ABSTRACT
Natural luteolysis involves multiple pulses of prostaglandin F2alpha (PGF) released by the nonpregnant uterus. This study investigated expression of 18 genes from five distinct pathways, following multiple low-dose pulses of PGF. Cows on Day 9 of the estrous cycle received four intrauterine infusions of 0.25 ml of phosphate-buffered saline (PBS) or PGF (0.5 mg of PGF in 0.25 ml of PBS) at 6-h intervals. A luteal biopsy sample was collected 30 min after each PBS or PGF infusion. There were four treatment groups: Control (n = 5; 4 PBS infusions), 4XPGF (4 PGF infusions; n = 5), 2XPGF-non-regressed (2 PGF infusions; n = 5; PGF-PBS-PGF-PBS; no regression after treatments), and 2XPGF-regressed (PGF-PBS-PGF-PBS; regression after treatments; n = 5). As expected, the first PGF pulse increased mRNA for the immediate early genes JUN, FOS, NR4A1, and EGR1 but unexpectedly also increased mRNA for steroidogenic (STAR) and angiogenic (VEGFA) pathways. The second PGF pulse induced immediate early genes and genes related to immune system activation (IL1B, FAS, FASLG, IL8). However, mRNA for VEGFA and STAR were decreased by the second PGF infusion. After the third and fourth PGF pulses, a distinctly luteolytic pattern of gene expression was evident, with inhibition of steroidogenic and angiogenic pathways, whereas, there was induction of pathways for immune system activation and production of PGF. The pattern of PGF-induced gene expression was similar in corpus luteum not destined for luteolysis (2X-non-regressed) after the first PGF pulse but was very distinct after the second PGF pulse. Thus, although the initial PGF pulse induced mRNA for many pathways, the second and later pulses of PGF appear to have set the distinct pattern of gene expression that result in luteolysis.
Keywords: corpus luteum, gene expression, hormone action, luteolysis, mRNA, prostaglandins
Repeated low doses of PGF produce distinct patterns of gene expression; the initial pulse increases expression of many genes, whereas second and later PGF pulses decrease expression of angiogenic and steroidogenic genes while further increasing genes involved in PGF production and the immune system.
INTRODUCTION
The corpus luteum (CL) is a transient endocrine gland involved in establishment and maintenance of pregnancy due to production of progesterone (P4). The new CL develops from cells that remain in the follicle following ovulation but is eventually composed of multiple, distinctive cell types including steroidogenic cells (small and large luteal cells) and non-steroidogenic cells (endothelial cells, pericytes, fibrocytes, and immune cells) [1,4]. In the absence of a viable embryo, functional and structural regression of the CL occurs. In many species, including the cow, regression of the CL is due to multiple pulses of prostaglandin F2α (PGF) released by the nonpregnant uterus [4,6]. There is substantial variability in the frequency and amplitude of PGF pulses associated with ruminant luteolysis, but typically, there are 48 discrete pulses that occur at 6- to 14-h intervals [7,9]. For example, in heifers evaluated at hourly intervals during the seventh day around luteolysis, there was complete luteolysis, as defined by a decrease in basal P4, after four distinct PGF pulses that occurred over ∼30 h [7]. Recent studies have mimicked the normal pattern of PGF pulses by infusing low doses of PGF into the uteri of heifers [10, 11]. The PGF metabolite patterns were similar to the patterns observed during natural luteolysis. In addition, the changes in circulating P4 concentrations and luteal blood flow showed patterns similar to those that occurred during natural luteolysis with an increase in circulating P4 and blood flow after the initial PGF pulse and a subsequent decrease in P4 and blood flow after the second, third, and fourth infusions of PGF [10, 11].
