Abstract
Salmonella is a globally widespread food-borne pathogen having major impact on public health. All motile serovars of Salmonella enterica of poultry origin are zoonotic, and contaminated meat and raw eggs are an important source to human infections. Information on the prevalence of Salmonella at farm/holding level, and the zoonotic serovars circulating in layer poultry in the South and South-East Asian countries including Bangladesh, where small-scale commercial farms are predominant, is limited. To investigate the prevalence of Salmonella at layer farm level, and to identify the prevalent serovars we conducted a cross-sectional survey by randomly selecting 500 commercial layer poultry farms in Bangladesh. Faecal samples from the selected farms were collected following standard procedure, and examined for the presence of Salmonella using conventional bacteriological procedures. Thirty isolates were randomly selected, from the ninety obtained from the survey, for serotyping and characterized further by plasmid profiling and pulsed-field gel electrophoresis (PFGE). Results of the survey showed that the prevalence of motile Salmonella at layer farm level was 18% (95% confidence interval 15–21%), and Salmonella Kentucky was identified to be the only serovar circulating in the study population. Plasmid analysis of the S. Kentucky and non-serotyped isolates revealed two distinct profiles with a variation of two different sizes (2.7 and 4.8 kb). PFGE of the 30 S. Kentucky and 30 non-serotyped isolates showed that all of them were clonally related because only one genotype and three subtypes were determined based on the variation in two or three bands. This is also the first report on the presence of any specific serovar of Salmonella enterica in poultry in Bangladesh.
Introduction
Salmonella is a major food-borne pathogen worldwide and contaminated poultry products, especially undercooked meat and raw eggs are important sources of it [1], [2]. The rationality in introducing statutory surveillance for Salmonella in poultry farms/holdings in the EU member countries and other developed parts of the world is to reduce human salmonellosis of poultry origin [3]–[6]. In contrast, monitoring for Salmonella in poultry is either of very primitive type or the need is completely ignored in developing countries because of resource constraints, and therefore, information on its prevalence is poorly documented, so is the consequence to the public health. The zoonotic Salmonella circulating in developing countries with the possible presence of antimicrobial resistance genes might have some global public heath impacts because of their transmissions to other countries beyond the geographical origin, by travellers or by trades [7]–[10], are impossible to prevent. Mitigation of the source(s) at the geographical origin should be the option to restrain a wider dissemination of the zoonotic serovars for which local knowledge on their prevalence is important.
Salmonella, a member of Enterobacteriaceae consists of two species – Salmonella enterica and Salmonella bongori. Salmonella enterica consists of six subspecies (ssp.) under which there are >2500 serovars [11] that can produce diseases in mammals including animals and humans, and a good number of them can be harboured by poultry without showing any clinical signs [12]–[14]. S. enterica ssp. enterica serovar Gallinarum-Pullorum is host specific and non-motile and produce clinical diseases with variable mortality only in chickens [15]. Only motile serovars for which poultry are known to be reservoirs are zoonotic. Among them, most frequently reported serovars in the United States are S. Typhimurium, S. Enteritidis, S. Newport, S. Heidelberg and S. enterica ssp. enterica 4, [5], 12: i:- [16], although persistency and prevalence of different serovars vary from place to place [12].
Eggs produced from layer farms are a major protein source for people in Bangladesh and the spent hens are also sold for consumption. Small-scale commercial farms (FAO-defined production system 3) [17] are predominating here as in the other South and South-East Asian countries where stocks range from several hundreds to a few thousands, kept in a semi-confined system with a minimum of biosecurity. In such a system (FAO-defined production system 3), unlike large-scale commercial production systems (FAO-defined production systems 1 and 2) seen in developed countries, the birds might be more vulnerable to become exposed to Salmonella. However, published information on the rate at which small-scale layer farms are harbouring the zoonotic Salmonella in the South and South-East Asian countries including Bangladesh is limited, if not absent. Here, we describe the prevalence of zoonotic Salmonella at layer farm level in Bangladesh, the circulating serovars and the molecular characterization of these isolates.
Materials and Methods
Ethics statement
Oral permission was taken from owner of each poultry farm while collecting faecal samples from the farm.
