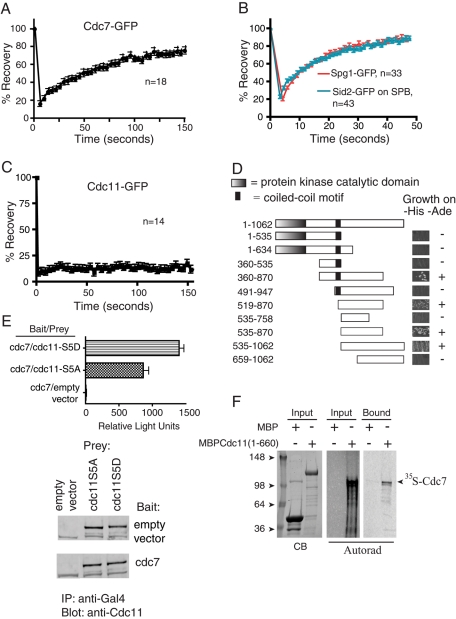
Cdc7 is recruited to SPBs primarily by activated GTP-bound Spg1 (Sohrmann et al., 1998). However, the observation that cdc7 overexpression rescued an spg1 null mutation suggested that Cdc7 might also interact with another SIN component at the SPB (Schmidt et al., 1997). We reasoned that if it did so, Cdc7 might display localization dynamics different than those of other SIN signaling components in fluorescence recovery after photobleaching (FRAP) experiments. Our previous FRAP studies had indicated that Spg1 and Sid2 had half-lives of recovery of less than 1 min, but the half-lives had not been measured precisely nor had we measured Cdc7-GFP dynamics at all (Morrell et al., 2004). We found that Cdc7-GFP had a significantly longer half-life of recovery during anaphase (67 ± 4.7 s) than either SPB-localized Sid2 (12.6 ± 0.7 s) or Spg1 (12.2 ± 0.9 s) (Figure 4, A and B). Cdc7 also had a significantly larger immobile fraction, a result that is consistent with the idea that Cdc7 interacts with a second SPB component. Given the above data showing that Cdc11 phosphostatus influenced Cdc7 dynamics, we wondered whether Cdc11 might be that component. Consistent with this possibility, we found that Cdc11-GFP is stably anchored at the SPB, because it does not recover appreciably after photobleaching (Figure 4C), as we had previously estimated (Morrell et al., 2004). Thus Cdc11 could represent a second SPB-interacting protein for Cdc7. Accordingly, directed two-hybrid analysis detected an association of the C-terminus of Cdc7 with the N-terminus of Cdc11 (Figure 4D). The interaction is likely sensitive to the phosphostatus of Cdc11, because the Cdc11-S5D mutant interacted more robustly with Cdc7 than did the Cdc11-S5A mutant (Figure 4E, top panel), although these bait proteins were produced at the same levels in cells (Figure 4E, bottom panel). MBP-Cdc11-(1-660) could also pull down Cdc7 produced in vitro (Figure 4F), suggesting that this association occurs directly. We therefore surmise that Cdc7-Cdc11 interaction contributes to the stabilization of Cdc7 SPB association and that this interaction is likely enhanced by Cdc11 phosphorylation.
FIGURE 4:
Cdc7 associates with Cdc11. (A–C) FRAP measurements of the indicated proteins are shown. Best-fit curves derived from the mean intensity values of n cells (Prism software) were used to calculate half-time of recovery. (D) Directed two-hybrid analysis between Cdc11-(1-660) and the indicated fragments of Cdc7. Growth on selective medium, which indicates a positive interaction, is shown and indicated on the right. (E) Top panel, ß-galactosidase activity (represented by relative light units) of strains with the indicated prey plasmids and bait plasmid Cdc7-(360-870). Bottom panel, immunoblot of Cdc11 mutant immunoprecipitations from yeast two-hybrid strains expressing the indicated plasmids as bait or prey. Abundances of Cdc11 proteins were determined by scanning with an Odyssey instrument and found to be equivalent. (F) Full-length [35S]-labeled Cdc7 was produced in a coupled transcription–translation system and incubated with MBP or MBP-Cdc11-(1-660) bound to amylose beads. [35S]-labeled Cdc7 bound to beads was detected by fluorography (right panel). Inputs into the reaction are shown in left and middle panels.