Many studies have focused on understanding the molecular mechanisms underlying the luteolytic process. Some of those studies evaluated changes in gene expression during natural luteolysis by removing the CL at specific days of the estrous cycle [12,14]. These studies provided substantial insight into the molecular changes associated with the later stages of luteolysis but could not provide detailed information for the changes associated with specific pulses of PGF (first, second, third) or during precisely defined stages of the luteolytic process. Many studies [14,20] have also investigated luteal regression by using a single injection of a supraphysiological dose of PGF. A single high dose of PGF suppressed steady-state concentrations of mRNA for specific genes that might have been involved in “luteotropic” functions, whereas there was an induction of mRNA for genes that might have been involved in functional or structural luteolysis [15, 16, 20,23]. For example, treatment with PGF decreased VEGFA, STAR, PTGFR, and LHCGR mRNA concentrations, but the same PGF treatments increased FAS, FASLG, TNF, IL1B, IFNG, EDN1, MMP1, and PGTS2 mRNA concentrations [15, 16, 20, 22,24]. Not surprisingly, molecular changes were not always consistent when results of studies of CL during natural luteolysis were compared to those of luteolysis induced by a single large PGF treatment. For example, patterns of luteal cytokine expression, such as CCL2, and luteal immune cell populations had quite different patterns during induced versus those during natural luteolysis [12, 14]. Recent studies have drawn particular attention to differences in luteal responses following a single supraphysiological dose of PGF compared to those after lower doses of PGF infused into the uterus, to more closely mimic natural luteal regression [10,12, 25,30]. Preliminary reports of the intraluteal changes that follow multiple low doses of PGF in the ovine CL have been presented [31,33], but, to our knowledge, no studies of the changes in gene expression that accompany multiple individual pulses of physiological doses of PGF in any species have been published.
This research used a 0.5-mg intrauterine infusion of PGF [11] in an attempt to mimic physiological pulses of PGF. In order to evaluate gene expression at multiple times after individual intrauterine PGF infusions, the previously described and validated method of ultrasound-guided luteal biopsy was used [34]. Three different treatment groups were evaluated after four different intrauterine infusions in this study, in order to provide information for the changes in concentrations of key mRNA that follow infusion of 4 pulses of PGF (4XPGF; expected full luteolysis), 2 pulses of PGF with two infusions of saline (2XPGF; expected partial luteolysis), and 4 infusions of saline (control, no luteolysis). Our main hypotheses were that an individual PGF pulse would induce a distinct pattern of gene expression that would vary by 1) whether the CL had been exposed to a previous PGF pulse (first vs. second vs. third vs. fourth) and 2) whether the CL underwent full or partial luteal regression (4XPGF vs. 2XPGF). Unexpectedly, some of the animals treated with two PGF infusions underwent complete luteal regression, whereas other animals did not undergo luteal regression after two PGF infusions, providing a more valid comparison for testing our second hypothesis that gene expression after individual PGF pulses would vary in CL that subsequently underwent complete luteal regression compared to CL that recovered after a similar PGF treatment. In order to test these two hypotheses, 18 different mRNA were evaluated in each luteal biopsy. Specific classes of mRNA were chosen to allow testing of these hypotheses including immediate early genes (jun proto-oncogene [JUN], FBJ murine osteosarcoma viral oncogene homolog [FOS], nuclear receptor subfamily 4, group A, member 1 [NR4A1], and early growth response 1 [EGR1]), steroidogenic genes (steroidogenic acute regulatory protein [STAR], cytochrome P450, family 11, subfamily A, polypeptide 1 [CYP11A1], nuclear receptor subfamily 5, group A, member 1 [NR5A1, formerly known as SF-1], and luteinizing hormone/choriogonadotropin receptor [LHCGR]), prostaglandin-related genes (prostaglandin-endoperoxide synthase 2 [PTGS2], prostaglandin-F synthase [PTGFS], hydroxyprostaglandin dehydrogenase 15-[NAD] [HPGD], prostaglandin F receptor [PTGFR]), immune-related genes (interleukin 1, beta [IL1B], interleukin 8 [IL8], Fas [FAS], Fas-ligand [FASLG]), and angiogenesis-related genes (vascular endothelial growth factor A [VEGFA], fibroblast growth factor 2 [FGF2]). Moreover, we evaluated the cellular localization for two of these genes (NR4A1 and PTGFR) and three of the proteins involved in P4 synthesis (STAR, CYP11A1, HSD3B7) by using in situ hybridization and immunohistochemistry, respectively, following intrauterine PGF infusions.
