The exon junction complex (EJC) allows the spliceosome to communicate with other cellular machinery. This study shows that assembled EJC cores are enriched in nuclear regions around speckles, called perispeckles. Speckles and perispeckles may represent specialized nuclear regions for messenger ribonucleoprotein maturation.
Abstract
The exon junction complex (EJC) is loaded onto mRNAs as a consequence of splicing and regulates multiple posttranscriptional events. MLN51, Magoh, Y14, and eIF4A3 form a highly stable EJC core, but where this tetrameric complex is assembled in the cell remains unclear. Here we show that EJC factors are enriched in domains that we term perispeckles and are visible as doughnuts around nuclear speckles. Fluorescence resonance energy transfer analyses and EJC assembly mutants show that perispeckles do not store free subunits, but instead are enriched for assembled cores. At the ultrastructural level, perispeckles are distinct from interchromatin granule clusters that may function as storage sites for splicing factors and intermingle with perichromatin fibrils, where nascent RNAs and active RNA Pol II are present. These results support a model in which perispeckles are major assembly sites for the tetrameric EJC core. This subnuclear territory thus represents an intermediate region important for mRNA maturation, between transcription sites and splicing factor reservoirs and assembly sites.
INTRODUCTION
The exon junction complex (EJC) is a multiprotein complex loaded onto mRNA as a consequence of splicing (Le Hir et al., 2000; Tange et al., 2004). Placed 20–24 nucleotides upstream of the splice junction, the EJC communicates splicing history to downstream events. Structurally, this RNA-binding complex is composed of more than a dozen proteins that have been classified into two groups: EJC “core” and “peripheral” factors (Tange et al., 2005; Le Hir and Seraphin, 2008). Four proteins form the EJC core: the constitutive heterodimer Magoh/Y14, the DEAD-box RNA helicase eIF4A3 (eukaryotic initiation factor 4AIII), and MLN51 (metastatic lymph node 51), also known as CASC3 and Barentsz (BTZ; Ballut et al., 2005; Tange et al., 2005). EJC cores accompany mRNAs to the cytoplasm, where the translation machinery removes them from open reading frames (Le Hir et al., 2001b; Dostie and Dreyfuss, 2002; Lejeune et al., 2002; Gehring et al., 2009b); they constitute binding platforms for peripheral factors that dynamically associate and dissociate during mRNA travel (Le Hir and Andersen, 2008; Buchwald et al., 2010; Bono and Gehring, 2011). Several functions are attributed to this complex, in both the nucleus and the cytoplasm. These include pre-mRNA splicing (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010), mRNA transport (reviewed in Giorgi and Moore, 2007, and Martin and Ephrussi, 2009), translation activation (Nott et al., 2004; Tange et al., 2005; Diem et al., 2007; Le Hir and Seraphin, 2008; Ma et al., 2008), and mRNA degradation via nonsense-mediated mRNA decay (NMD; reviewed in Isken and Maquat, 2008; Shyu et al., 2008; Stalder and Muhlemann, 2008; Rebbapragada and Lykke-Andersen, 2009).
The EJC core is not a preassembled complex, and core components are brought together by the splicing machinery. In vitro experiments have shown that the proteins eIF4A3, Magoh, and Y14 are recruited by the active spliceosome before exon ligation and form a precore complex on the mRNA (Bessonov et al., 2008; Reichert et al., 2002; Gehring et al., 2009a). In a second step that rapidly follows exon ligation, MLN51 joins this complex to generate the stable EJC core (Degot et al., 2004; Gehring et al., 2009a). The reconstitution and the crystal structure of the EJC core revealed the original mechanism by which it is stably bound to RNA in a sequence-independent manner (Ballut et al., 2005; Andersen et al., 2006; Bono et al., 2006). In the presence of ATP, the two conserved domains of eIF4A3 adopt a close conformation around the RNA, and this conformation is locked in by the Magoh/Y14 heterodimer (Figure 1A). The most conserved domain of MLN51, named SELOR, folds around eIF4A3 and also contributes to RNA binding. Of importance, all of these interactions are required to ensure the remarkably high stability of the complex (Figure 1A; Andersen et al., 2006; Bono et al., 2006). In vivo, the four components of the EJC core shuttle between the nucleus and the cytoplasm, but Magoh, Y14, and eIF4A3 are mainly nuclear, whereas MLN51 is predominantly cytoplasmic (Degot et al., 2002; Ferraiuolo et al., 2004; Kataoka et al., 2000; Le Hir et al., 2001a; Palacios et al., 2004; van Eeden et al., 2001). In this study, we used a combination of microscopy approaches to better understand how and where the EJC core is assembled in intact cells.
FIGURE 1:
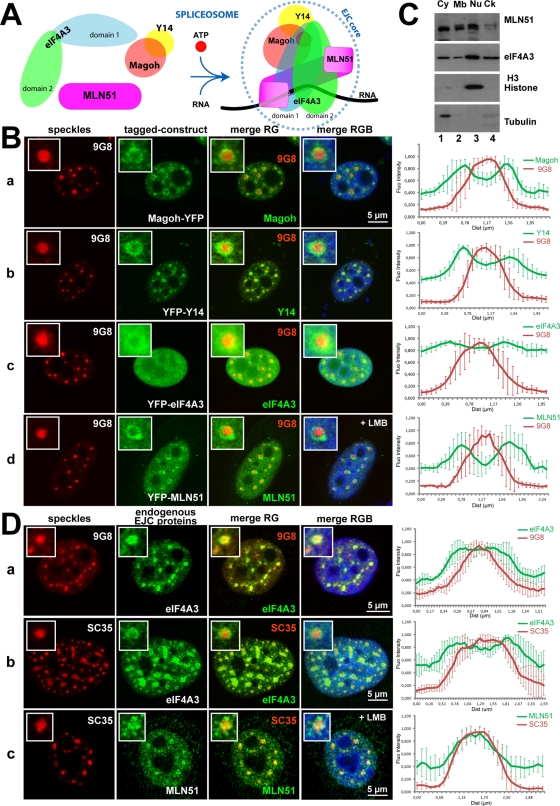
The EJC core factors are enriched in perispeckles. (A) Schematic representation of the EJC core assembly. Except for the constitutive Magoh/Y14 heterodimer, the four factors are not preassembled. As a consequence of splicing and in the presence of ATP and RNA, the EJC core is formed and stably anchored onto mRNA, mainly through direct contacts between eIF4A3 and MLN51 with RNA. (B) The core EJC factors are predominantly enriched at the periphery of nuclear speckles. The YFP-tagged EJC core constructs, Magoh (a), Y14 (b), eIF4A3 (c), and MLN51 (d), were transfected into HeLa cells, and the endogenous SR protein 9G8 was detected by immunofluorescence (red). Cells in d were treated with 20 ng/ml LMB for 5 h and then processed for immunofluorescence. Enlargements of selected speckles are shown with 3× magnification. (C) The core EJC factor MLN51 is also present in the nuclear fraction. Protein lysates from MCF7 cells were fractionated on ice into four collections, cytoplasmic (Cy), membrane-associated (Mb), nuclear (Nu), and cytoskeleton-containing (Ck) fractions, and then subjected to SDS–PAGE immunoblotting with specific antibodies against the core EJC factors MLN51 and eIF4A3. Anti–histone H3 and anti–α-tubulin were used for testing the purity of the nuclear and cytoplasmic fractions, respectively. Note that the absence of tubulin in the Ck fraction is due to the use of cold buffer, which dissociates microtubules. (D) Endogenous EJC proteins eIF4A3 and MLN51 are enriched in perispeckles. HeLa (a) and Sk-Br-3 (b) cells were costained with both anti-eIF4A3 (green) and anti-9G8 (red) antibodies (a) or with both anti-eIF4A3 (green) and anti-SC35 (red) antibodies (b). For MLN51 analysis, HeLa cells were incubated with LMB (20 ng/ml for 5 h) and then processed for immunofluorescence using anti-MLN51 (green) and anti-SC35 (red) antibodies (c). Insets show 4 and 2.5× magnifications in a–c. For immunofluorescence analysis (B, D), nuclei were counterstained with the Hoechst-33258 dye (blue; merge). Right, Mean fluorescence intensity line scans of the images, across five representative speckles, as described in Materials and Methods. Scale bar, 5 μm.
RESULTS
The EJC core factors are enriched at the periphery of nuclear speckles
With the exception of the proteins Magoh and Y14, which constitute a stable heterodimer (Fribourg et al., 2003), free eIF4A3, MLN51, and Magoh/Y14 proteins do not interact together in a preassembled EJC core (Figure 1A). Instead, the EJC core has an original mode of assembly that is governed by the spliceosome (reviewed in Bono and Gehring, 2011). To have a better view of the localization of EJC core and peripheral factors, we systematically tagged them with a fluorescent protein. When transfected into HeLa cells, the yellow fluorescent protein (YFP)–tagged EJC core factors Magoh and Y14 showed similar localization, with a characteristic doughnut-shaped accumulation around well-defined spherical nuclear speckles (Figure 1B, a and b).The YFP-tagged eIF4A3 (Figure 1B, c) was more diffusely distributed in the nucleus, with a weaker but significant enrichment around speckles. Nuclear speckles, also named “SC35 domains” or “splicing factor compartments,” are nuclear punctuate structures functioning as storage/assembly/modification compartments that supply splicing factors to active transcription sites (Hall et al., 2006; Lamond and Spector, 2003; Spector and Lamond, 2011). In cells, these structures can be traced using splicing factor labeling, including SC35/SRSF2 (serine/arginine–rich splicing factor 2) or 9G8/SRSF7 (serine/arginine–rich splicing factor 7; Carmo-Fonseca et al., 1991; Spector et al., 1991). We then looked at the localization of endogenous eIF4A3 and MLN51 in isolation and in untransfected cells. Consistent with previous reports, in both tested cell lines (HeLa and Sk-Br-3), speckles (traced here with 9G8 and SC35) have variable sizes and irregular shapes (Hall et al., 2006; Spector and Lamond, 2011). Endogenous eIF4A3 was found throughout 9G8- or SC35-labeled speckles, with enrichment at their periphery (Figure 1D, a and b). As mentioned before, MLN51 is a shuttling protein that is predominantly cytoplasmic (Degot et al., 2002; Macchi et al., 2003). Cell fractionation experiments confirmed that MLN51 is predominantly present in the cytoplasm but also showed that significant amounts are detected in the nucleus (Figure 1C). At steady state, our antibody was not sensitive enough to reliably detect MLN51 in the nucleus, and we thus studied its localization under crm-1 nuclear export blocking conditions (leptomycin B [LMB] treatment). After 5 h of treatment, MLN51 was enriched in the nucleus in all cells and gave a diffuse nucleoplasmic signal. In some cells, MLN51 accumulated at the periphery of SC35-positive speckles (Figure 1D, c). This labeling corresponded to MLN51 because, under LMB, a YFP-MLN51 fusion also localized at the periphery of nuclear speckles (Figure 1B, d).