Study population
In Bangladesh, there are two districts (out of 64 districts) where the percentages of commercial layer poultry farms are the highest [18]. They are Dhaka and Chittagong where the capital city and the 2nd largest city are located, respectively. By lottery we selected Chittagong, located in the South-East part, to conduct a cross-sectional survey for the prevalence of motile Salmonella at layer farm level. To provide state veterinary services to the public there are 64 districts and 481 sub-districts/Upazila (the lowest administrative unit in Bangladesh) livestock offices. We collected the list of all commercial layer poultry farms from the District Livestock Office. Using this list as the sampling frame, 500 farms were randomly selected. The sample size was estimated following the formula, n = Z2 1−α/2 p (1−p)/L2, where n = number of sample size, p = prevalence of the disease, Z1−α/2 = value of the standard normal distribution corresponding to a two-sided confidence level of 1−α/2 and L = maximum allowable error. Because the farm prevalence of Salmonella in any commercial production system had not previously been documented in Bangladesh or in any South-East Asian country, expected flock prevalence was considered as 50% with an allowable error on the estimate of L = 0.05 at 95% confidence level.
Collection of samples
The survey was conducted between July 2009 and June 2010. Each selected farm was physically visited once to collect pooled faecal samples and epidemiological information. Because most farms were single-housed we sampled one flock per farm. From five different locations in the farm, five naturally pooled faecal samples, each resulted from ∼30 cross-sectional pinches for achieving a total volume of about 200 g, were collected; no individual birds were sampled [19], [20]. Disposable plastic hand gloves were worn during sample collection. Each pooled sample was placed separately into a sterile plastic bag and transferred to the microbiology laboratory, Chittagong Veterinary and Animal Sciences University (ML-CVASU), Bangladesh at ambient temperature. After arrival at the laboratory the samples were stored at 5°C until examination.
Isolation and identification of Salmonella
At ML-CVASU, for each sample a slurry was created by mixing 200 g of faeces with 200 ml of buffered peptone water (BPW; CM0009; Oxoid Ltd., England), and 50 g of this mixture was inoculated into 200 ml of BPW and incubated at 37°C for 18 hours. After that, 0.1 ml of this culture was inoculated into Rappaport-Vassiliadis (RV) broth (02-379; Scharlau Chemie S. A, EU), and incubated at 42°C for 48 hours; then 0.010 ml of the fresh culture was streaked on to brilliant green (BG) agar (CM0329; Oxoid Ltd., England) and Xylose-Lysine Deoxycholate (XLD) agar (CM0469; Oxoid Ltd., England) surface, and incubated overnight at 37°C. Suspected colonies from both of the agar plates were transferred to triple-sugar-iron (TSI) agar (CM277; Oxoid Ltd., England) slant and incubated at 37°C for 24 hours. Typical reactions for Salmonella to TSI were regarded as the presence of Salmonella. A farm was considered presumptively Salmonella-positive when ≥1 of the 5 collected samples were diagnosed positive with Salmonella. With accruing one isolate per positive farm over the period of the survey a repository was maintained at −80°C at ML-CVASU. At the end of the survey all the isolates were shipped to the Department of Veterinary Disease Biology, University of Copenhagen, Denmark (DVDB-KU) using Stuart's transport medium (CM111; Oxoid Ltd., England) at normal temperature by a commercial courier service. Upon receiving the samples at DVDB-KU, the isolates were screened for confirmation of motile Salmonella. Here, each isolate was grown on Luria Bertani (LB) broth (240230; Difco, USA) at 37°C and 100 µl of the overnight culture, divided into three separate and equally-spaced drops, was inoculated on to the surface of Modified Semisolid Rappaport Vassiliadis (MSRV) medium (CM0910; Oxoid Ltd., England) supplemented with novobiocin (SR0161E; Oxoid Ltd., England) and incubated at 41.5°C for 24 to 36 hours. Any swarming growth observed on the MSRV plates was transferred to brilliant-green phenol-red lactose sucrose (BPLS) agar (1.07237.0500; Merck, Germany) by dipping an inoculating loop into the swarmed zone. Following overnight incubation at 37°C, suspected Salmonella colonies from BPLS agar plates were transferred into 5% citrated blood agar (Blood agar base; CM0055; Oxoid Ltd., England) and incubated at 37°C for 16 to 18 hours. Standard biochemical tests were performed to assess the growth for Salmonella and isolates showing typical reactions were confirmed serologically using anti-Salmonella polyvalent serum (SSI, Copenhagen, Denmark), and stored at −80°C using 15% glycerol.
Thirty randomly selected isolates were serotyped according to White-Kauffmann-Le Minor Scheme [11] at Statens Serum Institiut, Copenhagen, Denmark. We used CE marked (ISO) Salmonella antisera (SSI Diagnostica, Hillerød, Denmark) for serotyping. PBS (pH 7.38) was used as a control to check for autoagglutination of the individual antiserum.