MATERIALS AND METHODS
Reagents
TRIzol was purchased from Invitrogen (Invitrogen Life Technologies Inc., Carlsbad, CA). GoScript reverse transcriptase system, RNAse-free DNase I, dNTP set, and Taq DNA polymerase for PCR were purchased from Promega (Madison, WI). Maxima SYBR quantitative PCR (qPCR) Master Mix (2×) for real-time PCR was obtained from Fermentas Life Sciences (Thermo Scientific, Foster City, CA). Gel extraction kit and RNeasy mini-kit were obtained from Qiagen (Frederick, MD). Digoxigenin (DIG)-11-UTP, RNase inhibitor, T7 RNA polymerase anti-DIG-AP antibody, blocking reagent, and BM Purple AP substrate were purchased from Roche Applied Science (Mannheim, Germany). Specific oligonucleotide primers were synthesized by the Biotechnology Center of the University of Wisconsin-Madison. Steroidogenic acute regulatory protein (STAR); cytochrome P450, family 11, subfamily A, polypeptide 1 (CYP11A1); and hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 (HSD3B7) antibodies (anti-rabbit) were generous gifts from Dr. Leang-Shin Wu and Chich-Hsien Chiu from Taiwan. The PGF (Lutalyse) and intrauterine P4-releasing devices (Eazi-Breed CIDR) were gifts from Pfizer, Inc. (Pfizer Animal Health, New York, NY), and GnRH (Factrel, Fort Dodge, IA) was purchased. Other chemicals and reagents were obtained from Fisher Scientific (Kansas City, MO) or Sigma Chemical Company (St. Louis, MO).
Animals
All procedures were approved by the Animal Care and Use Committee of the College of Agriculture and Life Sciences at the University of Wisconsin-Madison. This experiment was conducted from October 2010 to December 2010, using cows housed at the University of Wisconsin dairy facilities. Nonpregnant, cyclic, lactating Holstein cows with normal estrous cycles were used for this experiment. Animals had ovulation synchronized with the Ovsynch procedure that involves an initial treatment with GnRH followed 7 days later by a PGF treatment, followed at 56 h by a second treatment with GnRH. Day of ovulation was determined by ultrasonography and designated Day 1. Biopsies were performed on Day 9.
Experimental Design
Cows with a mature CL on Day 9 were assigned randomly to three treatment groups, with unequal numbers in each group (1:2:1). All cows received four intrauterine infusions with either 0.25 ml of PBS or PGF (4XPGF; 0.5 mg of PGF in 0.25 ml of PBS) at 6-h intervals (PBS or PGF were infused into the greater curvature of the uterine horn ipsilateral to the CL). Control animals (n = 5) received 0.25 ml of PBS four times, once every 6 h. Cows receiving two infusions of PGF (2XPGF; n = 10) received two PGF treatments at first and third infusions, with PBS treatment at the other two infusions (PGF-6 h-PBS-6 h-PGF-6 h-PBS). It was assumed that the response by this group might be variable, and therefore, more cows were assigned to this treatment group. The cows receiving 4XPGF (n = 5) received four PGF treatments at 6-h intervals. All cows had an ultrasonography-guided biopsy of the CL, collected 30 min after each intrauterine PBS or PGF infusion, and only cows that had at least 10 mg of tissue collected at each of these biopsy sessions were used in the experiment.
Collection of Samples
Luteal biopsy samples were collected by the methods described by Tsai et al. [34]. Briefly, caudal epidural anesthesia was induced with 5 ml of lidocaine hydrochloride (Phoenix Pharmaceutical, Inc., St. Joseph, MO). An ultrasound probe with a needle guide was inserted in the vaginal fornix. The ovary containing the CL was positioned transrectally against the vaginal wall. The needle was then advanced through the vaginal wall and into the CL. The biopsy cutting blade was triggered with excised luteal tissue trapped in the specimen notch. The biopsy tool was removed, and the tissue was inspected to ensure that only luteal tissue was collected; the sample was then washed with PBS and weighed. Part of the biopsy sample was immediately frozen in dry ice and stored at −80°C for later isolation of RNA. The remaining biopsy tissue was fixed in a solution of 4% paraformaldehyde overnight at 4°C and then dehydrated using serial dilutions of methanol (25%, 50%, 75%, and 100%). Dehydrated samples were stored in 100% methanol at −20°C until evaluated by in situ hybridization or immunohistochemistry.