To rule out the possibility that the signals observed using the YFP-tagged EJC core factors constructs are a consequence of overexpression, low-expression vectors were designed and used for localization experiments (Varnai et al., 2007). Using these plasmids, we showed that the tagged EJC proteins were expressed at levels close to that of the endogenous proteins and that they localized in doughnut-shaped regions around nuclear speckles (Supplemental Figure S1). Therefore we can conclude that the nuclear region around nuclear speckle is not a storage site for overexpressed proteins.
Collectively, these localization experiments show that the four EJC core factors are localized in and around speckles; they are enriched in doughnut-shaped regions at the periphery of speckles. This territory resembles the “perispeckle” region, previously described as a nuclear subdomain where Y14 and the NXF1 export factor were found to interact in a bimolecular fluorescence complementation assay (Schmidt et al., 2006). Therefore we will use the term “perispeckle” hereafter to describe this region of the nucleus.
Peripheral EJC factors, including the apoptosis and splicing-associated protein complex, are predominantly enriched in perispeckles
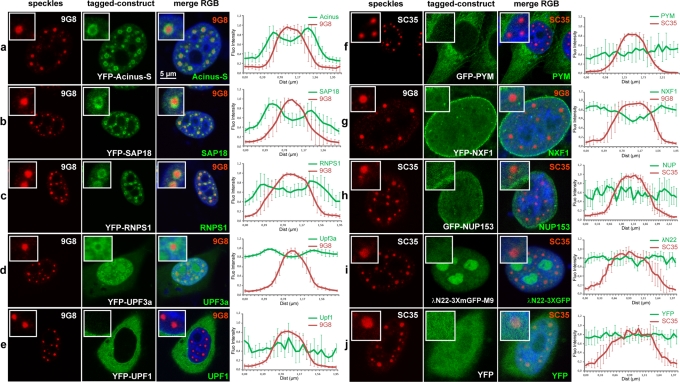
We next looked at the localization of two different classes of EJC peripheral factors (Figure 2). Factors of the first class are believed to bind the EJC core in the nucleus and include the splicing factors Acinus-S, SAP18, and RNPS1, which together constitute the nuclear trimeric apoptosis and splicing-associated protein (ASAP) complex (Schwerk et al., 2003; Tange et al., 2005), the export factor NXF1 (Huang et al., 2003; Luo et al., 2001; Schmidt et al., 2006), and the NMD factor Upf3b (Lykke-Andersen et al., 2000). The ASAP proteins, present in active spliceosome (Bessonov et al., 2008), are expected to associate with the EJC core very early after mRNA synthesis (Tange et al., 2005), whereas NXF1 and Upf3b are supposed to be recruited early after mRNA release from the spliceosome (Le Hir et al., 2001b). Of interest, the three ASAP proteins transiently expressed showed an accumulation in perispeckles as factors of the EJC core (Figure 2, a–c). Upf3a presented a more diffuse nuclear localization but was also enriched in perispeckles (Figure 2d). In contrast, the export factor NXF1/TAP was predominantly localized at the nuclear envelope (Figure 2g). The second class of EJC peripheral factors includes factors important for EJC functions that are not supposed to associate with the EJC in the nucleus but rather in the cytoplasm: the NMD factor UPF1, which associates with the UPF complex (Behm-Ansmant and Izaurralde, 2006), and the EJC disassembly factor PYM, which interacts with the heterodimer Magoh/Y14 (Diem et al., 2007). Consistently, both UPF1 and PYM were mainly cytoplasmic (Figure 2, e and f). In addition, nuclear export inhibition was not sufficient to enrich UPF1 and PYM at the perispeckle region (Supplemental Figure S2). In parallel experiments, the localization of additional fluorescent protein–tagged control constructs was undertaken. The transfection of the empty YFP vector results in a diffuse nuclear and cytoplasmic distribution of the YFP protein (Figure 2j). The nuclear pore complex protein NUP153 tagged with green fluorescent protein (GFP) is enriched at the nuclear envelope, without signal enhancement in the speckle and perispeckle regions (Figure 2h). In agreement with previous observations (Daigle and Ellenberg, 2007), the λN22-3XGFP-M9 probe remained in the nucleus, with some nucleolar enrichment (Figure 2i). Taken together, these data show that peripheral EJC factors known to associate with the core in the nucleus, especially factors from the ASAP complex, are in and out speckles with an enrichment in perispeckles.
FIGURE 2:
A subset of nuclear peripheral EJC factors accumulates in perispeckles. HeLa cells were transfected with various constructs encoding fluorescence-tagged fusion proteins (green) and colabeled with the endogenous SR protein 9G8 (a–e, g) or SC35 (f, h–j; red). Peripheral EJC factors that are known to associate either in the nucleus (Acinus-S, SAP18, RNPS1, Upf3a, NXF1) or in the cytoplasm (Upf1, PYM) were analyzed. Different constructs were also used as negative controls and include the YFP protein alone, the nucleoporin NUP153, and the reporter gene λN22-3XmGFP-M9, which is specifically targeted to nucleoli. Nuclei were counterstained using the Hoechst-33258 dye (blue). Insets are centered on a representative speckle and show 3× (b–e, i), 2.5× (a, f, h, j), and 2× (g) magnifications. Right, mean fluorescence intensity line scans of the images, using line sections on five selected speckles, as described. The factors constituting the ASAP complex, Acinus-S, RNPS1, and SAP18 proteins, are enriched in perispeckles (a–c), as well as in the NMD factor Upf3a, which is partially enriched in this area (d). In contrast, the cytoplasmic NMD factor Upf1 and the EJC disassembly protein PYM are mainly in the cytoplasmic compartment (e, f). Finally, the nuclear export factor TAP (NXF1) is both diffuse in the nucleoplasm and associated with the nuclear envelope (g). Scale bar, 5 μm.
EJC assembly–deficient mutants are not enriched in perispeckles
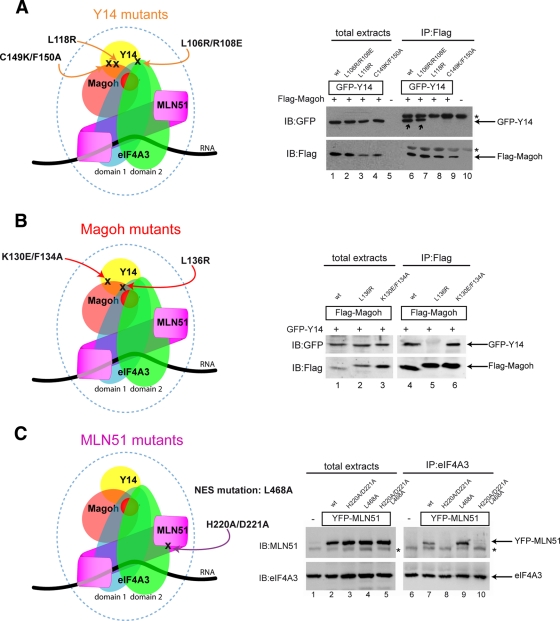
Because the EJC is dynamically assembled in the nucleus, the enrichment of its components in perispeckles could correspond to either storage sites for unassembled factors or sites where assembled EJC would transiently accumulate, for instance, if splicing and EJC assembly occur there. To distinguish between these possibilities, we analyzed the localization of several EJC assembly–defective mutants. For this purpose, we generated mutants for Y14, Magoh, and MLN51 proteins that are unable to integrate the EJC complex in vitro or in cell extracts (EJC-defective) and/or to trigger NMD using tethering experiments (NMD-defective; Fribourg et al., 2003; Ballut et al., 2005; Gehring et al., 2005). Three Y14 mutants unable to form the EJC core were tested: the Y14-C149K/F150A and Y14-L118R mutants, which cannot interact with Magoh, and the Y14-L106R/R108E mutant, which can interact with Magoh but not with eIF4A3 (Figure 3A; Fribourg et al., 2003; Gehring et al., 2005; Andersen et al., 2006; Bono et al., 2006). Similarly, two Magoh mutants were analyzed: the L136R point mutant, which cannot bind Y14, and K130E/F134A, which can bind Y14 but not eIF4A3 (Fribourg et al., 2003; Gehring et al., 2005; Figure 3B). With regard to MLN51 (Figure 3C), two key motifs were chosen for mutagenesis: the SELOR module with the H220A/D221A mutation, which abrogates its interaction with eIF4A3, thus preventing EJC assembly (Ballut et al., 2005), and the NES consensus region with the L468A mutation, which prevents nuclear export (Macchi et al., 2003). First, we verified the binding specificities of the mutants with respect to the core factor interactions by coimmunoprecipitation in HeLa cells. Consistent with previous report (Gehring et al., 2005), Flag-Magoh coprecipitated wild-type (wt) YFP-Y14 and YFP-Y14-L106R/R108E but not YFP-L118R and YFP-Y14-C149K/F150A mutants (Figure 3A, compare lanes 6 and 7 to lanes 8 and 9). Similarly, GFP-Y14 was coimmunoprecipitated with wt Flag-Magoh and Flag-Magoh K130E/F134A but not with Flag-Magoh L136R (Gehring et al., 2005; Figure 3B, lanes 4–6). Finally, endogenous eIF4A3 coprecipitated wt YFP-MLN51 and the NES mutant YFP-MLN51 L468A but not the YFP-MLN51-H220A/D221A mutant (Ballut et al., 2005; Gehring et al., 2005) and not the double mutant YFP-MLN51-H220A/D221A/L468A (Figure 3C, lanes 7–10).