Plasmid profiling
Plasmid was isolated according to the alkaline-lysis method described by Kado and Liu [21] with minor modifications [22]. Plasmids in E. coli 39R861 [23] and E. coli V517 [24] were used as references for standard plasmid sizes. The sizes of plasmids were estimated by calculating the migration of plasmid mobility relative to that of the reference plasmids [25].
Genotyping of the isolates by pulsed-field gel electrophoresis (PFGE)
PFGE was performed following the standardized CDC PulseNet protocol [26] to determine the genetic diversity and relatedness among the isolates. Overnight culture of bacteria grown on brain heart infusion (BHI) broth (CM1135; Oxoid Ltd., England) was used. Genomic DNA was prepared using 1% agarose (SeaKem® gold agarose, Lonza, Rockland, ME USA) and embedded DNA was digested using 60U of the restriction enzyme XbaI (R0145L; New England BioLabs Inc.) for 14 hours at 37°C. The DNA fragments were isolated by electrophoresis in 0.5× TBE buffer using CHEF DR III (Bio-Rad Laboratories, Hercules, California, USA) system at 14°C with initial switch time 2.2 sec, final switch time 63.8 sec, current 6 V/cm, included angle 120 and run time 19 hours. S. Braenderup H9812 was used as a reference strain and as standard size marker [27]. The gel was stained with 1% ethidium bromide (E1510; Sigma-Aldrich, USA) solution for 30 minutes and destained in deionzed water for 3 times with 20 minutes interval. Using UV transillumination, gel image was captured by GelDoc EQ system with Quantity One® (Version 4.2.1) software (Bio-Rad Laboratories, Hercules, California, USA) and obtained images were saved in TIF format in computer. The analysis of the fingerprints was performed using GelCompar®II (version 4.6) software (Applied Maths, Belgium). Dice coefficient with a band position tolerance of 1% and 0.5% optimization level were used to determine similarity between fingerprints. The unweighted pair group method with arithmetic averages (UPGMA) was applied to produce the dendrogram. The DNA restriction patterns of the isolates were interpreted according to the criteria described by Tenovar et al. [28].
Statistical analysis
All epidemiological data were entered into a spreadsheet of Microsoft Excel 2003 and transferred to statistical software SPSS (version 13.0, 2006) for windows (SPSS Inc., Chicago, IL). Farm-prevalence of Salmonella was calculated as the number of positive farms divided by total number of farms investigated. The difference in prevalence among different variables was shown using a χ2 test.
Results
Prevalence of Salmonella at layer farm level
An overall farm prevalence of motile Salmonella along with the prevalence seen with the variables: flock size, age, feed with animal protein, source of protein, use of antibiotics, and season are shown in Table 1. Of the 500 farms investigated 90 were positive for motile Salmonella, giving a prevalence of 18% (95% confidence interval (CI) 15–21%). The prevalence varied among the flock sizes; the farms of >1 but ≤2 thousand birds had the lowest prevalence compared with other groups (p<0.001). There was no significant difference observed on the prevalence of Salmonella in the farms having birds of different age groups (P = 0.48). None but one farm had a history of Salmonella vaccination. Commercially available feed or self-made feed, by mixing raw ingredients purchased from the local markets, were fed to the birds. The farms that had the history of using any kind of animal protein ingredient had a proportionately higher prevalence, although statistically borderline insignificant (P = 0.08) and the prevalence was ∼3 times higher (p<0.001) where fish meal was used compared with other protein sources. No significant difference was observed on the prevalence of Salmonella between farms using or not using antibiotics in feed as feed additives. The prevalence of Salmonella varied proportionately according to different seasons, although statistically not significant (P = 0.9).
Table 1. Prevalence of motile Salmonella in commercial layer poultry farms in Bangladesh, 2009–2010 (n = 500).
| No. farms investigated | No. positive with Salmonella | Prevalence (%) | ||
| Flock size* | ≤1000 | 155 | 39 | 25.2 |
| 1001–2000 | 195 | 17 | 8.7 | |
| 2001–4000 | 85 | 17 | 20.0 | |
| >4000 | 65 | 17 | 26.2 | |
| Age of birds (wks) | ≤20 | 135 | 21 | 15.6 |
| 21–40 | 135 | 24 | 17.8 | |
| 41–60 | 105 | 17 | 16.2 | |
| >60 | 125 | 28 | 22.4 | |
| Feed with animal protein | Yes | 281 | 60 | 21.4 |
| No | 58 | 7 | 12.1 | |
| Unknown | 161 | 23 | 14.3 | |
| Source of protein* | Fish meal | 187 | 50 | 26.7 |
| Others | 70 | 6 | 8.6 | |
| Unknown | 243 | 34 | 14.0 | |
| Use of antibiotics in feed | Yes | 107 | 22 | 20.6 |
| No | 279 | 56 | 20.1 | |
| Unknown | 114 | 12 | 10.5 | |
| Season | Summer (March–May) | 114 | 23 | 20.2 |
| Rainy (June–August) | 110 | 20 | 18.2 | |
| Autumn (September–November) | 125 | 22 | 17.6 | |
| Winter (December–February) | 151 | 25 | 16.6 | |
| Overall | 500 | 90 | 18 |
P<0.001.