Sid2 contributes to SIN symmetry breaking
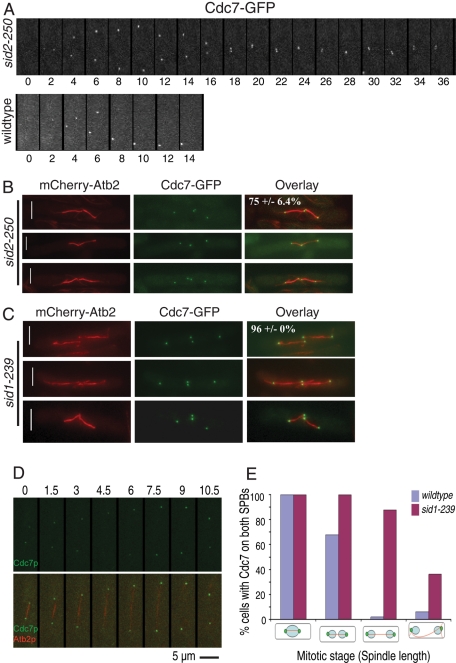
Given that Sid2-mediated phosphorylation of Cdc11 affected the behavior of Cdc7 at the SPB, we predicted that Cdc7 dynamics would be influenced by the loss of Sid2 activity. Thus Cdc7-GFP was imaged by time-lapse microscopy in sid2-250 mutant cells that had been synchronized in G2 prior to temperature shift and imaging. Unexpectedly, we found that Cdc7 SPB localization never became asymmetric as cells proceeded through anaphase, as it did in wild-type cells (Figure 5A, compare top and bottom panels). This was surprising, because previous studies reported that Cdc7-HA localizes asymmetrically in the sid1-239 and sid2-250 mutants (Sparks et al., 1999; Guertin et al., 2000). While we cannot explain the apparent discrepancy, it is possible that fixation led to some loss or masking of the HA epitope in previous studies. Cdc7 also did not become asymmetric in cdc11-S5A (n = 4) or cdc11-S5D (n = 11) mutants containing the sid2-250 mutation (unpublished data). Furthermore, symmetric Cdc7 localization was detected during the second anaphase in the sid2-250 mutant (Figure 5B). Because Sid2 activity depends upon Sid1 (Sparks et al., 1999), we tested whether Cdc7 became asymmetric in a sid1 mutant strain. As in the case with sid2-250 mutants, we found that Cdc7 remained symmetrically localized during anaphase in fixed sid1-239 cells arrested at the restrictive temperature (Figure 5C). Time-lapse imaging further confirmed that Cdc7 was either symmetric all through mitosis or the generation of Cdc7 asymmetry was delayed until mitotic spindle breakdown in sid1-239 cells (Figure 5, D and E). These results indicate that Sid1 and Sid2 are required for generating SIN asymmetry.
FIGURE 5:
Sid2 is required for SIN asymmetry. (A) Still images captured every 2 min from a movie of a representative cdc7-GFP sid2-250 cell (total cells examined = 10) held at 36ºC, or wild-type cells at 36ºC (total cells examined = 5). (B) sid2-250 cdc7-GFP mCherry-atb2 cells were shifted to 36ºC for 5 h and fixed with methanol. At least 50 anaphase cells in three separate experiments were examined to determine the indicated percentage of SPB pairs with Cdc7 at both SPBs. (C) sid1-239 cdc7-GFP mCherry-atb2 cells were shifted to 36ºC for 5 h and fixed with methanol. At least 50 anaphase cells in three separate experiments were examined to determine the indicated percentage of SPB pairs with Cdc7 at both SPBs. (D and E) Time-lapse imaging of Cdc7 and mCherry-α-tubulin in sid1-239 cells. sid1-239 cdc7-GFP mCherry-atb2 cells were shifted to 36ºC for 3 h and then still images were captured every 1.5 min at 36°C using a spinning-disk confocal microscope and the percentage of cells with asymmetric Cdc7 localization was quantified.