FIGURE 3:
Mutants EJC factors display specific binding requirements with their core EJC partners. EJC-defective mutants Y14, Magoh, and MLN51 were generated through directed mutagenesis and were characterized by coimmunoprecipitation. (A) Left, positions of the Y14 mutations in the context of the core EJC; right, binding properties of Y14 mutants. HeLa cells were cotransfected with Flag-Magoh and GFP-Y14 wild type (wt), GFP-Y14 L106R/R108E, GFP-Y14 L118R, or GFP-Y14 C149K/F150A, as indicated. Cells extracts were subjected to immunoprecipitation with anti-Flag antibody. Immunoprecipitated complexes were immunoblotted with anti-GFP (top) and anti-Flag (bottom) antibodies. Expression of both Flag- and GFP-tagged constructs was confirmed by analyzing whole total extracts. Asterisk indicates immunoglobulin G chains. Note that only Y14 wt and Y14 L106R/R108E (lanes 6 and 7) coprecipitate with Magoh (black arrows). (B) Left, positions of the Magoh mutations in the context of the core EJC; right, binding properties of Magoh mutants. HeLa cells were cotransfected with GFP-Y14 and Flag-Magoh wt, Flag-Magoh L136R, or Flag-Magoh K130E/F134A, as indicated. Proteins were immunopurified using anti-Flag antibody and then immunoblotted with anti-GFP (top) and anti-Flag (bottom) antibodies. Efficiencies of the coexpression were analyzed on total extracts. Note that Magoh wt and Magoh K130E/F134A interact with Y14 (lanes 4 and 6). (C) Left, position of the MLN51 mutation in the context of the core EJC (H220A/D221A) with or without the NES motif mutation; right, binding properties of MLN51 mutants. HeLa cells were transfected with YFP-MLN51 wt, YFP-MLN51 H220A/D221A, YFP-MLN51 L468A, or YFP-MLN51 H220A/D221A/L468A, as indicated. Endogenous eIF4A3 was immunoprecipitated using a specific anti-eIF4A3 antibody, and immunoprecipitated complexes were then immunoblotted with anti-MLN51 (top) and anti-eIF4A3 (bottom) antibodies. Transfection efficacy was assessed by immunoblotting with anti-MLN51 antibody on total extracts. Asterisk indicates the expression of the endogenous MLN51 protein. Note that MLN51 wt is coprecipitated with eIF4A3 and that mutation in the NES motif L468A enhances binding between MLN51 and eIF4A3 (lanes 7 and 9).
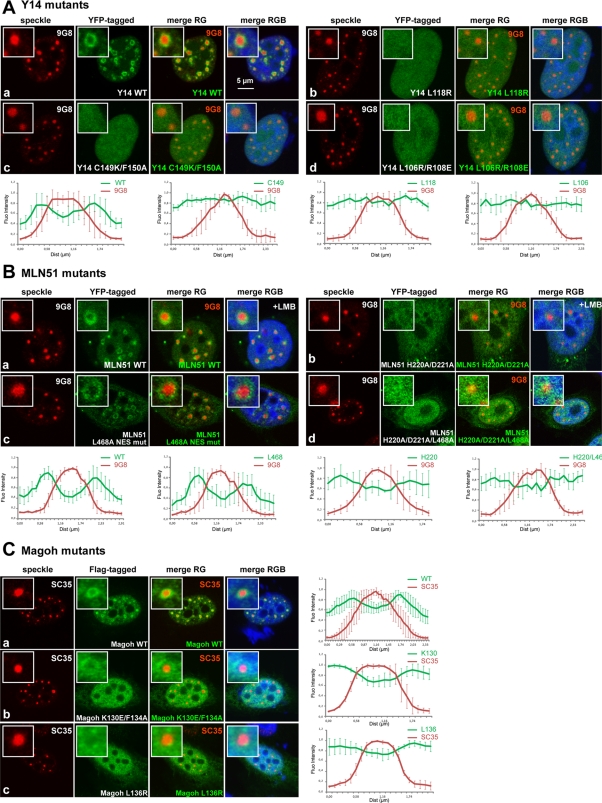
Next we looked at the localization of these mutants in HeLa cells. Although the wt YFP-Y14 protein was concentrated in perispeckles (Figure 4A, a), all EJC assembly–defective Y14 mutants were diffusely distributed in the nucleus (Figure 4A, b–d). Similar results were observed with EJC assembly–defective Magoh mutants (Figure 4C, b and c) and with the EJC-defective MLN51 mutants bearing the mutation in the SELOR module YFP-MLN51-H220A/D221A and YFP-MLN51-H220A/D221A/L468A (Figure 4B, b and d). Thus, all the EJC assembly–defective mutants tested were distributed throughout the nucleoplasm, including speckles, and not significantly enriched in perispeckles.
FIGURE 4:
EJC-defective mutants fail to localize at the perispeckle region. (A, B) HeLa cells were transfected with wt and mutant YFP-tagged Y14 (A) and MLN51 (B) constructs (green), and the endogenous SR protein 9G8 was detected by immunofluorescence (red). LMB treatment (20 ng/ml for 5 h) was done in the context of MLN51 wt and H220A/D221A mutant (B, a, b). Both EJC-defective Y14 mutants (A, b–d) and MLN51 mutants (B, b, d) are diffuse in the nucleoplasm. (C) HeLa cells were transfected with wt and mutant Flag-tagged Magoh constructs and then stained using anti-Flag (green) and anti-SC35 (red). Only Magoh wt is enriched at the perispeckle. For immunofluorescence analysis, nuclei were counterstained using the Hoechst-33258 dye (blue; merge). (A–C) Enlargements of selected speckles are shown with 3× magnification, and line-scan analyses (right) were performed as previously described. Scale bar, 5 μm.
To confirm the localization of MLN51 in perispeckles without using LMB, we looked at the distribution of the YFP-MLN51-L468A mutant, which lacks a functional NES signal. Of interest, this construct was significantly enriched at the perispeckle territory (Figure 4B, c). In addition, as mentioned, simultaneous mutations of the NES and SELOR motifs resulted in a diffuse nuclear localization of the protein, with no accumulation at the perispeckle (Figure 4B, d). These data indicate that MLN51 transits in the nucleus and that its enrichment at the perispeckle requires its association with the EJC.
Taken together, these data show that EJC assembly–defective mutants are uniformly distributed in and out speckles and no longer enriched in perispeckles. They support the notion that perispeckles are not storage sites for free EJC core factors but represent regions enriched in EJC-bound mRNAs.
Fluorescence resonance energy transfer imaging indicates that the four EJC core factors interact in living cells
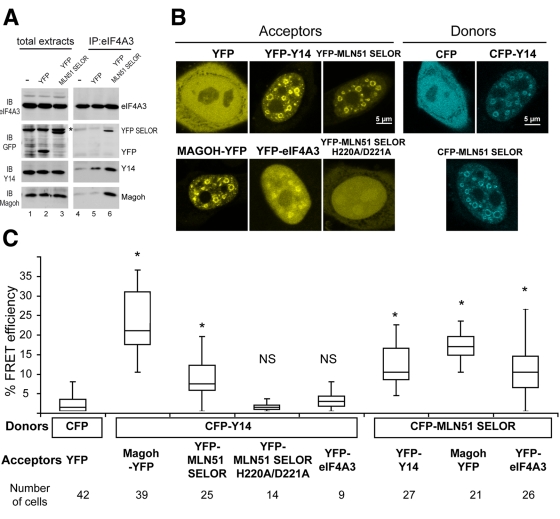
To substantiate this model, we used fluorescence resonance energy transfer (FRET) to detect protein–protein interactions in live cells. We used the sensitized emission method, which exploits the ability of a donor fluorophore to transfer some energy from its excited state to an adjacent acceptor fluorophore, inducing energy emission. This energy transfer is distance and orientation dependent; therefore we used two distinct settings. In a first set of experiments, the cyan fluorescent protein (CFP) tag of Y14 served as the fluorescence donor and the YFP tags of Magoh, MLN51-SELOR, MLN51-SELOR-H220A/D221A, and eIF4A3 served as fluorescence acceptors. In the second setting, the CFP-MLN51-SELOR was used as the donor molecule and the YFP tags of Y14, Magoh, and eIF4A3 were acceptors of fluorescence. On the basis of biochemical and localization studies, we used the SELOR module of MLN51 to design this assay. In fact, this motif is essential and sufficient to stabilize the EJC core (Degot et al., 2004; Ballut et al., 2005; Stroupe et al., 2006; Andersen et al., 2006) and is predominantly localized in the nucleus in the absence of LMB treatment. To show that YFP-SELOR is part of the tetrameric core, the EJC was immunoprecipitated using an antibody against eIF4A3. In agreement with previous studies (Degot et al., 2004; Ballut et al., 2005; Andersen et al., 2006; Bono et al., 2006), the tetrameric EJC core is stabilized by the presence of YFP-SELOR (Figure 5A). In live cells, the fusion proteins showed doughnut-shaped signals characteristic of perispeckle enrichment (Figure 5B), as observed before (Figure 1A). The YFP-eIF4A3 fusion construct was more diffuse in the nucleus, with some enrichment in perispeckles (Figure 5B). Compared with the signals obtained with the untagged control CFP and YFP proteins, a strong FRET signal was observed for CFP-Y14 and Magoh-YFP. This strong FRET signal was expected given their heterodimeric state (Fribourg et al., 2003). In addition, among the tested pairs, a significant FRET efficiency was observed between CFP-Y14 and the YFP-tagged Magoh and MLN51 and between CFP-MLN51 and the YFP-tagged Y14, Magoh, and eIF4A3 (Figure 5C). Confirming immunoprecipitation and localization experiments (Figures 3C, 4B, and 5B), the MLN51 H220A/D221A mutation abrogated FRET (Figure 5C). No significant FRET signal was measured between CFP-Y14 and YFP-eIF4A3 (Figure 5C). Because fluorophore orientation is crucial for energy transfer, we speculated that this conformation was not appropriated and could interfere with the peculiar configuration of the helicase eIF4A3.