Serotyping and plasmid profiling
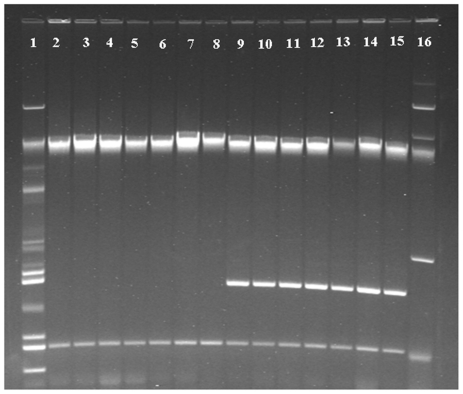
The results of serotyping revealed that all of the 30 isolates belonged to the serovar S. Kentucky. We examined all the 90 isolates by plasmid profiling and the results of 14 S. Kentucky isolates are shown in Figure 1. Irrespective of serotypic identity two distinct profiles with two different sizes of plasmid were seen; 53 isolates harbored only one plasmid of 2.7 kb and 37 had two plasmids of 2.7 and 4.8 kb. Among the 30 S. Kentucky isolates 18 had one plasmid (2.7 kb) each and the others each had two plasmids (2.7 and 4.8 kb).
Figure 1. Plasmid profiles of 14 of the 30 Salmonella Kentucky isolates from commercial layer poultry farms in Bangladesh, 2009–2010.
Lane 2–15 for S. Kentucky; Lane 1 and 16 are plasmid size markers in Escherichia coli strains V517 and 39R861, respectively (90 isolates: 30 Salmonella Kentucky and 60 non-serotyped were investigated).
PFGE genotyping
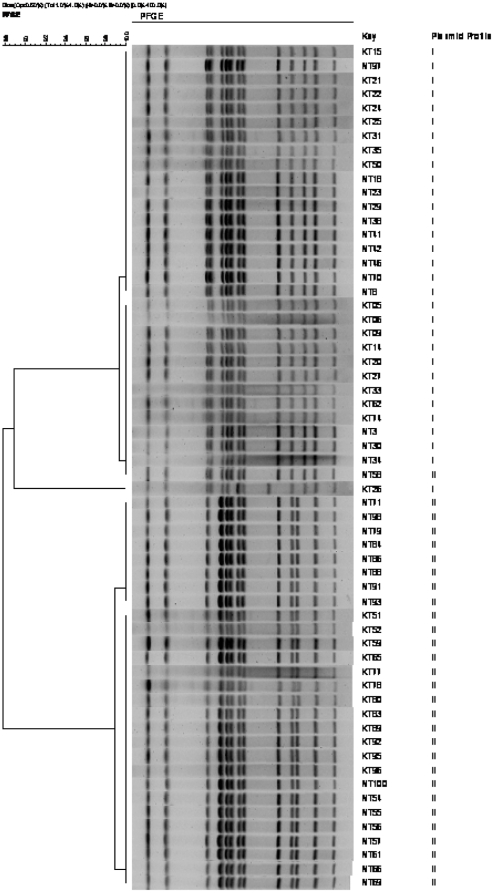
PFGE typing demonstrated that all the S. Kentucky isolates were closely related, displaying a common band pattern. Three subtypes were identified based on the variations in two or three bands among the isolates tested. In addition to 30 S. Kentucky isolates, 30 randomly selected non-serotyped isolates were also subjected for genotyping and the results showed that their fingerprint patterns were similar to the S. Kentucky isolates. The dendrogram showing the restriction fingerprint pattern of the 60 isolates is illustrated in Figure 2.
Figure 2. Dendrogram showing the cluster analysis on the basis of XbaI-PFGE of the 30 Salmonella Kentucky and 30 non-serotyped isolates obtained from commercial layer poultry farms in Bangladesh, 2009–2010.
Dice coefficient was used to perform similarity analysis, and clustering was performed by using unweighted pair-group method with arithmetic means (UPGMA) with 1% band position tolerance and 0.5% optimization parameter. KT, S. Kentucky; NT, Non-serotyped.