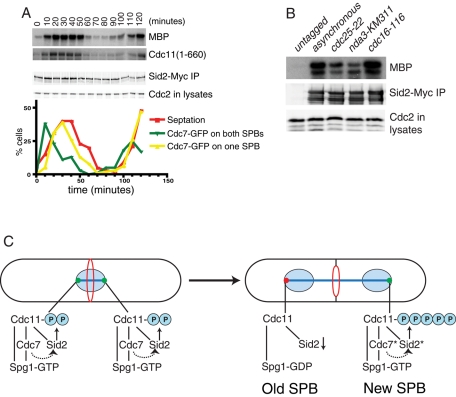
Sid2 activity peaks with SIN asymmetry
Although it has been established that Sid2 activity requires Cdc7 function (Sparks et al., 1999), it has not previously been tested how Sid2-Mob1 activity relates to the development of Cdc7 asymmetry, which is generally considered a reflection of SIN activation. Thus we monitored Sid2 kinase activity in synchronized cdc7-GFP cells. Sid2-Mob1 kinase activity was very low in interphase and spindle checkpoint–arrested cells (Figure 6, A and B), consistent with the previous observation that Cdc11 phosphorylation does not occur at these times (Krapp et al., 2003). Sid2 activity peaked with the development of asymmetric Cdc7 SPB localization as expected (Krapp et al., 2004b; Wolfe and Gould, 2005), but it was detected earlier (Figure 6A). This result is consistent with the finding that the SIN participates in contractile ring assembly prior to anaphase (Hachet and Simanis, 2008) and the possibility that Cdc11 begins to be phosphorylated by Sid2 before anaphase B ensues.
FIGURE 6:
Sid2 kinase activity correlates with Cdc7 asymmetry. (A) Cells were synchronized by centrifugal elutriation and samples were collected at the indicated intervals. Cell cycle progression was monitored by determining the septation index and the appearance of Cdc7-GFP at one or two SPBs. Sid2-Myc was also immunoprecipitated from samples collected at each time point and assayed for activity toward MBP-Cdc11-(1-660) and MBP. The amount of Sid2 in each kinase assay was determined by immunoblotting, and the amount of protein in each lysate was controlled by the amount of Cdc2 determined by immunoblotting with anti-PSTAIRE. (B) Sid2-Myc was immunoprecipitated from the indicated strains and assayed for activity toward MBP. The amount of Sid2 immunoprecipitated and the amount of protein in each lysate prior to immunoprecipitation was determined by immunoblotting. (C) Speculative model of Cdc11 phosphoregulation. The dotted arrow indicates the activation of Sid2 by Cdc7 through Sid1, which is not shown. Asterisks indicate more active forms of Cdc7 and Sid2 kinases.
DISCUSSION
Biological signaling pathways typically contain positive- and negative-feedback loops (Brandman and Meyer, 2008). In this paper, we have presented evidence that the ultimate protein kinase of the SIN pathway, Sid2, feeds back to control the function of upstream elements, the first demonstration of positive-feedback control in the SIN. We report that one target of Sid2-Mob1 in this feedback loop is the SIN scaffold component, Cdc11. Sid2-Mob1 phosphorylates Cdc11 on multiple sites to affect Cdc7 function and the robustness of SIN signaling. We also discovered that Sid2-Mob1 feeds back to control the establishment of Cdc7 asymmetry, which correlates with effective SIN signaling and timely SIN silencing (Sohrmann et al., 1998; Garcia-Cortes and McCollum, 2009; Singh et al., 2011).