FIGURE 5:
The core EJC proteins interact in vivo in the nucleus. (A) HeLa cells were transfected with YFP-SELOR or with the YFP empty vector, as indicated. Endogenous eIF4A3 was immunoprecipitated using a specific anti-eIF4A3 antibody, and immunoprecipitated complexes were analyzed by immunoblotting with anti-eIF4A3, anti-GFP, anti-Y14, and anti-Magoh antibodies. Transfection efficacy was assessed on total extracts with anti-GFP antibody. The asterisk indicates a nonspecific band. Note that the tetrameric EJC core is stabilized by the presence of YFP-SELOR (lane 6). (B) HeLa cells were transfected with YFP- (left) or CFP-tagged (right) constructs as indicated and monitored by live imaging. Representative images of YFP and CFP signals are confocal z-sections. Scale bar, 5 μm. (C) FRET assays were designed with distinct CFP-donor/YFP-acceptor pairs: left, control with CFP and YFP unfused proteins; middle, CFP-Y14 and Magoh-YFP, YFP-SELOR, YFP-eIF4A3; right, CFP-SELOR and YFP-Y14, Magoh-YFP, YFP-eIF4A3. FRET efficiencies between CFP and YFP fusion proteins were measured by the sensitized emission method (median for n cells as indicated), as mentioned in Materials and Methods. Box-and-whiskers plots are of mean FRET efficiencies ± SD between CFP and YFP pairs. p values were obtained from Mann–Whitney statistical test; *p < 0.001.
Collectively, these experiments revealed the presence of the tetrameric EJC core, including MLN51, in the nucleus of living cells. These results reinforce the notion that perispeckles are sites containing messenger ribonucleoprotein (mRNP)–bound EJCs rather than storage sites for unassembled subunits.
Ultrastructural characterization of perispeckles using correlative light and electron microscopy
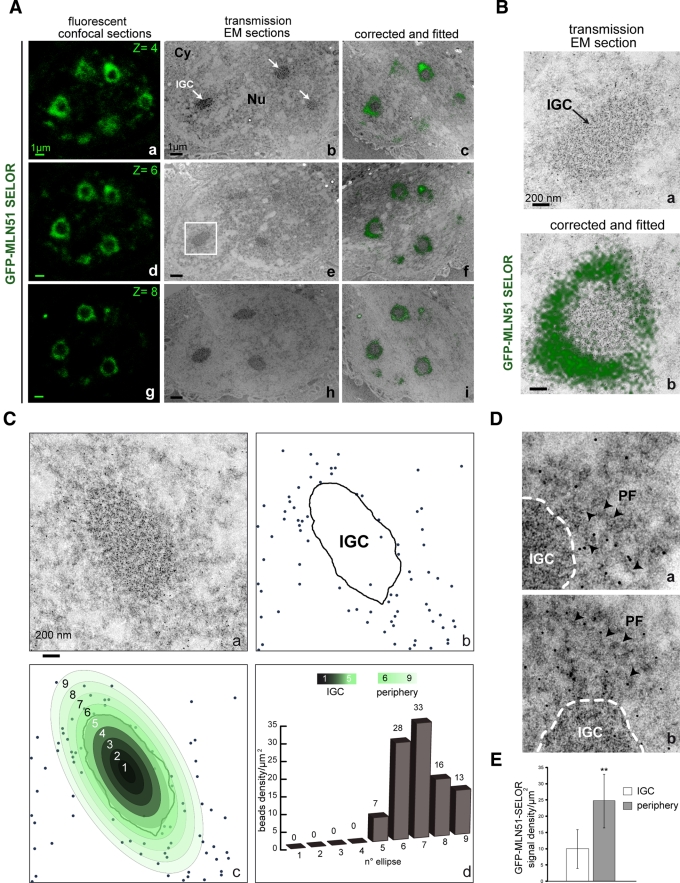
At the ultrastructural level, speckles comprise clusters of 20- to 25-nm granules connected by a thin fibril. These structures are called interchromatin granule clusters (IGCs), and they function as storage, modification, and assembly sites for many RNA-processing factors (Spector et al., 1983; Fakan et al., 1984; Thiry, 1995; Puvion and Puvion-Dutilleul, 1996; Spector and Lamond, 2011). Structures called perichromatin fibrils (PFs) can be observed at the outer periphery of IGCs and in the nucleoplasm. These fibrillar structures (3–5 nm in diameter) of intermediate density were previously shown to contain nascent and polyadenylated RNAs (polyAs) and are believed to represent a major site of transcription and splicing (Fakan, 1994; Puvion and Puvion-Dutilleul, 1996). No specific nuclear structure was previously described that could correspond to the perispeckle region. To determine whether perispeckles overlap with IGCs or PFs, we used a correlative microscopic approach to determine the localization of a fluorescent signal on ultrathin electron microscopy (EM) sections (Spiegelhalter et al., 2010; Supplemental Figure S3). To do so, we seeded MCF7 cells on a micropatterned Aclar support and transfected them with GFP-MLN51-SELOR. Because Y14, Magoh, and eIF4A3 are nuclear-resident proteins, we used the SELOR domain of MLN51 to mark the perispeckle territory (Degot et al., 2004). As for the FRET experiments, we reasoned that the SELOR module will better reflect the presence of the assembled core than the full-length protein. The SELOR module lacks accessory motifs functioning independently of the EJC core, including the N-terminal coil-coiled motif, the NES, and the C-terminal region involved in stress granule assembly (Degot et al., 2002; Macchi et al., 2003; Baguet et al., 2007). Moreover, the isolated SELOR module is sufficient to stably assemble the EJC core complex on RNA (Ballut et al., 2005). The fluorescent signals corresponding to GFP-SELOR were acquired before fixation, whereas transmission EM pictures were taken after high-pressure freezing and embedding (Supplemental Figure S3). Fluorescent confocal acquisitions first acquired were fitted with EM pictures on three serial sections (Figure 6A). On images representing fitted confocal and EM sections, the fluorescent doughnut-shaped signals were at the outer periphery of dense, granular structures characteristic of IGCs (Figure 6, A and B).
FIGURE 6:
Ultrastructural characterization of the perispeckle region by CLEM. MCF7 cells were transiently transfected with GFP-MLN51-SELOR construct and then processed for microscopy analysis and immunogold labeling by CLEM. (A) GFP-MLN51 SELOR is present at the periphery of IGCs. Left, three representative images of GFP signal are live confocal z-sections (a, d, g). Middle, transmission EM images were acquired from thin sections (b, e, h). Right, fitting between light and electron microscopy acquisitions was performed after EM image compression (c, f, i). Nuclear speckles, also called IGCs, are indicated with white arrows (b) inside the nucleus (Nu). A thin cytoplasmic strip is observable (Cy). Scale bar, 1 μm. (B) The GFP signal fits at the periphery of the nuclear speckle. A selected area in A, e (white square), was enlarged with transmission EM section (a) and the corresponding fitted image (b). (C) The GFP signal is enriched in the close vicinity of nuclear speckles. (a) A nonfitted view of a representative IGC immunogold labeled with GFP-specific antibody. (b) Schematic representation of the GFP-specific immunogold labeling; the black line indicates the IGC territory, and gold beads are highlighted with blue dots. (c) Radial distribution of the labeling densities within and around the IGC territory; the central IGC domain is separated into five concentric ellipses and is surrounded by four similar and peripheral circles (pseudocolored from dark gray to light green). Each ring is independently analyzed to determine the radial variation in labeling density. (d) Histogram representing the labeling density of the GFP-MLN51 SELOR signal in concentric nuclear ellipses per surface unit in μm2. From left to right, bars depict the IGC center to the peripheral environment. (D) Gold particles are associated with perichromatin fibrils. (a, b) Enlarged regions showing the distribution of the GFP labeling around the IGC (dashed line). Gold particles overlap with filamentous structures in a less granular region, corresponding to perichromatin fibrils (PF, arrowheads). (E) The GFP-MLN51 SELOR signal is enriched at the periphery of IGC. Graph representing the preferential distribution of GFP-MLN51 SELOR at the periphery of IGC. The mean labeling density of gold particles per surface unit in μm2, which marks the presence of GFP-SELOR, was quantified inside IGCs vs. periphery ± standard deviations. The p value was obtained from Student's statistical t test; **p < 0.01.
To gain more details about the localization of GFP-MLN51-SELOR with respect to nuclear architecture, we performed immunogold labeling on ultrathin sections using anti-GFP antibody coupled to gold particles (Figures 6C and S3). To quantify the radial variation of the labeling density in and out speckles, the IGC domain was contoured and the gold particles (blue dots) were counted in concentric ellipses with fixed intervals (Figure 6C, b and c). Very few gold particles were found in the chromatin or in nucleoli (Figure 6C and unpublished data). In addition, the labeling was not homogeneously distributed over the delimited subnuclear territory (from ellipse 1 to 9) since the density decreased toward the center of the IGC (Figure 6C, b–d). The gold particle density per surface area was 7 particles/μm2 (p/μm2) in the most external ring of the IGC territory and increased to 33 p/μm2 at the outer periphery of the IGC (ellipse 7). The 2.5-fold increase in labeling of the outer IGC (25 p/μm2) is significant as compared with the inner IGC (10 p/μm2; Figure 6E). Of interest, the gold labels were often arranged in regions of intermediate density, along some wide and distinct fibers (Figure 6D, a and b, arrowheads). This fiber-like patterning corresponding to PFs thus overlaps with the perispeckle region. To get more insight into the relationships between the EJC and the nuclear architecture, we also investigated the distribution of Acinus. Acinus is a peripheral EJC factor that associates with the core in the nucleus (Tange et al., 2005). The correlative light and electron microscopy (CLEM) methodology was similarly applied to MCF7 cells overexpressing the YFP-tagged Acinus protein (Supplemental Figure S4). Consistent with the localization of YFP-Acinus by light microscopy (Figure 2A), the YFP signal corresponding to Acinus was enriched at the outer periphery of IGC; little signal was observed inside the IGC (Supplemental Figure S4A). The patterning of YFP-Acinus was then assessed by immunogold labeling using an antibody specific for the YFP tag. Most labeling was found at the outer IGC periphery (Supplemental Figure S4B and unpublished data). A significant 6.4-fold increase of particle density per μm2 was measured between the speckle and the peripheral zone. Of importance, the PFs close to IGC limits were also decorated with gold particles (Supplemental Figure S4).