Discussion
In this survey the prevalence of motile Salmonella in commercial layer poultry farms in Bangladesh is 18%. Surprisingly, all of the 30 isolates investigated for serotyping belonged to the serovar S. Kentucky, and the fingerprint pattern of the other 30 non-serotyped isolates in PFGE analysis unraveled a similar identity, illustrating a high dissemination of S. Kentucky in layer farms in Bangladesh. However, because we investigated only one isolate per Salmonella-positive farm the circulation of other serovar(s) can not entirely be addressed. This is also the first report on the presence of any specific serovar in poultry in Bangladesh, and to the authors' knowledge, S. Kentucky has probably never been reported before in layer poultry from any other South and South-East Asian countries, although a report on prevalence of Salmonella belonging to serogroups B and D in poultry in a selected area in Bangladesh [29] is available.
We performed serotyping of the 30 isolates at Statens Serum Institut, which is a national reference laboratory in Denmark. The laboratory holds an accreditation according to DS/EN ISO/IEC 17025∶2000 for all the analyses. Therefore, we believe that the serotyping of the isolates reflects the true result. In addition, PFGE genotyping results echo the similar identity of the isolates.
Although RV broth was initially used, MSRV medium was used later as selective enrichment to ensure the growth of only motile Salmonella.
The dominance of one serovar over others in a particular geographical area is not uncommon [30], [31], although the presence of more than one zoonotic serovar of poultry origins have been reported frequently from well-structured surveillance carried out in the developed world [16], [32]. However, circulation of only one serotype in a spatial area is probably a rarity. Historically, S. Typhimurium and S. Enteritidis are two widely reported common zoonotic serovars associated with poultry, but none of them were found in this study. Other serovars have also been reported in different parts of the world – S. Paratyphi B var. Java in the Netherlands [33], S. Hiduddify in Nigeria [34], S. Infantis in Hungary [35], S. Hadar, S. Heidelberg, S. Manhattan and S. Virchow in Algeria [36], indicating temporal increase of a particular serovar in a specific region.
Although large flock size constitutes a potential risk factor for occurrence of Salmonella [37], [38], we observed an inconsistent result – farms with smallest flock size (<1000 birds) and the largest (>4000 birds) and the second largest (2001–4000 birds) had a prevalence of >20% while the farms of the second smallest flock size (1001–2000 birds) had the lowest prevalence, 9% (Table 1). It is hard to explain why such variations occur, however, because the prevalence was the same for the farms having flocks of the four different age groups (Table 1), a common exposure source for Salmonella to them might be a possibility, not just increasing excretion frequencies from carrier birds housed.
S. Kentucky was first reported by Edwards [39]. Poultry is considered a reservoir of this serovar, [7] which apparently is becoming more common [16] in chickens. It's presence in layer farms has been documented [31], [36], and the fraction of this serovar in broiler chickens has increased from 25% in 1997 to nearly 50% in 2006 in relation to top serovars identified in the USA [40]. Recently a particular clone of S. Kentucky acquiring a virulence plasmid from avian pathogenic Escherichia coli (APEC) has been described [41]. S. Kentucky was previously reported as a less successful pathogen in relation to human illness [8], however, evolving resistance in this serovar to multiple antibiotics, especially ciprofloxacin [7]–[9], [42], [43] are posing a new threat to public health.
Plasmid profiling is one of the earliest molecular tools used for subtyping Salmonella [44]. Two distinct profiles which shared a common small plasmid (2.7 kb) were seen in this study among the isolates, not consistent with Majtán et al. [9] who reported two large size plasmids in S. Kentucky isolates of human origin. Plasmid free strains of S. Kentucky have also been documented [43]. The plasmid contents of bacteria may change over time during storage [45]. For genotyping of Salmonella, PFGE is considered to be a gold standard test to investigate the epidemiology of outbreaks including source identification [46]. The band differences, seen in PFGE analysis, among the isolates investigated could be due to a single genetic event with a point mutation or an insertion or deletion [28], however, the overall results reveal that they all are clonally related. However, S. Kentucky has previously been shown to be highly clonal concerning population structure [47], and more discriminatory typing methods, e.g. multiple-locus variable-number tandem-repeats analysis (MLVA) may add new information to the epidemiology of the infection.
Animal protein sources used in poultry feed have been documented to be reservoirs of many serovars including S. Kentucky [48]–[50]. In this study, the prevalence was three times higher where fish meal was used. Experience on farm visits suggests that this raw ingredient is purchased from the local markets by the farmers themselves to produce low-cost feed by mixing with other ingredients. Animal protein added locally produced feeds are also used, but their Salmonella-free status is questionable because of absence of any structured surveillance and regulatory legislation.