Cdc7 is necessary for Sid2 activity (Sparks et al., 1999), and this dependency might explain why changes in Cdc11 SDS-gel mobility were observed previously in cdc7 mutants (Krapp et al., 2003). However, we cannot exclude the possibility that Cdc7 participates directly in Cdc11 phosphoregulation, although this was not detected previously (Krapp et al., 2003). Our evidence that Sid2 is responsible for a significant fraction of mitotic Cdc11 phosphorylation is consistent with the previous observation that SIN-dependent Cdc11 phosphorylation does not occur in spindle checkpoint–arrested cells (Krapp et al., 2003), a condition in which we have shown that Sid2 is inactive. Also, Cdc11 phosphorylation requires Sid4 function, evidence that it occurs at SPBs (Krapp et al., 2003), and we have shown here and previously that Sid2 forms a physical complex with Cdc11 (Morrell et al., 2004), which is constitutively SPB-localized (Krapp et al., 2001; Tomlin et al., 2002). We conclude that Sid2-Mob1 is active at the SPBs to phosphorylate Cdc11, as well as at the site of division.
On the basis of our characterization of SIN dynamics in cdc11 phosphomutants, we propose the following model for the role of Cdc11 phosphorylation in SIN signaling (Figure 6C). Spg1 and Cdc7 become activated at both SPBs during metaphase. This leads to Sid2 activity on both SPBs and phosphorylation of Cdc11. However, phosphatases, such as the recently described SIN-inhibitory complex (SIP) that promotes Cdc11 dephosphorylation and is present at SPBs at this time (Singh et al., 2011), limit the extent of Cdc11 phosphorylation. Upon anaphase initiation in wild-type cells, the limited amount of Cdc11 phosphorylation diminishes at the old SPB, due to increased phosphatase activity at that pole (which is enriched in SIP; Singh et al., 2011), and this promotes the recruitment of the Cdc16-Byr4 GAP complex for Spg1 (Furge et al., 1998) that binds preferentially to hypophosphorylated Cdc11 (Krapp et al., 2003). This then contributes to loss of Cdc7 from the old SPB. As more active Cdc7 accumulates at the new SPB, so do active Sid1 and Sid2, leading to maximal levels of SIN signaling. Increased Cdc11 phosphorylation resulting from increased Sid2 activity at the new SPB would promote the progressive accumulation of Cdc7 at the new SPB (Garcia-Cortes and McCollum, 2009). We propose that cdc11-5A cells break Cdc7 symmetry more rapidly than wild-type cells because Cdc11 phosphorylation is not present at all to help retain Cdc7 at the old SPB. The new SPB also cannot develop a robust positive-feedback loop in the absence of Cdc11 phosphorylation. As a result, cytokinesis takes longer in cdc11-5A cells and is sensitive to perturbation. Indeed, if Cdc7 function is further compromised by mutation, cytokinesis fails completely in cdc11-S5A cells. It may be that the SIN uses a “feedback-first” strategy to ensure switch-like activation and achieve a bistable state, as do other cell cycle signaling networks (reviewed in Ferrell, 2011). In contrast with cdc11-S5A, the cdc11-S5D mutant behaved similarly to wild-type cdc11, which suggests that it does not represent either the fully phosphorylated or dephosphorylated form of the protein. Indeed, it is worth noting that aspartates cannot provide docking sites for 14-3-3 proteins (Chen et al., 2008; Roberts-Galbraith et al., 2010) and Cdc11-S5D, therefore, cannot be fully mimicking the Sid2-phosphorylated state. Our model would predict that if Cdc11-S5D were fully phosphomimetic, the SIN would be activated constitutively.