This correlative approach shows that perispeckles represent nuclear territories distinct from but topologically related to IGCs and supports the hypothesis that perispeckles represent regions where EJC cores are assembled.
Perispeckles are enriched in polyA mRNAs and transcripts undergoing splicing and are close to regions containing active RNA polymerase II
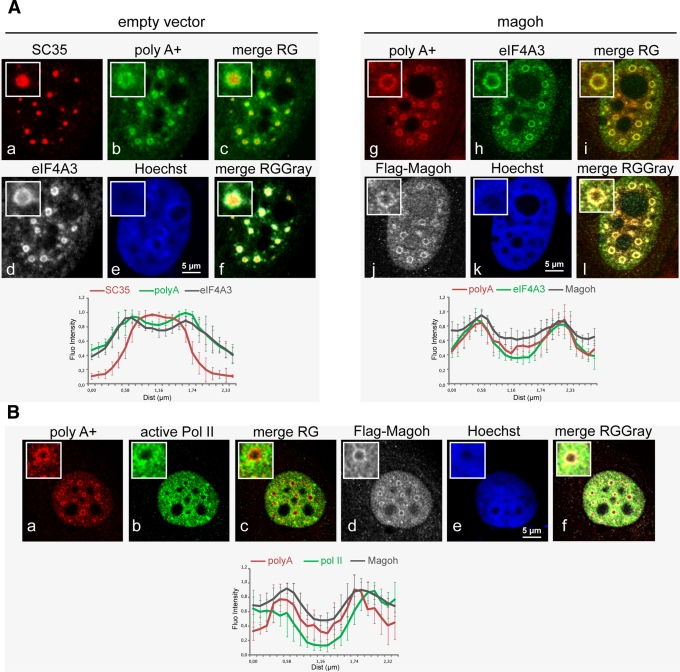
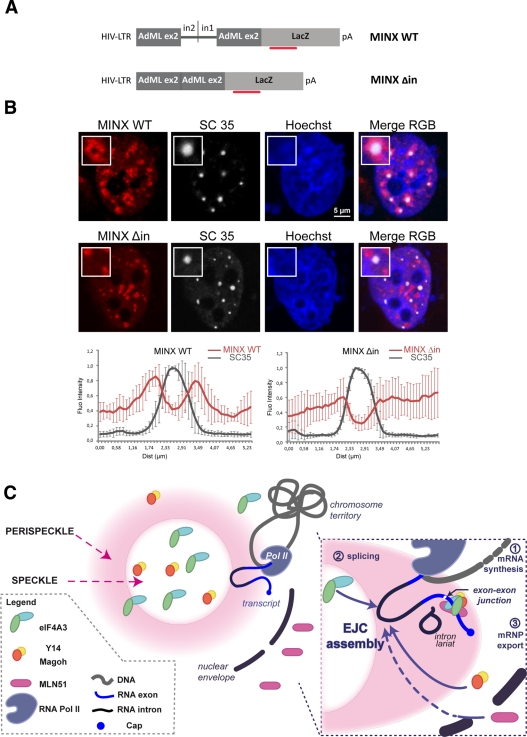
Because PFs contain both polyA RNAs and nascent transcripts (Fakan, 1994; Puvion and Puvion-Dutilleul, 1996), we next examined whether assembled EJC colocalized with either of these markers. Given that eIF4A3, Y14, and Magoh are nuclear-resident proteins, we reasoned that eIF4A3 and Magoh colocalizing signals indicate the presence of assembled EJC rather than the localization of the free subunits or of other complexes. A triple labeling between polyA RNAs, endogenous eIF4A3, and Flag-Magoh showed that they were enriched in doughnut-shaped regions surrounding regular and round speckles (Figure 7A). Line-scan analyses on different speckles highlighted the similar position of the three signals (Figure 7A, magoh, g–l). In transfected cells, eIF4A3 and polyA mRNAs are predominantly detected in perispeckles (Figure 7A, d,h and b,g).The enrichment of both markers is independent of Magoh overexpression (Figure 7A, d and h). It has been shown that cotranscriptional splicing can occur around speckles (Hall et al., 2006; Hu et al., 2008; Brown et al., 2008). Recruitment of some EJC proteins at the site of transcription and splicing was previously documented in mammalian cells. By following transcription using a β-globin gene reporter cell line, Custodio et al. (2004) showed the accumulation of two factors from the core complex Magoh and Y14, together with the core spliceosomal components (U small nuclear ribonucleoproteins [snRNPs]) at transcription sites, thus providing evidence that Magoh and Y14 can cotranscriptionally bind to mRNPs. Magoh and Y14 are recruited before splicing completion (Bessonov et al., 2008; Reichert et al., 2002; Gehring et al., 2009a); therefore it is possible that their cotranscriptional recruitment precedes the formation of the tetrameric EJC core. We then sought to determine the position of perispeckle region with respect to transcription sites. To do so, we performed a triple labeling between Flag-Magoh and polyA mRNAs to mark perispeckles and endogenous active RNA polymerase II (Pol II) detected with the specific antibody against its phosphorylated C-terminal domain (Pol II, H14; Sutherland and Bickmore, 2009). Active polymerases were excluded from nucleoli and from the central region of speckles and were distributed in the nucleoplasm. However, sites of enrichment in active Pol II intermingled with perispeckles (Figure 7B). Line scans highlighted the vicinity between active Pol II on one side and polyA RNAs and Flag-Magoh on the other (Figure 7B). We sought to determine the position of perispeckles with respect to spliced and unspliced transcripts. To do so, we used reporter genes containing (MINX WT) or lacking (MINX Δin) functional splice sites (Schmidt et al., 2011). The MINX sequence is an artificial derivative from the adenovirus genome (Zillmann et al., 1988; Spiluttini et al., 2010). It is composed of two exons flanking an intron, and this splicing unit is followed by the β-galactosidase coding sequence (Figure 8A). Fluorescence in situ hybridization (FISH) using specific probes corresponding to the LacZ sequence (Figure 8A) was used to localize both reporter transcripts, and nuclear speckles were labeled using endogenous SC35 (Figure 8B). Both reporters were efficiently transcribed, as shown by the nuclear accumulation of LacZ transcripts. Although both transcripts were absent from the core of nuclear speckles, they were differently distributed in the nucleus (Figure 8B). Transcripts synthesized from the MINX WT construct were enriched at the periphery of nuclear speckles, whereas transcripts corresponding to MINX Δin were present as random clusters in the nucleus (Figure 8B).
FIGURE 7:
Perispeckles contain polyA mRNAs and overlap with sites of active Pol II. (A) EJC core factors and polyA RNA colocalize at the perispeckle. HeLa cells were transfected with an empty vector (empty vector, a–f) or a Flag-Magoh construct (magoh, g–l) and then processed for combined immunofluorescence and RNA FISH. Fluorescent Alexa Fluor 488 or Cy3-oligo-d(T) probe were hybridized for polyA RNA detection (green in empty vector, b; red in magoh, g). The EJC core components Magoh and eIF4A3 were stained with anti-Flag (white in magoh, j) and anti-eIF4A3 (white in empty vector, d; green in magoh, h) antibodies, respectively. The splicing factor SC35 is detected by immunofluorescence (red in empty vector, a). (B) Active RNA polymerase II is close to perispeckle. HeLa cells were transfected with Flag-Magoh construct (magoh) and then processed for combined immunofluorescence RNA FISH. A fluorescent Cy3-oligo-d(T) probe was hybridized for polyA RNA detection (red). The EJC core component Magoh and active RNA polymerase II were stained with anti-Flag (white) and anti–Pol II H14 (green) antibodies, respectively. In A and B, nuclei were counterstained with the Hoechst-33258 dye (blue). Insets show a representative speckle with 2.5× magnification. Scale bar, 5 μm. Line-scan graphs were analyzed as described in the legend to Figure 1.
FIGURE 8:
Intron-containing RNAs concentrate around nuclear speckles. (A) Scheme of the MINX constructs. The plasmid comprises the LTR-HIV promoter, followed by the MINX sequence fused to the LacZ coding sequence. The MINX sequence consists of adenovirus major late gene sequences (AdML): an exon2/intron2 sequence fragment is fused to a fragment of intron1/exon2 sequence (MINX WT). For MINX Δin, the fused intron2/intron1 is excised. The red bar represents the position of the FISH probes. (B) MINX WT is recruited at the periphery of nuclear speckles. HeLa cells were cotransfected with either MINX WT or MINX Δin and Tat-expressing vector. Cells were then processed for combined immunofluorescence LacZ FISH. A pool of fluorescent Cy3-LacZ probes was hybridized for MINX reporter detection (red). The endogenous SC35 protein was stained with anti-SC35 (white) antibody. Nuclei were counterstained with the Hoechst-33258 dye (blue). Insets show a representative speckle with a 2.5× magnification. Line-scan graphs were analyzed as previously mentioned. Scale bar, 5 μm. (C) Model for the assembly of the EJC core. Unassembled Magoh/Y14 and eIF4A3 are in the nucleus, whereas MLN51 is in the cytoplasm. Nascent pre-mRNAs exiting from Pol II (step1) are processed into mRNP (step2) in perispeckles. Following spliceosome assembly, eIF4A3 and Magoh/Y14 are recruited onto the RNA. After splicing completion, MLN51 joins the complex. The stable EJC core recruits peripheral factors (step 3).
Taken together, these results show that transcripts undergoing splicing are preferentially localized around speckles and support the notion that perispeckles represent EJC-bearing, mRNA-enriched regions intermingled with transcription sites.
DISCUSSION
The experiments presented here support the idea that the EJC core is assembled in the nucleus and marks discrete subnuclear territories containing spliced mRNAs. These doughnut-shaped territories are at the periphery of 9G8- and SC35-positive nuclear speckles and were termed perispeckles (Schmidt et al., 2006).