P4 Analyses
To determine changes in P4 concentrations during and after treatment, blood samples were collected from coccygeal vessels before treatment and every 3 h after treatment for 24 h. Blood samples were then collected every 6 h from 24 to 48 h after treatment. Blood samples were allowed to clot and were then centrifuged at 3000 rpm for 20 min. Sera were stored at −20°C until assayed for P4. Concentrations of P4 were quantified using a competitive ELISA method described by Tsai and Wiltbank [17].
RNA Isolation, RT Reaction, and Quantitative PCR
Ten milligrams of luteal tissue were minced with a scalpel and homogenized in TRIzol. Total RNA was then extracted using the manufacturer's protocol (TRIzol; Invitrogen). Integrity of RNA was verified by agarose gel electrophoresis and by optical density at 260/280 nm of 2 ± 0.1 with NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE). Two micrograms of total RNA were treated with DNase I to eliminate genomic DNA contamination, and RNA was then reverse transcribed in the presence of both random hexamer and oligo(dT) primers in equal volumes, using GoScript kit according to the manufacturer's protocol. To clarify RT specificity, all components of the RT kit without RNA (RT negative) were also analyzed.
Sequences for primers were designed using Primer3 from the NCBI (http://www.ncbi.nlm.nih.gov/) gene database or were derived from published studies [16, 22, 35,37]. Primer pair sequences and product sizes are shown in supplemental Table S1 (available online at www.biolreprod.org). All PCR reactions were set up as follows: 5 μl of SYBR Green Master Mix (2×), 2.5 pmol of each primer, 0.5 μl of cDNA, and ddH2O to bring the final volume to 10 μl. Thermal cycling was done by initially incubating the mixture at 50°C for 2 min with subsequent denaturation at 95°C for 10 min. This was followed by 40 cycles of denaturation, annealing, and amplification (95°C for 30 sec, 60°C for 1 min, 72°C for 30 sec). All reactions were carried out on an Opticon2 real-time PCR system (Bio-Rad Life Science, Foster City, CA). Melting curve analysis was performed as follows: 95°C for 1 min, followed by fluorescence measurement performed at 1° increments between 55°C and 95°C. In each run, negative controls with no cDNA template and RT negative controls were included. To verify reaction specificity, amplification products were evaluated after separation on a 2% agarose gel. All samples were evaluated in duplicate for each cDNA. For the determination of the dynamic range and amplification efficiencies of real-time PCR assays for each gene product, amplifications were performed with specific primers in duplicate with a serial dilution of specific pooled cDNA. Efficiencies of qPCR for amplifications were between 95% and 105% for all genes.
In Situ Hybridization and Immunohistochemistry
The first and fourth biopsy samples were evaluated from biopsies in the control and 4XPGF groups for NR4A1 and PTGFR mRNAs by using in situ hybridization. Tissue sectioning and in situ hybridization were conducted as previously described [38] with modifications. Briefly, luteal tissue sections were embedded in a low-melting agarose and cut into 28-μm-thick sections by using a vibrating microtome. Primer sequences for NR4A1 (forward: 5′-ggcatggtgaaggaagttgt-3′; reverse: 5′-cgatgttaatacgactcactataggg-catgtcggtctgtgatgagg-3′; product size: 559 bp) and PTGFR (forward: 5′-ttgccaactggaagaagacc-3′; reverse: 5′-cgatgttaatacgactcactatagggtgagacctgccttgtctgtg-3′; product size: 666 bp) were used to generate DIG-11-UTP-labeled RNA probe templates. After hybridization with DIG-labeled riboprobes, sections were incubated with anti-DIG antibody that was coupled to alkaline phosphatase. To prevent nonspecific binding of anti-DIG-AP antibody, the antibody was preincubated with a protein powder prepared from bovine luteal tissue according to the method described by Abler et al. [38]. To visualize bound probes, BM Purple was used as an alkaline phosphatase chromagen to detect antibody-bound DIG-labeled riboprobes.