We found that in addition to binding Spg1, Cdc7 directly associates with Cdc11. Furthermore, we have found that the Cdc11 phosphorylation state influences Cdc7 dynamics at the SPB. These findings are consistent with previous studies showing that overproduction of Cdc7 rescues the spg1-null mutant and that there is an SPB-targeting domain in Cdc7 independent of the Spg1-binding domain (Schmidt et al., 1997; Mehta and Gould, 2006). This dual strategy of Cdc7 recruitment appears to be conserved. The pathway analogous to the SIN in Saccharomyces cerevisiae is called the mitotic exit network, and it was recently found that two independent mechanisms also govern the recruitment of S. cerevisiae Cdc15, the orthologue of S. pombe Cdc7, to SPBs (Rock and Amon, 2011). One factor is the activation of Tem1, the homologue of Spg1, and the other factor is the protein kinase activity of Cdc5, the homologue of Plo1. Interestingly, it was speculated that Nud1, the S. cerevisiae homologue of Cdc11, might be the direct or indirect target of Cdc5's activity in promoting Cdc15 SPB association (Rock and Amon, 2011), as was reported previously for Cdc11 (Schmidt et al., 1997; Krapp et al., 2003, 2004a). Thus, it is likely that the mechanism we propose here for Cdc11 phosphoregulation is conserved.
How could phosphorylation of Cdc11 affect its function? It might be that phosphorylated Cdc11 adopts a different conformation than the dephosphorylated form. In this case, 14-3-3 protein binding would protect phosphates from phosphatase action and stabilize the phosphorylated conformation, which might interact better with other SIN components such as Cdc7. Structural studies will be required to test this possibility. Alternatively, 14-3-3 protein binding might be involved more directly in stabilizing Cdc11 protein–protein interactions and SIN signaling.
Several factors might explain how initial Sid2 activity, which we hypothesize begins equally at both SPBs during metaphase (Figure 6C), could become asymmetric. As mentioned above, Cdc11 dephosphorylation at the old SPB is likely to be a factor and result from the asymmetric action of one or more PP2A phosphatase complexes that have an impact on Cdc11 phosphostatus and SIN signaling (Le Goff et al., 2001; Krapp et al., 2003; Lahoz et al., 2010; Singh et al., 2011). Additionally, Cdk1 activity may be involved. In S. cerevisiae, Cdk1 inhibits S. cerevisiae Cdc15 (Cdc15 is analogous to S. pombe Cdc7) (Jaspersen and Morgan, 2000) and shows asymmetric localization to SPBs during anaphase (Konig et al., 2010). If the analogous mechanism operates in S. pombe, relief of Cdk1-mediated Cdc7 inhibition at the new SPB may contribute to initial asymmetric Sid2 activation, which would then promote a strong positive-amplification loop on that SPB (Figure 6C). This possibility is consistent with previous studies showing that Cdc7 asymmetry and maximum SIN activation depend upon Cdk1 inhibition (Guertin et al., 2000; Chang et al., 2001; Dischinger et al., 2008).
We have shown that Cdc7 asymmetry requires Sid2 function and that Cdc11 is a Sid2 target. However, Cdc7 becomes asymmetrically localized to SPBs in cdc11 phosphomutants, indicating that additional Sid2 targets must exist that contribute to the positive-feedback loop. Candidates for additional Sid2 substrates include Cdc7 itself; the Spg1 GAP component Byr4, which is a heavily phosphorylated protein (Song et al., 1996; Krapp et al., 2008; Johnson and Gould, 2011); the Polo kinase Plo1, which is essential for SIN signaling (Ohkura et al., 1995; Bahler et al., 1998a; Mulvihill et al., 1999); and one or more phosphatase complexes that influence the establishment of SIN asymmetry and SIN signaling (Le Goff et al., 2001; Krapp et al., 2003; Lahoz et al., 2010; Singh et al., 2011). Further study will be required to define the complete repertoire of Sid2 substrates influencing the establishment of asymmetric SIN signaling.