It had been unclear whether MLN51 is part of the nuclear core of the EJC or whether it joins the EJC after the nuclear export of the spliced mRNA. Indeed, MLN51 functions as a nucleocytoplasmic shuttling protein but exhibits predominantly cytoplasmic localization (Degot et al., 2002; Macchi et al., 2003). Several observations argue in favor of a nuclear association of MLN51 within the EJC core. Indeed, MLN51 was reported to be a bona fide component of the spliced nuclear mRNPs in proteomic analysis of spliceosomal complexes and can be loaded on the mRNA during in vitro splicing reactions (Degot et al., 2004; Tange et al., 2005; Zhang and Krainer, 2007; Herold et al., 2009). In addition, MLN51, via its conserved SELOR module, participates in the stabilization of the core complex by increasing eIF4A3 affinity for RNA (Ballut et al., 2005) and directly binding RNA in the context of the assembled core (Andersen et al., 2006; Bono et al., 2006). Using subcellular fractionation, we showed the presence of low concentration of MLN51 in the nucleus. In addition, under conditions in which the crm1-nuclear export pathway is inhibited with LMB, MLN51 was predominantly in perispeckles. Consistently, the YFP-MLN51/L468A construct bearing a mutation in the consensus NES motif was localized in perispeckles in the absence of treatment. Endogenous MLN51 was nuclear in LMB-treated cells, and, for a small proportion of cells, enrichment was observable at the periphery of nuclear speckles. These results are consistent with a nuclear association of MLN51 in the EJC core and show that localization of the endogenous MLN51 protein involves a complex regulatory scheme, involving both nuclear localization signals and NES. This shuttling mechanism is responsible for the low concentration of MLN51 in the nucleus. As previously proposed, the low concentration of MLN51 might prevent the formation of the stable EJC core inadvertently on RNA without the splicing process to orchestrate its assembly (Tange et al., 2005).
Besides MLN51, the three other EJC core proteins, Y14, Magoh, and eIF4A3, are nuclear-resident proteins. To further study whether perispeckles represent sites enriched in EJC-bound mRNPs rather than storage sites for free subunits, we looked at the localization of specific EJC core mutants that cannot be assembled in the complex. Structural and mechanistic studies have identified binding interfaces required to build the EJC core (Fribourg et al., 2003; Ballut et al., 2005; Gehring et al., 2005; Andersen et al., 2006; Shibuya et al., 2006). EJC-defective Y14, Magoh, and MLN51 mutants were diffusely present in the nucleoplasm. In contrast to their wild-type counterparts, these mutants were present in and out speckles without any enrichment at their periphery, indicating that perispeckles are not storage sites for free EJC components. We propose that perispeckles represent sites enriched in EJC-bound mRNPs. To test this hypothesis, interaction between EJC core factors was measured in situ in living cells by a FRET microscopic approach. A significant FRET efficiency was measured between Magoh, Y14, eIF4A3, and MLN51-SELOR. Taken together, these experiments thus indicate that perispeckles represent nuclear territories enriched in EJC cores.
The architectural organization of the nucleus is essential for the coordination of the different steps involved in gene expression, including transcription and mRNA processing (Matera et al., 2009; Moore and Proudfoot, 2009). The role of nuclear speckles in splicing and gene expression is still a subject of debate (Han et al., 2011). This domain has been considered as a storage site for splicing factors, which are recruited to a subset of active genes for cotranscriptional splicing (reviewed in Lamond and Spector, 2003). Other studies are in favor of an active role of this domain in gene expression. These studies showed that a subset of actively transcribed genes or coexpressed genes cluster together around, or associate with, nuclear speckle domains (Shopland et al., 2003; Hall et al., 2006; Brown et al., 2008; Hu et al., 2008, 2010; Dias et al., 2010; Zhong et al., 2009). In addition, the observation that splicing factors are rapidly exchanged in speckles while the domain is relatively immobile suggests a role of these domains in supplying splicing complexes (Phair and Misteli, 2000). Recent studies highlighted the role of a protein called Son and the long, noncoding transcript named MALAT1 in the structure and function of nuclear speckles. The SR-related protein Son was shown to function as a scaffold for the proper organization of nuclear speckle components, and its depletion was associated with mitotic arrest (Sharma et al., 2010). MALAT1 was shown to play a critical role in the localization and activity of SR-splicing factors (Tripathi et al., 2010; Bernard et al., 2010). In addition, Son was directly involved in the nuclear localization of MALAT1 (Tripathi et al., 2010). The Son and MALAT1 components are interconnected and cooperate to maintain the functional and morphological integrity of nuclear speckles. Taken together, these reports support the view that speckles have an active role during gene expression.
By transmission microscopy, the core region of nuclear speckles corresponds to IGCs, which do not show gene activity, whereas its periphery contains PFs with nascent RNAs, indicative of active transcription (Fakan et al., 1984; Spector et al., 1991) and considered as the main support of splicing (Visa et al., 1993; Fakan, 1994; Puvion and Puvion-Dutilleul, 1996). Both regions are functionally connected, since disruption of IGCs perturbs coordination between transcription and pre-mRNA splicing (Sacco-Bubulya and Spector, 2002). We characterized the perispeckles at the ultrastructural level with respect to the nuclear architecture. For this purpose, we used a correlative microscopic approach that enables the precise localization of a fluorescent signal on ultrathin EM sections. The fluorescence-tagged SELOR-MLN51 signal was ring-shaped and excluded from typical granular IGCs on serial ultrathin sections. To gain more details on its localization, we performed immunogold labeling on serial ultrathin sections. Gold particles were significantly more numerous at the periphery of IGCs versus their core regions: the enrichment peaked at the proximal periphery of IGCs. Of more interest, we found that this signal intermingled with regions of intermediate density containing PFs. Similarly, the peripheral nuclear EJC factor Acinus harbored this pattern distribution. These ultrastructural studies show that the perispeckle territory, traced with two EJC factors, is distinct but topologically related to IGCs and correlates with the presence of PFs.
How spliceosomal components are recruited and assembled onto nascent transcripts in living cells remains unclear. Recently the use of FRET microscopy revealed that splicing complexes can be differently localized in the nucleus. Interactions of the splicing factors SF2/ASF and SC35 with snRNPs are preferentially localized in nuclear speckles and in the nucleoplasm, respectively (Ellis et al., 2008). Whereas nonassembled spliceosomal snRNPs concentrate in Cajal bodies, polyA RNAs concentrate at sites colocalized with snRNP- and SC35-rich domains (Carter et al., 1993; Xing et al., 1993; Sleeman et al., 2003). Consistently, Hall et al. (2006) noted that the polyA signal demarcates a slightly larger “speckle” region, which was believed to overlap with PFs. Other studies also showed that the speckle domain has a substructure (Mintz and Spector, 2000) and contains a variety of proteins, some of which are not involved in splicing (Saitoh et al., 2004). The identity of this “speckle shell” is not yet characterized (Hall et al., 2006), but it shares many features with the perispeckle region. Very recently, the visualization of both cotranscriptional and posttranscriptional splicing was achieved in living cells using a single-molecule approach. This study showed that the released transcripts are not randomly distributed in the nucleus but are attracted to nuclear speckles (Vargas et al., 2011). Analogous observations were made by injection of intron-containing transcripts and in situ hybridization (Dias et al., 2010) and using MINX gene-cassette models (this study). The fact that spliced mRNAs tend to cluster around speckles is consistent with our results indicating that the EJC cores are assembled at these places.
In conclusion, the EJC plays fundamental roles in mRNA biogenesis by allowing the spliceosome to communicate with many other key machineries. However, it had been unclear where the tetrameric core is assembled in the cell. In this study, we show that 1) endogenous and transfected core EJC components are enriched in perispeckles, which consist of doughnut-shaped regions that surround nuclear speckles; 2) assembly-defective mutants fail to accumulate in perispeckles and remain diffusely located in the nucleoplasm; 3) perispeckles contain fully assembled EJC cores; 4) perispeckles are distinct from interchromatin granule clusters, which may principally function as storage and assembly sites for many splicing factors, but intermingle with perichromatin fibrils, which are sites of mRNA synthesis and splicing; and 5) perispeckles are close to sites of active RNA Pol II. Perispeckles appear to be intermediary regions between sites of transcription and sites of storage/maturation and assembly for RNA-processing factors. The following model can be proposed (Figure 8C): nascent pre-mRNAs exiting from active polymerases localize in the perispeckle region and, following spliceosome assembly, recruit eIF4A3 and the Magoh/Y14 heterodimer. After splicing completion, MLN51 joins and stabilizes the EJC core, which then recruits peripheral factors to control mRNA fate. Our data do not exclude that mRNA transcription, splicing, and EJC assembly can occur in other nucleoplasmic locations, but they reveal that a major location for these activities is the perispeckle. In the future, it will be extremely interesting to determine whether other mRNP maturation or assembly steps occur in perispeckles.
MATERIALS AND METHODS
Plasmid constructs
The vectors encoding MLN51 and truncated versions of the protein fused to the enhanced yellow fluorescent protein (EYFP) were previously described (Baguet et al., 2007; Degot et al., 2004). CFP-fused MLN51 constructs were obtained as follows. PCR fragments corresponding to MLN51/1-703 (full-length) and MLN51/137-283 (SELOR) were obtained by PCR using pCR3.1-MLN51 construct as template and the following primers: AEG200 (5′-GAGACAATTG CGTTCTCCGT AAGATGGCGG AC-3′) as forward primer (fp) and AGF274 (5′-CAATTGTTAA CTGGAACCCC TGCTTACAAC-3′) as reverse primer (rp) and AGF268 (5′-AACAATTGGG ACACCAAAAG CACTGTGACT-3′) as fp and AGF272 (5′-CAATTGTTAG CGATGAGACT TGTTTAGCCG-3′) as rp, respectively. The cDNA fragments were digested by MunI and inserted in frame into the EcoRI site of the pECFP-C1 expression vector (Clontech, Mountain View, CA). The YFP-SELOR-HDAA mutant was previously described (Ballut et al., 2005). The complete open reading frames (ORFs) of wild-type and mutants Y14/RNA-binding motif protein 8 (RBM8; NM_005105; Degot et al., 2004; Gehring et al., 2005) were digested by MunI and inserted in frame into the EcoRI site of the pEYFP-C1 and pECFP-C1 and pEGFP-C1 plasmids. The complete ORFs of wild-type and mutant Magoh (NM_018048) were inserted into Flag-tagged vector (Fribourg et al., 2003; Degot et al., 2004). The ORFs of UPF1 (NM_002911), UPF3 (NM_080632), Magoh (NM_002370), Acinus-S (NM_074977), SAP18 (NM_005870), and eIF4A3 (NM_014740) were obtained by RT-PCR using Flag-tagged, pMS2-tagged, or pET28a constructs (Ballut et al., 2005; Chamieh et al., 2008; Degot et al., 2004; Tange et al., 2005) and cloned into pmCHERRY-N1, pEYFP-N1, and pEGFP-N1 plasmids. The MLN51-H220A/D221A mutant was obtained from pEYFP-MLN51 vector by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Stratagene, Santa Clara, CA) using the primer HDAA (5′-GAGGGTCGCT GGGAGGCGGC CAAATTCCGG GAAGAT-3′), which replaces a histidine (H220) and an aspartic acid (D221) by two alanines. The MLN51-H220A/D221A, MLN51-L468A, and MLN51-H220A/D221A/L468A mutants were obtained from pEYFP-MLN51 vector by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit) using the primers HDAA (5′-GAGGGTCGCT GGGAGGCGGC CAAATTCCGG GAAGAT-3′) and L468A (5′-GAGCAAGATG TGGCACAAGC GAACAGAATT GGAGTCCG-3′), which replace a histidine (H220) and an aspartic acid (D221) by two alanines and a leucine (L468) by an alanine, respectively. The ORFs of RNPS1 (NM_006711), NXF1 (NM_006362), and PYM (NM_032345) were isolated by PCR from HeLa or MCF7 polyA RNAs and cloned into pEYFP-C1 plasmids. Plasmids encoding GFP-tagged control constructs, NUP153, and the mRNA probe p4XLamdbaN22-3XmEGFP-M9 were purchased from EUROSCARF (Oberusel, Germany). The MINX WT and MINX Δin reporter genes were used as described previously (Schmidt et al., 2011).