Although prevalence of motile Salmonella was significantly associated with use of feed containing animal protein sources, especially fish meal, this practice was however not a commonality for the S. Kentucky positive farms. The farmers in the study area buy fish meal or other feed ingredients from the local markets where birds and eggs of different farms are also sold. The same vehicles are used for transportation of birds, eggs and feeds between the farms and the markets, and in most cases, these vehicles remain contaminated with faeces, and non-disinfected. The use of the same vehicles between farms and markets for transportation of birds, eggs and feeds, and the access of the products of the farms to the same local markets were two practice commonalities. Different degrees of faecal contaminations of vehicles and frequencies of market visits could have some contributory roles in the farm-positivity for S. Kentucky. This speculation however needs to be verified in future investigations.
The present study provides novel information on the prevalence of Salmonella, and circulation of a clonally related genotype of S. Kentucky in commercial layer poultry farms in Bangladesh. The high prevalence of this genotype should concern the authorities that it can be transmitted to humans by contaminated eggs. Tracing of the probable sources of the genotype is important to minimize its zoonotic risks within and outside of the country for which more molecular epidemiological studies are required to screen commercial feeds, raw feed ingredients, especially fish meal, and breeder farms, because S. Kentucky can also be vertically transmitted.
In conclusion, the prevalence of motile Salmonella in small-scale commercial layer poultry farms in Bangladesh is 18%, and only one serovar, S. Kentucky has been demonstrated so far. Based on plasmid profiling and PFGE analysis it is evident that the circulating isolates of the serovar is clonally related. This may suggest a common source of origin but more discriminatory typing methods may be able to add more information as to the epidemiology of S. Kentucky in Bangladesh. The high prevalence of the serovar might be attributable to some common sources of spread, not known from this study, but its emergence and persistency might have public health impacts in Bangladesh and beyond.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: These authors have no support or funding to report.
References
- 1.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, et al. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol. 2011;77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez S, Hofacre CL, Lee MD, Maurer JJ, Doyle MP. Animal sources of salmonellosis in humans. J Am Vet Med Assoc. 2002;221:492–497. doi: 10.2460/javma.2002.221.492. [DOI] [PubMed] [Google Scholar]
- 3.Wegener HC, Hald T, Wong LF, Madsen M, Korsgaard H, et al. Salmonella control programs in Denmark. Emerg Infect Dis. 2003;9:774–780. doi: 10.3201/eid0907.030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EEC. Council Directive 92/117/EEC of 17 December 1992 concerning measures for protection against specified zoonoses and specified zoonotic agents in animals and products of animal origin in order to prevent outbreaks of food-borne infections and intoxications. 1992. Available: http://ec.europa.eu/food/fs/sfp/mr/mr07_en.pdf. Accessed: 2011 Oct 25.
- 5.Keery I. Salmonella Enteritidis control programs in the Canadian poultry industry. 2010. Available: http://www.agf.gov.bc.ca/lhmr/pubs/se_control_programs0910.pdf. Accessed: 2011 Oct 25.
- 6.Cozens M. Salmonella enteritidis monitoring and certification program, Queensland. 2008. Available: http://www.dpi.qld.gov.au/documents/Biosecurity_GeneralAnimalHealthPestsAndDiseases/Salmonella-enteritidis-program.pdf. Accessed: 2011 Oct 25.
- 7.Weill FX, Bertrand S, Guesnier F, Baucheron S, Grimont PAD, et al. Ciprofloxacin-resistant Salmonella Kentucky in Travelers. Emerg Infect Dis. 2006;12:1611–1612. doi: 10.3201/eid1210.060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collard JM, Place S, Denis O, Rodriguez-Villalobos H, Vrints M, et al. Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J Antimicrob Chemother. 2007;60:190–192. doi: 10.1093/jac/dkm114. [DOI] [PubMed] [Google Scholar]
- 9.Majtán V, Majtán T, Majtán J, Szabóová M, Majtánová L. Salmonella enterica serovar Kentucky: antimicrobial resistance and molecular analysis of clinical isolates from the Slovak Republic. Jpn J Infect Dis. 2006;59:358–362. [PubMed] [Google Scholar]
- 10.Aarestrup FM, Hendriksen RS, Lockett J, Gay K, Teates K, et al. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis. 2007;13:726–731. doi: 10.3201/eid1305.061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimont PAD, Weill F-X. Antigenic Formulae of the Salmonella Serovars (9th edition). 2007. WHO Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France. Available: http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089. Accessed: 2010 Feb 10.