For immunohistochemistry, sections were deparaffinized in xylene, rehydrated, and boiled in 10 mM sodium citrate (pH 6.0) for 20 min. Tissues were washed with a solution containing 25 mM Tris-HCl, pH 7.5, 140 mM NaCl, 2.7 mM KCl, and 0.1% Tween-20 (TBSTw), and nonspecific binding sites were blocked for 1 h in TBSTw containing 1% blocking reagent (Roche Diagnostics, Indianapolis, IN), 5% normal goat serum, and 1% bovine serum albumin fraction 5 (RGBTw). Tissue samples were incubated overnight at 4°C with primary antibodies diluted in RGBTw as follows: 1:60 dilution of rabbit anti-STAR, 1:80 dilution of rabbit anti-CYP11A1, and 1:80 dilution of rabbit anti-HSD3B7. After several washes with TBSTw, tissues were incubated for 1 h with RGBTw containing 1:250 dilution of Dylight488-conjugated goat anti-rabbit immunoglobulin G (Jackson Immunoresearch, West Grove, PA). The secondary antibodies were preincubated with luteal protein powder for at least 1 h to prevent nonspecific binding. Labeled tissue sections were counterstained with 4′, 6-diamidino-2-phenylindole dilactate and mounted in anti-fade medium (PBS containing 80% glycerol and 0.2% N-propyl gallate).
Brightfield images were captured using Nikon Eclipse E80i. Fluorescent images were captured using a Nikon Eclipse E600 compound microscope and were merged using NIS elements imaging software (Nikon Instruments Inc., Melville, NY).
Statistical Analysis
Glyceraldehyde-3 phosphate dehydrogenase (GAPDH) was used to normalize the expression of target genes. The use of GAPDH as a housekeeping gene has been previously validated [36], and we further validated the consistency of steady-state concentrations for this gene in the CL in preliminary experiments (unpublished results). This preliminary experiment indicated similar expression of GAPDH after multiple biopsies of the same animal and similar concentrations in the different treatment groups and different collection times in the present experiment. Mean cycle threshold (Ct) values from the first biopsy of the control animals were used as the reference points, and Ct values for each biopsy were used to calculate the fold changes from the reference points for each biopsy (including original control values at first biopsy) using reference point Ct values according to the 2−ΔΔCt methods described by Livak and Schmittgen [39]. Fold-change values that reflected the steady-state mRNA concentrations were then analyzed by one-way ANOVA using the general linear model procedure of the Statistical Analysis System (version 9.2). Differences among the means were separated by Fischer least significant difference test. Data were considered statistically significant when P values were lower than 0.05. Data in text are expressed as fold change from controls (increased) or percentage decrease as compared to controls.
RESULTS
Concentrations of Circulating P4 after Treatments
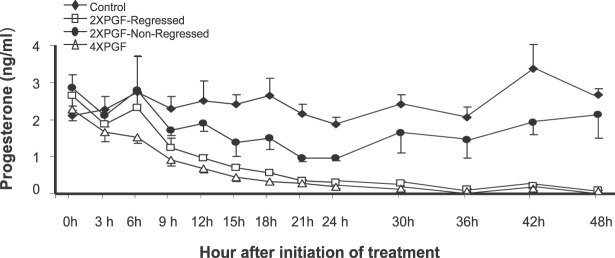
Circulating P4 concentrations were similar in all groups before treatments, although numerically, the control group was lowest, and the 2XPGF-non-regressed and 2XPGF-regressed groups were highest (Fig. 1). Circulating concentrations of P4 in control animals did not change significantly during or by 48 h after the intrauterine PBS treatments. P4 concentrations in the 4XPGF group decreased significantly compared to those in the control group by 9 h after first PGF infusion and at all subsequent time points (P < 0.05). Cows in the 2XPGF group could be divided into two distinct groups. One group of 2XPGF cows (n = 5) had luteal regression that was similar to that in the 4XPGF group with decreasing P4 by 9 h after first PGF treatment and at all subsequent times (P < 0.05). Another group of 2XPGF cows (n = 5) could be classified as not regressing during the experiment. Despite initial decreases in circulating P4 in this group, P4 concentrations returned to normal and were not significantly different from those of controls at 30 to 48 h after initial PGF treatment. Nevertheless, both 2XPGF groups showed a numerical decline in P4 after the saline infusion at 6 h, suggesting that endogenous PGF may have been released at this time due to the initial PGF infusion. The 2XPGF-regressed cows continued the downward trend for P4 from 9 to 12 h, even though no further PGF treatments were given, whereas the 2XPGF-non-regressed cows recovered P4 levels between 9 and 12 h after the first PGF infusion. The 2XPGF-non-regressed group remained intermediate between the control and two groups of cows that regressed following PGF infusion throughout the remainder of the sampling period.