Cell culture and transfections
HeLa, MCF7, and Sk-Br3 cells provided by the American Type Culture Collection (Rockville, MD) were maintained in DMEM supplemented with 5% of fetal calf serum (FCS) for HeLa and in 10% FCS for MCF7 and Sk-Br3. For immunofluorescence, 5000–10,000 cells were plated on glass coverslips in 24-well plates and transfected using jetPEI transfection reagent (Polyplus-transfection SA, Illkirch, France) with 1 μg of various plasmids. For immunoprecipitations, HeLa cells grown in 10-cm dishes were either transfected with 10 μg of different YFP-MLN51 constructs or cotransfected with 5 μg of Flag-Magoh and 5 μg of YFP-Y14 plasmids. To inhibit Crm1-dependent nucleocytoplasmic transport, HeLa cells were incubated, 24 h after transfection, for 1 h with 10 μg/ml cycloheximide, and then 20 ng/ml leptomycin B (Sigma-Aldrich, St. Louis, MO) was added for 5 h.
Immunofluorescence microscopy
After 24 h of culture, cells were washed with phosphate-buffered saline (PBS), fixed for 5 min at room temperature in 4% paraformaldehyde in PBS, and permeabilized for 10 min with 0.1% Triton X-100 in PBS. After blocking in 1% bovine serum albumin (BSA) in PBS, cells were incubated at room temperature with the primary antibodies for 1 h. Cells were washed three times in PBS and incubated 1 h with Cy3- and Alexa Fluor 488–conjugated appropriate secondary antibody (1:1000). Cells were washed three times in PBS, and nuclei were counterstained with Hoechst-33258 dye. Slides were mounted in Vectashield (Polysciences, Warrington, PA). For RNA FISH, fixed cells were incubated in 2× saline–sodium citrate (SSC)–20% formamide for 5 min and hybridized overnight at 37°C with hybridization buffer (20 ng/ml fluorescent oligo d(T) probe [40 Ts], 2× SSC, 20% formamide, 30 μg/ml Escherichia coli tRNA, 0.02% RNase-free BSA, 2 mM vanadyl-ribonucleoside complex, 1% dextran sulfate). Cells were washed two times for 30 min at room temperature in 2× SSC–20% formamide prior to immunostaining. In situ hybridization and immunofluorescence for transcripts containing LacZ sequences (MINX WT and MINX Δin) were performed as previously described (Schmidt et al., 2011).
Image acquisition and data analysis
Observations were made with confocal fluorescence microscopes (Leica SP1 and Leica SP2 UV, 63×, numerical aperture 1.4; Leica Microsystems, Wetzlar, Germany). Images were confocal sections ranging from 0.2 to 0.3 μm in thickness. Image analyses were done using ImageJ (National Institutes of Health, Bethesda, MD) and Photoshop (Adobe, San Jose, CA) software. Quantitative analysis of fluorescence signals over the speckle territory was performed in ImageJ with the line-scan function. Within ImageJ, a line of length around ∼2 μm was selected centered on a representative speckle. The PlotProfile feature of ImageJ was used to record the pixel intensities along the selected line. This process was repeated for five representative speckles. The data for each profile were then exported into Excel software (Microsoft, Redmond, WA) in order to be normalized with a higher pixel intensity value for each channel. For a given channel, the mean and SD (n = 5 speckles) were calculated. Graphs were plotted and analyzed with Excel.
Generation and analysis of low-expression YFP-tagged constructs
To generate constructs expressing low levels of EJC proteins, we used a strategy adapted from Varnai et al. (2007), by exchanging the cytomegalovirus promoter with the herpes simplex virus thymidine kinase (TK) promoter. The minimal TK promoter was amplified by PCR using the pRL-TK vector from Promega (Madison, WI; nucleotides 610–1029) as template and the synthetic oligonucleotides 5′-AGATCTATTA ATATGATGAC ACAAACCCCG CCCAGCGTCTT-3′ and 5′-AGATCTTCTA GACTATAGTG AGTCGTATTA AGTACTCTAGC-3′ as fp and rp primers, respectively. This DNA fragment was digested using AseI and XbaI and inserted into the AseI and NheI sites in the following plasmids: pEYFP-N1 empty, pEYFP-MLN51, pEYFP-MLN51/137-283 (SELOR), pEYFP-Y14, pEYFP-Magoh, and pEYFP-eIF4A3. To analyze the level of expression of these constructs, HeLa cells were transfected and analyzed by immunoblot and immunofluorescence. Immunofluorescence allowed estimating transfection efficiency by directly counting the number of GFP-positive cells. For immunoblot analysis, transfected cells were washed with 1× PBS twice and lysed in RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM ethylene glycol tetraacetic acid, 1% NP-40, 1% sodium deoxycholate, 1% SDS) supplemented with 0.5 mM dithiothreitol and Complete Protease Inhibitor (Roche, Neuilly sur Seine, France). Nearly equal amounts of proteins (20 μg) were separated on 7–14% SDS–PAGE and transferred onto nitrocellulose membranes. Membranes were blocked with milk 3% in 1× PBS, Tween-20 0.1%, and incubated overnight at 4°C with anti-Magoh (#2D2, 1/500), anti-Y14 (#3H4, 1/250), anti-eIF4A3 (#2625, 1/5000), anti-MLN51 (#1608; 1/2000) and anti-GFP (1/2000) antibodies. Signals were acquired using the LAS Quant 4000 System (GE Healthcare, Uppsala, Sweden) and quantified using ImageJ. For Magoh, Y14, MLN51, and eIF4A3, the signals corresponding to the YFP fusions were measured and adjusted according to the transfection efficiency, which was 50%. The signals corresponding to the endogenous protein were measured on the same lane. Signals corresponding to the endogenous proteins were set to 1. The YFP-SELOR construct expression level was measured indirectly. This truncated protein form cannot be detected using our anti-MLN51 antibody; therefore its expression was measured using the YFP signal and quantified by comparison with the YFP-MLN51 signal.
Immunoprecipitation
Flag-Magoh complexes or eIF4A3-containing complexes were immunoprecipitated from RNaseA-treated HeLa cell lysates (20 μg/ml, 15min on ice) with M2 anti-Flag agarose (Sigma-Aldrich) and anti-eIF4A3 rabbit polyclonal antibody bound to protein A/protein G–coupled agarose beads, respectively; the incubation was achieved at 4°C for 2 h for M2 anti-Flag and overnight for anti-eIF4A3 in lysis buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5% Triton X-100 supplemented with Complete Protease Inhibitor). Beads were washed four times in lysis buffer. Precipitated complexes were eluted with SDS-sample buffer and separated in SDS-polyacrylamide 12% gels by gel electrophoresis (PAGE).
Antibodies
To generate the anti-eIF4A3 rabbit polyclonal antibody (#2625), the synthetic peptides corresponding to residues 1–20 of the human eIF4A3 protein sequence were coupled to ovalbumin and injected into New Zealand rabbits. The same peptide was used to generate a mouse monoclonal antibody (#2E5) as described before (Moog-Lutz et al., 1997). The recombinant Magoh/Y14 heterodimer, produced as in Ballut et al. (2005), was used to generate two monoclonal antibodies (#2D2 against Magoh and #3H4 against Y14). The anti-9G8 and anti–histone H3 were from the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC; Illkirch, France) mouse monoclonal antibody facility. The anti-MLN51 antibodies were the anti-MLN51 Ct (#1608) described previously (Degot et al., 2002) and the CASC3 polyclonal antibody from ProteinTech (Chicago, IL). The anti–activated RNA polymerase II (CTD-Ser-5 phosphospecific) H14 antibody, anti-SC35, and anti-tubulin antibodies were from Covance Research Products (Princeton, NJ), Sigma-Aldrich. and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. For immunoprecipitation, YFP-tagged proteins and Flag-Magoh were detected using the anti-GFP (IGBMC mouse monoclonal antibody facility) and the anti-Flag rabbit polyclonal (Sigma-Aldrich) antibodies, respectively. For Western blotting, horseradish peroxidase–conjugated anti-mouse or anti-rabbit antibodies were used as secondary antibodies (Jackson ImmunoResearch, West Grove, PA). For immunofluorescence, secondary Cy3-, Cy5-, and Alexa 488–conjugated, affinity-purified donkey anti-rabbit and anti-mouse were purchased from Jackson ImmunoResearch and Invitrogen/Molecular Probes (Eugene, OR), respectively.