- 12.Gast RK. Serotype-specific and serotype-independent strategies for preharvest control of food-borne Salmonella in poultry. Avian Dis. 2007;51:817–828. doi: 10.1637/8090-081807.1. [DOI] [PubMed] [Google Scholar]
- 13.Lutful Kabir SM. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int J Environ Res Public Health. 2010;7:89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poppe C. Wray C, Wray A, editors. Salmonella infections in the domestic fowl. Salmonella in Domestic Animals. 2000. pp. 107–132. CAB International.
- 15.Shivaprasad HL. Saif YM, editor. Pullorum Disease and Fowl Typhoid. Diseases of Poultry, 11th Ed. 2003. pp. 568–582. Iowa State Press, Ames, Iowa.
- 16.Centers for Disease Control and Prevention (CDC) Salmonella Surveillance: Annual Summary, 2006. 2008. US Department of Health and Human Services, CDC. Atlanta, GA. Available: http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2006/salmonellaannualsummary2006.pdf. Accessed: 2011 Apr 20.
- 17.FAO and OIE in collaboration with WHO. A global strategy for the progressive control of highly pathogenic avian influenza (HPAI). 2005. Available: http://www.fao.org/avianflu/documents/HPAIGlobalStrategy31Oct05.pdf. Accessed: 2011 Jan 5.
- 18.Dolberg F. Poultry sector country review: Bangladesh. Food and Agriculture Organization of the United Nations, Rome. 2008. Available: ftp://ftp.fao.org/docrep/fao/011/ai319e/ai319e00.pdf. Accessed: 2010 Jul 11.
- 19.European Commission. Baseline study on the prevalence of Salmonella in laying flocks of Gallus gallus in the EU: Technical specifications. 2004. SANCO/34/2004 Rev 3. Available: http://ec.europa.eu/food/food/biosafety/salmonella/tech_spec_sanco-34-2004_rev-3_en.pdf. Accessed: 2010 Jan 15.
- 20.Carrique-Mas JJ, Davies RH. Sampling and bacteriological detection of Salmonella in poultry and poultry premises: a review. Rev Sci Tech. 2008;27:665–677. doi: 10.20506/rst.27.3.1829. [DOI] [PubMed] [Google Scholar]
- 21.Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen JE. Wray C, Wray A, editors. Molecular typing of Salmonella. Salmonella in Domestic Animals. 2000. pp. 429–446. CAB International.
- 23.Threlfall EJ, Rowe B, Ferguson JL, Ward LR. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J Hyg (Lond) 1986;97:419–426. doi: 10.1017/s0022172400063609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 25.Rochelle PA, Fry JC, Day MJ, Bale MJ. An accurate method for estimating sizes of small and large plasmids and DNA fragments by gel electrophoresis. J Gen Microbiol. 1985;132:53–59. doi: 10.1099/00221287-132-1-53. [DOI] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention (CDC) One-Day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). 2009. Available: http://www.pulsenetinternational.org/SiteCollectionDocuments/pfge/5%201_5%202_5%204_PNetStand_Ecoli_with_Sflexneri.pdf. Accessed: 2011 Apr 15.
- 27.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahmud MS, Bari ML, Hossain MA. Prevalence of Salmonella serovars and antimicrobial resistance profiles in poultry of Savar area, Bangladesh. Foodborne Pathog Dis. 2011;8:1111–1118. doi: 10.1089/fpd.2011.0917. [DOI] [PubMed] [Google Scholar]
- 30.Pointon A, Sexton M, Dowsett P, Saputra T, Kiermeier A, et al. A baseline survey of the microbiological quality of chicken portions and carcasses at retail in two Australian states (2005 to 2006). J Food Prot. 2008;71:1123–1134. doi: 10.4315/0362-028x-71.6.1123. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Payne JB, Santos FB, Levine JF, Anderson KE, et al. Salmonella populations and prevalence in layer feces from commercial high-rise houses and characterization of the Salmonella isolates by serotyping, antibiotic resistance analysis, and pulsed field gel electrophoresis. Poult Sci. 2007;86:591–597. doi: 10.1093/ps/86.3.591. [DOI] [PubMed] [Google Scholar]
- 32.European Food Safety Authority (EFSA) The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. The EFSA Journal. 2007;130:352. [Google Scholar]