FIG. 1.
Effect of intrauterine PGF or PBS treatment on circulating P4 concentration in cows. Data are presented as means ± SEM.
Effect of PGF on mRNA for Immediate Early Genes (JUN, FOS, NR4A1, EGR1)
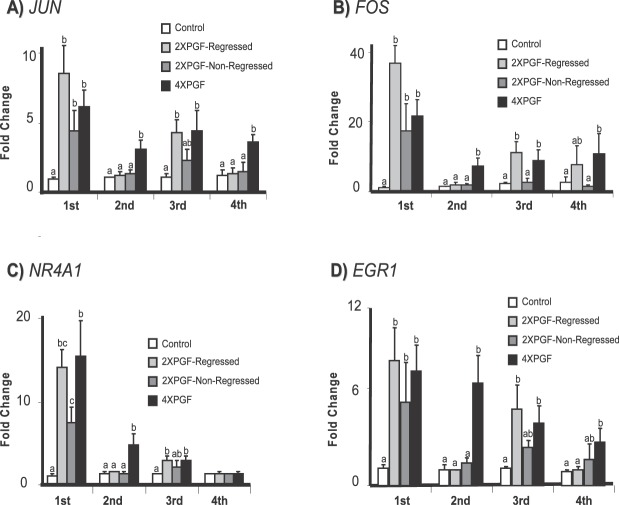
Steady-state concentrations of mRNA for immediate early genes (JUN, FOS, NR4A1, EGR1) in the biopsy samples taken during the treatments are shown in Figure 2. Concentrations of mRNA for JUN, FOS, NR4A1, and EGR1 increased significantly at the first biopsy (30 min after PGF infusion) for all three treatments groups that received PGF infusion. The increase after the initial PGF treatment generally exhibited the greatest magnitude compared to subsequent increases. All three PGF treatments exhibited similar increases for JUN (6.2 ± 1.2-fold for 4XPGF, 8.6 ± 1.9-fold for 2XPGF-regressed, 4.3 ± 1.4-fold for 2XPGF-non-regressed), FOS (21.5 ± 4.7-fold for 4XPGF, 36.7 ± 5-fold for 2XPGF-regressed, 17.5 ± 7.5-fold for 2XPGF-non-regressed), and EGR1 (7.1 ± 1.6-fold for 4XPGF, 7.7 ± 2.05-fold for 2XPGF-regressed, 5.1 ± 2.6-fold for 2XPGF-non-regressed); however, NR4A1 had a greater increase in 4XPGF (15.5 ± 4.2-fold) than 2XPGF-non-regressed (7.6 ± 1.8-fold), with 2XPGF-regressed being intermediate (14.2 ± 2.3-fold). At the second luteal biopsy only the 4XPGF group had increased mRNA concentrations for JUN, FOS, NR4A1, and EGR1, while the other three groups that were all treated with PBS at 30 min before this biopsy showed no differences in concentrations for any of these mRNA. At the third biopsy, the 4XPGF and 2XPGF-regressed groups had increased mRNA for all four immediate early genes; whereas the 2XPGF-non-regressed group, despite receiving PGF prior to this biopsy, similar to the 4XPGF and 2XPGF-regressed cows, appeared to have a reduced response to this treatment. At the fourth biopsy, there were increased mRNA for JUN, FOS, and EGR1 in the 4XPGF group but not in the other three groups. Surprisingly, there was no increase in NR4A1 after the fourth PGF treatment (Fig. 2C).
FIG. 2.
Effect of intrauterine PGF or PBS treatment on steady-state mRNA concentrations for A) JUN (jun proto-oncogene), B) FOS (FBJ murine osteosarcoma viral oncogene homolog), C) NR4A1 (nuclear receptor subfamily 4, group A, member 1), and D) EGR1 (early growth response 1). Data are shown as fold changes ± SEM. Columns with different letters (a, b, c) at each biopsy indicate differences; P < 0.05. First, second, third, and fourth indicate biopsy times.