Subcellular fractionation
The presence of MLN51 in the nucleus was analyzed by subcellular fractionation in both HeLa and MCF7 cells and gave similar results. Fractionation of cultured cells was done using the Proteo-Extract Subcellular Proteome Extraction kit (Calbiochem, La Jolla, CA) according to the manufacturer's instructions. Aliquots of subcellular extracts representing cytoplasm, membrane, nucleus, and cytoskeletal elements were separated in SDS-polyacrylamide 6–10% gels by gel electrophoresis (PAGE), blotted, and analyzed.
Fluorescence resonance energy transfer
FRET exploits the capacity of a higher-energy fluorophore (donor) to transfer energy when excited to a lower-energy fluorophore (acceptor) when these two molecules are closer than 10 nm (reviewed in Wouters et al., 2001). Thus, the binding of two fluorescent protein–tagged constructs can be detected by measuring the emission intensity of the acceptor fluorophore after exciting the donor fluorophore. FRET between EJC core components was measured using the Sensitized Emission FRET wizard in Leica MP confocal software. This software calculates FRET efficiency by the method of Gordon et al. (1998). Each symbol starts with an uppercase letter representing the filter set—A, for the acceptor filter set; D, for the donor filter set; and F, for the FRET filter set. The second letter is lowercase and indicates which fluorochromes are present in the specimen—a, for the acceptor only; d, for the donor only; and f, for both donor and acceptor present. The third letter is lowercase and indicates the signal from only one of the fluorochromes when both fluorochromes are present—a, d, and f indicate acceptor, donor, and FRET signal, respectively.
Ffd = Dfd × (Fdd/Ddd) refers to only the donor signal when both donor and acceptor are present.
Ffa = Afa × (Faa/Aaa) refers to only the acceptor signal when both donor and acceptor are present.
FRET efficiency = (Fff – Ffd – Ffa)/Afa
Owing to spectral bleedthrough between donor and acceptor emission channels, FRET can only be confirmed after background subtraction. So, appropriate background subtraction is determined from a series of control images involving donor-only (CFP), acceptor-only (YFP), and FRET samples. Because sensitized emission FRET is nondestructive, it lends itself well to time-lapse imaging of live cells.
In practice, HeLa cells were plated onto 35-mm glass-bottom dishes (MatTek, Ashland, MA). Cells were transfected with CFP-Y14, Magoh-YFP, YFP-SELOR, YFP-SELOR-HDAA, or eIF4A3-YFP plasmids (1.5 μg of donor, 1.5 μg of acceptor). The day after transfection, live imaging was performed on a fluorescence microscope (Leica TCS MP-AOBS, 63×, numerical aperture 1.4) equipped with an incubation chamber at 37°C (PeCon GmbH, Erbach, Germany). Cells with identical expression level of the fluorophores were chosen. The reference images from control cells expressing donor-only (CFP), acceptor-only (YFP), and FRET images from cells expressing both were therefore collected simultaneously under identical optical conditions. FRET efficiency was calculated in the region of interest (ROI), including nuclear speckles, perispeckles, and a portion of the nucleoplasm. Statistical analysis was performed by using the Mann–Whitney test (http://faculty.vassar.edu/lowry/VassarStats.html). Values correspond to three independent experiments. Statistics were acquired by comparing each pair to the YFP/CFP empty plasmid pair using the Mann–Whitney test, and an asterisk indicates p < 0.001.
Correlative light and electron microscopy and immunogold labeling
MCF7 cells were grown on supports coated with type I collagen and suitable for high-pressure freezing (HPF) as described in Spiegelhalter et al. (2010). Briefly, a laser-micropatterned Aclar film was mounted onto gold-plated live-cell carriers (16707897, Leica Microsystems). This system makes it possible to select cells and store the exact position of the cell of interest with the reference coordinates. Cells at 70% confluency were transfected with 1 μg GFP-SELOR. For acquisitions and further freezing steps, the live-cell carriers were mounted on the rapid loader of the HPF machine (EMPACT-2, Leica Microsystems). Acquisitions were performed using a fluorescence confocal microscope (Leica TCS MP-AOBS, 63×, numerical aperture 1.4; Leica Microsystems). Cryoprotection is achieved by dipping the samples into a solution of 20% of BSA in the culture medium. The loader was removed from the confocal setup and transferred to the EMPACT-2 for HPF. After freezing, samples were collected in liquid nitrogen. For immunogold analysis, dehydration, and infiltration with Lowicryl HM20, procedures were as previously described (Spiegelhalter et al., 2010). After embedding with resin and removal of the carriers, serial thin sections (60 nm) were cut on an ultramicrotome (Cryotim; Diatome, Biel, Switzerland) and were collected on Formvar-carbon–coated nickel slot grids. Immunogold labeling was performed on the EM IGM automated system (Leica Microsystems). Rabbit polyclonal anti-GFP (ab6556; Abcam, Cambridge, United Kingdom) was revealed with 10-nm, gold-coupled protein A (EM Lab, Utrecht University, Utrecht, Netherlands). Sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (Philips CM12; Philips, Eindhoven, Netherlands). Images were acquired with an Orius 1000 charge-coupled device camera (Gatan, Pleasanton, CA). Fitting and compression sectioning were resolved using Fiji software (ImageJ). Quantification of gold particles was performed to appreciate the enrichment of the GFP signal in distinct subnuclear regions. The number of gold particles per μm2 was quantified for 10 speckles for each immunolabeling experiment. The ROIs were delimited, and their surfaces were measured using ImageJ. Gold particles were finally manually counted within each ROI, and the graphical data set is presented as mean ± SEM. Statistics were acquired by comparing particle distributions between peripheral and core ROI using Student's t test; **p < 0.01 and ***p < 0.0001, respectively.
Supplementary Material
Acknowledgments
We thank S. Chan and E. Soutoglou for critical reading of the manuscript, Y. Schwab, B. Seraphin, M. Thiry, G. Pierron, and the members of the Molecular and Cellular Biology of Breast Cancer team (IGBMC) for helpful advice and discussion, P. Eberling, G. Duval, and M. Oulad for technical assistance, and B. Pradet-Balade, M. C. Robert, J. Stevenin, N. Dreumont, H. Moine, and M. Moore for reagents. Both A.B and E.D. received allocations from the Ministère de l'Enseignement Supérieur et de la Recherche (France). In addition, E.D. had a fellowship from the Fondation pour la Recherche Médicale and A.B. had a fellowship from the Association pour la Recherche sur le Cancer. We also acknowledge funds from the Institut National de Santé et de Recherche Médicale, the Centre National de la Recherche Scientifique, the Université de Strasbourg, the Agence Nationale de la Recherche Program Blanc, Alsace Contre le Cancer, and the Ligue Nationale Contre le Cancer.
Abbreviations used:
- CFP
cyan fluorescent protein
- CLEM
correlative light and electron microscopy
- EJC
exon junction complex
- FISH
fluorescence in situ hybridization
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- IGC
interchromatin granule cluster
- LMB
leptomycin B
- mRNP
messenger ribonucleoprotein
- NMD
nonsense-mediated mRNA decay
- PF
perichromatin fibrils
- SELOR
speckle localizer and RNA binding
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0040) on March 14, 2012.
REFERENCES
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Izaurralde E. Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring N. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci USA. 2010;107:10050–10055. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Custodio N, Carvalho C, Condado I, Antoniou M, Blencowe BJ, Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 2004;10:622–633. doi: 10.1261/rna.5258504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods. 2007;4:633–636. doi: 10.1038/nmeth1065. [DOI] [PubMed] [Google Scholar]
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Degot S, Regnier CH, Wendling C, Chenard MP, Rio MC, Tomasetto C. Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene. 2002;21:4422–4434. doi: 10.1038/sj.onc.1205611. [DOI] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Ellis JD, Lleres D, Denegri M, Lamond AI, Caceres JF. Spatial mapping of splicing factor complexes involved in exon and intron definition. J Cell Biol. 2008;181:921–934. doi: 10.1083/jcb.200710051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J Cell Biol. 1984;98:358–363. doi: 10.1083/jcb.98.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009a;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009b;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001a;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001b;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Macchi P, Kroening S, Palacios IM, Baldassa S, Grunewald B, Ambrosino C, Goetze B, Lupas A, St Johnston D, Kiebler M. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J Neurosci. 2003;23:5778–5788. doi: 10.1523/JNEUROSCI.23-13-05778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz PJ, Spector DL. Compartmentalization of RNA processing factors within nuclear speckles. J Struct Biol. 2000;129:241–251. doi: 10.1006/jsbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Moog-Lutz C, Tomasetto C, Regnier CH, Wendling C, Lutz Y, Muller D, Chenard MP, Basset P, Rio MC. MLN64 exhibits homology with the steroidogenic acute regulatory protein (STAR) and is over-expressed in human breast carcinomas. Int J Cancer. 1997;71:183–191. doi: 10.1002/(sici)1097-0215(19970410)71:2<183::aid-ijc10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIFS4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Puvion E, Puvion-Dutilleul F. Ultrastructure of the nucleus in relation to transcription and splicing: roles of perichromatin fibrils and interchromatin granules. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P, Spector DL. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol. 2011;193:819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Richter K, Berger AB, Lichter P. In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. J Cell Biol. 2006;172:373–381. doi: 10.1083/jcb.200503061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Prasad J, Degenhardt K, Erdjument-Bromage H, White E, Tempst P, Kidd VJ, Manley JL, Lahti JM, Reinberg D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol Cell Biol. 2003;23:2981–2990. doi: 10.1128/MCB.23.8.2981-2990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Stroupe ME, Moore MJ. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12:360–374. doi: 10.1261/rna.2190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter C, Tosch V, Hentsch D, Koch M, Kessler P, Schwab Y, Laporte J. From dynamic live cell imaging to 3D ultrastructure: novel integrated methods for high pressure freezing and correlative light-electron microscopy. PLoS One. 2010;5:e9014. doi: 10.1371/journal.pone.0009014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Stroupe ME, Tange TO, Thomas DR, Moore MJ, Grigorieff N. The three-dimensional architecture of the EJC core. J Mol Biol. 2006;360:743–749. doi: 10.1016/j.jmb.2006.05.049. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M. The interchromatin granules. Histol Histopathol. 1995;10:1035–1045. [PubMed] [Google Scholar]
- Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol. 2001;154:511–523. doi: 10.1083/jcb.200105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci USA. 2007;104:11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Zapp ML, Berget SM. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988;8:814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.