- 33.van Pelt W, van der Zee H, Wannet WJB, van de Giessen AW, Mevius DJ, et al. Explosive increase of Salmonella Java in poultry in the Netherlands: consequences for public health. Euro Surveill. 2003;8:31–35. doi: 10.2807/esm.08.02.00398-en. [DOI] [PubMed] [Google Scholar]
- 34.Raufu I, Hendriksen RS, Ameh JA, Aarestrup FM. Occurrence and characterization of Salmonella Hiduddify from chickens and poultry meat in Nigeria. Foodborne Pathog Dis. 2009;6:425–430. doi: 10.1089/fpd.2008.0150. [DOI] [PubMed] [Google Scholar]
- 35.Nógrády N, Tóth A, Kostyák A, Pászti J, Nagy B. Emergence of multidrug-resistant clones of Salmonella Infantis in broiler chickens and humans in Hungary. J Antimicrob Chemother. 2007;60:645–648. doi: 10.1093/jac/dkm249. [DOI] [PubMed] [Google Scholar]
- 36.Bouzidi N, Aoun L, Zeghdoudi M, Bensouilah M, Elgroud R, et al. Salmonella contamination of laying-hen flocks in two regions of Algeria. Food Res Int. 2011 doi: 10.1016/j.fooders.2011.05.027. [Google Scholar]
- 37.Mollenhorst H, van Woudenbergh CJ, Bokkers EGM, de Boer IJM. Risk factors for Salmonella enteritidis infections in laying hens. Poult Sci. 2005;84:1308–1313. doi: 10.1093/ps/84.8.1308. [DOI] [PubMed] [Google Scholar]
- 38.Namata H, Méroc E, Aerts M, Faes C, Abrahantes JC, et al. Salmonella in Belgian laying hens: An identification of risk factors. Prev Vet Med. 2008;83:323–336. doi: 10.1016/j.prevetmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Edwards PR. A new Salmonella type: Salmonella Kentucky. J Hyg (Lond) 1938;38:306–308. doi: 10.1017/s0022172400011177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.USDA. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). 2008. Veterinary isolates final report, slaughter isolates, 2006. USDA, Washington, DC. Available: http://www.ars.usda.gov/sp2UserFiles/Place/66120508/NARMS/narms_2006/NARMS2006.pdf. Accessed: 2011 Sep 20.
- 41.Johnson TJ, Thorsness JL, Anderson CP, Lynne AM, Foley SL, et al. Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS ONE. 2010;5(12):e15524. doi: 10.1371/journal.pone.0015524. doi: 10.1371/journal.pone.0015524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infect Dis. 2011;204:675–684. doi: 10.1093/infdis/jir409. [DOI] [PubMed] [Google Scholar]
- 43.Turki Y, Mehri I, Cherif H, Najjari A, Aissa RB, et al. Epidemiology and antibiotic resistance of Salmonella enterica serovar Kentucky isolates from Tunisia: The new emergent multi-drug resistant serotype. Food Res Int. 2011 doi: 10.1016/j.foodres.2011.03.044. [Google Scholar]
- 44.Mayer LW. Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin Microbiol Rev. 1988;1:228–243. doi: 10.1128/cmr.1.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsen JE, Brown DJ, Baggesen DL, Bisgaard M. Stability of plasmids in five strains of Salmonella maintained in stab culture at different temperatures. J Appl Bacteriol. 1994;77:155–159. doi: 10.1111/j.1365-2672.1994.tb03059.x. [DOI] [PubMed] [Google Scholar]
- 46.Heir E, Lindstedt BA, Nygård I, Vardund T, Hasseltvedt V, et al. Molecular epidemiology of Salmonella Typhimurium isolates from human sporadic and outbreak cases. Epidemiol Infect. 2002;128:373–382. doi: 10.1017/s0950268802007045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xi M, Zheng J, Zhao S, Brown EW, Meng J. An enhanced discriminatory pulsed-field gel electrophoresis scheme for subtyping Salmonella serotypes Heidelberg, Kentucky, SaintPaul, and Hadar. J Food Prot. 2008;71:2067–2072. doi: 10.4315/0362-028x-71.10.2067. [DOI] [PubMed] [Google Scholar]
- 48.Rose N, Beaudeau F, Drouin P, Toux JY, Rose V, et al. Risk factors for Salmonella enterica subsp. enterica contamination in French broiler-chicken flocks at the end of the rearing period. Prev Vet Med. 1999;39:265–277. doi: 10.1016/s0167-5877(99)00002-1. [DOI] [PubMed] [Google Scholar]
- 49.Barbour EK, Nabbut NH, Hinners SW. Distribution of paratyphoids on Saudi Arabian poultry farms and pathogenicity studies of predominant serotypes. Avian Dis. 1983;27:616–622. [PubMed] [Google Scholar]
- 50.Papadopoulou C, Carrique-Mas JJ, Davies RH, Sayers AR. Retrospective analysis of Salmonella isolates recovered from animal feed in Great Britain. Vet Rec. 2009;165:681–688. [PubMed] [Google Scholar]