MATERIALS AND METHODS
Strains and media
S. pombe strains (Table S2) were grown in yeast extract (YE) medium or minimal medium with appropriate supplements (Moreno et al., 1991). Transformations were performed by the lithium acetate method or electroporation (Keeney and Boeke, 1994; Gietz et al., 1995). Epitope-tagged strains were constructed as described previously (Wach et al., 1994; Bahler et al., 1998b) so that open reading frames were tagged at the 3′ end of endogenous loci with the GFP-KanR or a TAP-KanR cassette. Appropriate tagging was confirmed by PCR and immunoblotting. Strain construction and tetrad analysis were accomplished through standard methods.
TAP and MS analysis
Proteins were purified by TAP as previously described (Tasto et al., 2001) and subjected to MS analysis as previously detailed (McDonald et al., 2002; Roberts-Galbraith et al., 2009).
In vitro kinase and phosphatase assays
The Sid2-Mob1 kinase complex was purified by immunoprecipitation with anti-Myc antibodies from sid2-Myc13 cdc16-116 cells that had been shifted to 36°C for 2.5–4 h. Kinase assays were performed as described (Sparks et al., 1999). Relative kinase activity was measured by determining the incorporation of 32P into MBP-Cdc11-(1-660) with a scintillation counter, dividing by the amount of MBP-Cdc11-(1-660), and then dividing by the amount of Sid2 present in the immunoprecipitates. Relative protein abundance was determined on an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Immunoprecipitated Cdc11-GFP variants were incubated in the presence of λ-phosphatase at 30°C for 30–45 min in phosphatase assay buffer (50 mM imidazole, pH 6.9, 1 mM EDTA, 1 mM dithiothreitol). Reactions were terminated by the addition of SDS sample buffer and boiling for 5 min. Proteins were resolved by SDS–PAGE and detected by immunoblotting.
Microscopy
GFP- or RFP-tagged proteins were visualized using an Ultraview LCI spinning-disk confocal microscope (Perkin Elmer-Cetus, Waltham, MA) equipped with a 100×/1.40 numerical aperture (NA) Plan-Apochromat oil immersion objective and a 488-nm argon ion laser (GFP) or a 594-nm helium neon laser (mCherry and RFP). For time-lapse images, cells were placed on a hanging-drop glass slide containing YE agar, covered with a coverslip, and sealed using Valap (a Vaseline, lanolin, and paraffin mixture). Images were captured on a charge-coupled device camera (Orca-ER; Hamamatsu Photonics, Hamamatsu City, Japan) and processed using MetaMorph 7.1 software (Molecular Devices, Sunnyvale, CA). Time-lapse images were obtained at an interval of 1.5, 2, or 2.5 min. For temperature-sensitive strains, cells were treated and imaged in two ways. First, cells were grown at 25ºC and separated by lactose gradient centrifugation. The smallest cells (representing those in G2 phase) were collected, resuspended in fresh YE, and shifted to 36ºC for 1.5 h prior to being mounted on a 36ºC heated objective. The objective temperature was controlled by a Bioptech Objective Heater (Butler, PA). Slides, coverslips, tips, and immersion oil used for the samples processing were prewarmed in a Boekel (Trevose, PA) incubator at 36ºC. Second, cells were grown at 25ºC and shifted to 36ºC. After 3 h, the cells were imaged using a microscopic setup equipped with a temperature-controlled chamber.
FRAP experiments were performed on a Zeiss LSM 510 confocal microscope equipped with a 63×/1.40 NA Plan-Apochromat oil immersion objective (Thornwood, NY). A 0.6-µm-diameter circle around the SPB was bleached with a sequence of 20 (Sid2-GFP, Spg1-GFP) or 15 (Cdc7-GFP) 100% intensity laser iterations, and images were then taken every 0.5 or 3 s, respectively. Fluorescence intensities were analyzed using Zeiss 510 LSM software, and intensity values were normalized against a second unbleached area of equal size to correct for overall bleaching. Normalized data were plotted using Prism (Irvine, CA). Mobile fractions and half-time of recovery values (t1/2) were calculated from the best-fit curve equation as the difference in final and bleach intensity and the time to reach half of the intensity after photobleaching, respectively. Student's t test and SE were calculated to determine significant differences. Differences reported as significant have p values less than 0.05 and nonoverlapping SE intervals.