Effect of PGF on mRNA for Steroidogenic Genes (STAR, CYP11A1, NR5A1, LHCGR)
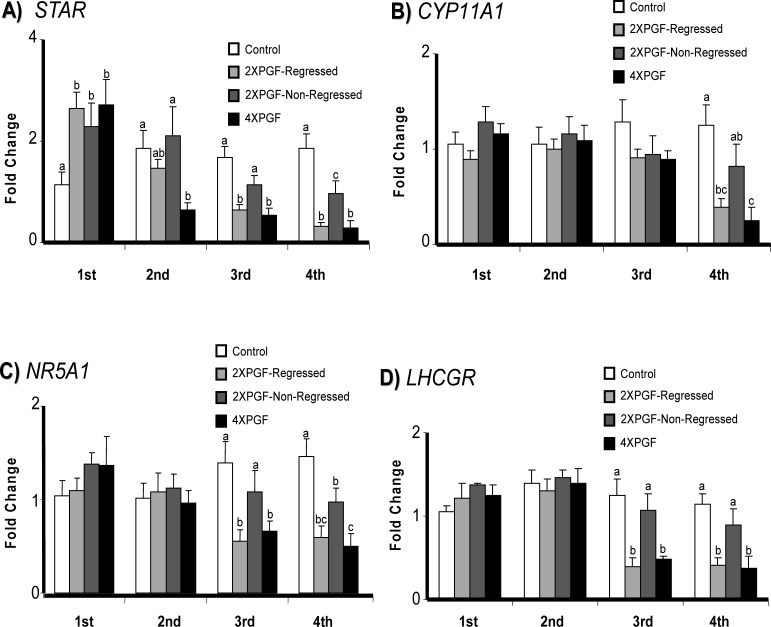
The steady-state concentrations of luteal mRNA for various steroidogenic genes (STAR, CYP11A1, NR5A1, and LHCGR) are shown in Figure 3. Surprisingly, STAR mRNA expression was increased at the first biopsy for all three treatments groups that received PGF infusion. In contrast, expression of STAR was decreased (66%) at the second biopsy in the 4XPGF group and decreased in both the 4XPGF (69%) and 2XPGF-regressed (63%) groups at the third biopsy. At the fourth biopsy, there was a decreased expression of STAR mRNA for the 4XPGF (84%) and 2XPGF-regressed (82%) groups, with the 2XPGF-non-regressed group having an intermediate value for STAR mRNA (49%). Messenger RNA concentrations for NR5A1 and LHCGR were decreased at the third and fourth biopsies for 2XPGF-regressed and 4XPGF compared to those of control. At the fourth biopsy, the 2XPGF-non-regressed group had decreased mRNA concentration for NR5A1 but no change in LHCGR and CYP11A1 mRNA. Meanwhile, mRNA expression for CYP11A1 was decreased only at the fourth biopsy in the two groups that had luteal regression, 2XPGF-regressed and 4XPGF.
FIG. 3.
Effect of intrauterine PGF or PBS treatment on steady-state mRNA concentrations for A) STAR (steroidogenic acute regulatory protein), B) CYP11A1 (cytochrome P450, family 11, subfamily A, polypeptide 1), C) NR5A1 (nuclear receptor subfamily 5, group A, member 1), and D) LHCGR (luteinizing hormone/choriogonadotropin receptor). Data are shown as fold changes ± SEM. Columns with different letters (a, b, c) at each biopsy indicate differences; P < 0.05. First, second, third, and fourth indicate biopsy times.