sid2-250 cdc7-GFP mCherry-atb2 cells were shifted to 36ºC for 3 or 5 h. They were then fixed for 6 min with 100% methanol (prechilled at −20ºC) and observed with an Olympus (Center Valley, PA) IX71 microscope equipped with a Photometrics (Tucson, AZ) CoolSNAP ES camera.
Immunoprecipitations and immunoblotting
Cell pellets were frozen in a dry ice/ethanol bath and lysed by bead disruption in HEN buffer with inhibitors (Krapp et al., 2001). Proteins were immunoprecipitated from various amounts of protein lysates using anti-GFP (Roche, Indianapolis, IN) or anti-Myc (9E10) followed by Protein G Sepharose beads (GE Healthcare, Waukesha, WI).
For immunoblotting, proteins were resolved by 8% or 10% SDS–PAGE, transferred by electroblotting to a polyvinylidene difluoride membrane (Immobilon P; Millipore, Bedford, MA) and incubated with the set of primary antibodies indicated at 1 μg/ml. Primary antibodies were detected with secondary antibodies coupled to Alexa Fluor 680 (Invitrogen, Carlsbad, CA) or IRDye800 (LI-COR) and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Phosphoamino acid analysis and tryptic peptide mapping
[32P]-labeled proteins were subjected to partial acid hydrolysis and tryptic digestion while bound to polyvinylidene fluoride membrane and analyzed as described in McCollum et al. (1999) and references therein.
Expression of labeled and recombinant fusion proteins
MBP, MBP-fusion proteins, GST, and GST-fusion proteins were produced in BL21 bacterial cells and purified from bacterial lysates using amylose beads (New England Biolabs, Ipswich, MA) or GST bind resin (Novagen, Billerica, MA), as specified by the manufacturers. Proteins were also produced in vitro in the presence of [35S]methionine (Trans-label; ICN Pharmaceuticals, Irvine, CA) via the TNT Coupled Reticulocyte lysate system (Promega, Madison, WI). Purified MBP fusion proteins bound to amylose resin were mixed with [35S]-labeled proteins in binding buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 2 mM EDTA, 0.1% bovine serum albumin, and 0.1% NP-40) and incubated at 4ºC for 1 h. The beads were washed once with 100 μl binding buffer before being resolved by SDS–PAGE.
Yeast two-hybrid analyses
Yeast two-hybrid assays were done as described using S. cerevisiae strain PJ69-4A and the pGBT9 and pGAD vectors (James et al., 1996). ß-Galactosidase reporter enzyme activity in the two-hybrid strains was measured with the Galacto-Star chemiluminescent reporter assay system according to the manufacturer's instructions (Tropix, Bedford, MA), with the exception that cells were lysed by glass bead disruption. Each sample was measured in triplicate. Results were normalized by subtracting the activity associated with the empty prey vector.
Molecular biology techniques
All plasmid constructions were performed by standard molecular biology techniques. All DNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). All sequences were PCR-amplified with TaqPlus Precision (Stratagene, Santa Clara, CA) according to the manufacturer's protocol. Site-directed mutagenesis was carried out using Quickchange (Stratagene) according to the manufacturer's protocols.
Supplementary Material
Acknowledgments
We are grateful to Alyssa Johnson for comments on the manuscript and to Sam Wells and Charles Day for technical advice on FRAP experiments. FRAP experiments were performed through the use of the VUMC Cell Imaging Shared Resource (supported by National Institutes of Health grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126). This work was supported by the Singapore Millennium Foundation and the Howard Hughes Medical Institute, of which K.L.G. is an investigator.
Abbreviations used:
- 2D-LC MS/MS
two-dimensional liquid chromatography tandem mass spectrometry
- CR
contractile ring
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- HA
hemagglutinin
- MBP
myelin basic protein
- NA
numerical aperture
- RFP
red fluorescent protein
- SIN
septation initiation network
- SIP
SIN-inhibitory complex
- SPB
spindle pole body
- TAP
tandem affinity purification
- YE
yeast extract
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0792) on March 14, 2012.
REFERENCES
- Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998a;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998b;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Morrell JL, Feoktistova A, Gould KL. Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol Cell Biol. 2001;21:6681–6694. doi: 10.1128/MCB.21.19.6681-6694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V. Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J Cell Sci. 2008;121:843–853. doi: 10.1242/jcs.021584. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F- actin at mitosis [correction published in Cell (1997) 89, 1185] Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Simple rules for complex processes: new lessons from the budding yeast cell cycle. Mol Cell. 2011;43:497–500. doi: 10.1016/j.molcel.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Cortes JC, McCollum D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J Cell Biol. 2009;186:739–753. doi: 10.1083/jcb.200902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 2004;18:1007–1021. doi: 10.1101/gad.296204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J. 2011;30:341–354. doi: 10.1038/emboj.2010.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig C, Maekawa H, Schiebel E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol. 2010;188:351–368. doi: 10.1083/jcb.200911128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Cano E, Simanis V. Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr Biol. 2003;13:168–172. doi: 10.1016/s0960-9822(02)01417-3. [DOI] [PubMed] [Google Scholar]
- Krapp A, Cano E, Simanis V. Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett. 2004a;565:176–180. doi: 10.1016/j.febslet.2004.03.098. [DOI] [PubMed] [Google Scholar]
- Krapp A, Collin P, Cano Del Rosario E, Simanis V. Homeostasis between the GTPase Spg1p and its GAP in the regulation of cytokinesis in S. pombe. J Cell Sci. 2008;121:601–608. doi: 10.1242/jcs.022772. [DOI] [PubMed] [Google Scholar]
- Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004b;14:R722–730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Krapp A, Schmidt S, Cano E, Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Lahoz A, Alcaide-Gavilan M, Daga RR, Jimenez J. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics. 2010;186:1261–1270. doi: 10.1534/genetics.110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X, Buvelot S, Salimova E, Guerry F, Schmidt S, Cueille N, Cano E, Simanis V. The protein phosphatase 2A B′-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 2001;508:136–142. doi: 10.1016/s0014-5793(01)03047-2. [DOI] [PubMed] [Google Scholar]
- Li C, Furge KA, Cheng QC, Albright CF. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J Biol Chem. 2000;275:14381–14387. doi: 10.1074/jbc.275.19.14381. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Gould KL. Phosphorylation of the myosin-II light chain does not regulate the timing of cytokinesis in fission yeast. J Biol Chem. 1999;274:17691–17695. doi: 10.1074/jbc.274.25.17691. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR., III Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- Mehta S, Gould KL. Identification of functional domains within the septation initiation network kinase, Cdc7. J Biol Chem. 2006;281:9935–9941. doi: 10.1074/jbc.M600160200. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell JL, et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr Biol. 2004;14:579–584. doi: 10.1016/j.cub.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol Biol Cell. 1999;10:2771–2785. doi: 10.1091/mbc.10.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, III, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Amon A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011;25:1943–1954. doi: 10.1101/gad.17257711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Singh NS, et al. SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr Biol. 2011;21:1968–1978. doi: 10.1016/j.cub.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Mach KE, Chen CY, Reynolds T, Albright CF. A novel suppressor of ras1 in fission yeast, byr4, is a dosage- dependent inhibitor of cytokinesis. J Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, Hayes McDonald W, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Tomlin GC, Morrell JL, Gould KL. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol Biol Cell. 2002;13:1203–1214. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wolfe BA, Gould KL. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.