Effect of PGF on mRNA for Prostaglandin-Related Genes (PTGS2, PTGFS, HPGD, PTGFR)
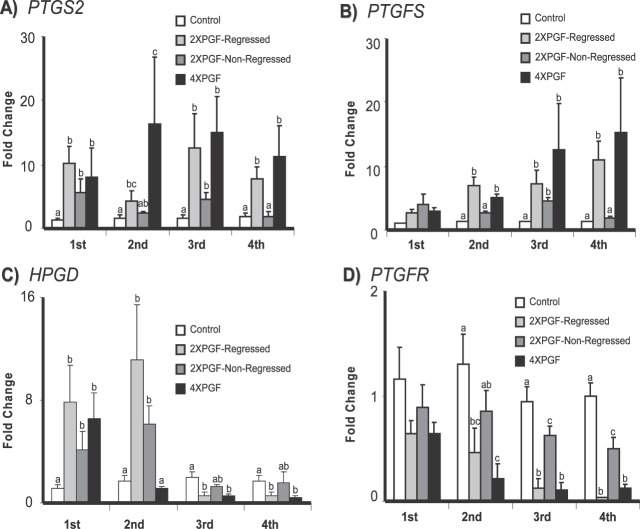
The steady-state concentrations of mRNA for prostaglandin-related genes (PTGS2, PTGFS, HPGD, and PTGFR) are shown in Figure 4. There were dramatic increases in mRNA concentrations for PTGS2 at all four biopsies taken from 2XPGF-regressed and 4XPGF groups. The 2XPGF-non-regressed CL also had an increase in PTGS2 mRNA at the first and third biopsies but not at the second and fourth biopsies. Expression of PTGFS increased at the second biopsy and remained at higher concentrations in both 2XPGF-regressed and 4XPGF groups; however, PTGFS increased only at the third biopsy for 2XPGF-non-regressed group.
FIG. 4.
Effect of intrauterine PGF or PBS treatment on steady-state mRNA concentrations for A) PTGS2 (prostaglandin-endoperoxide synthase 2), B) PTGFS (prostaglandin F synthase), C) HPGD (hydroxyprostaglandin dehydrogenase 15-[NAD]), and D) PTGFR (prostaglandin F receptor). Data are shown as fold changes ± SEM. Columns with different letters (a, b, c) at each biopsy indicate differences, P < 0.05. First, second, third, and fourth indicate biopsy times.
The pattern for HPGD expression was very different from our expectations (Fig. 4C). At the first biopsy, there was a similar increase in steady-state mRNA concentrations for HPGD in all three treatment groups (6.5 ± 2.05-fold for 4XPGF, 7.9 ± 2.8-fold for 2XPGF-regressed, 4.1 ± 1.5-fold for 2XPGF-non-regressed). However, at the second biopsy, mRNA concentrations for HPGD remained elevated in the 2XPGF-regressed and 2XPGF-non-regressed groups, but HPGD mRNA decreased to control concentration levels for the 4XPGF group. At the third and fourth biopsies, HPGD mRNA concentrations were much lower than those of controls in the 2XPGF-regressed and 4XPGF groups but not in the 2XPGF-non-regressed group.
Concentrations of mRNA for PTGFR showed numerical decreases at the first biopsy (44% and 45%) and significant decreases compared to controls at the second (65% and 84%), third (87% and 89%), and fourth (97% and 90%) biopsies in the regressed groups (2XPGF-regressed and 4XPGF, respectively). The 2XPGF-non-regressed group had decreased PTGFR mRNA only at the third and fourth biopsies, and the decreases were much less dramatic than those observed in the regressed groups (34% at third, 52% at fourth biopsies).
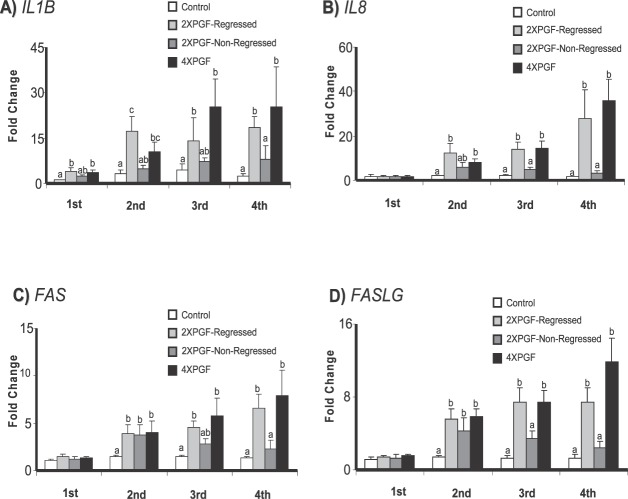
Effect of PGF on mRNA for Immune-Related Genes (IL1B, IL8, FAS, FASLG)
The concentrations of mRNA for immune-related genes (IL1B, IL8, FAS, and FASLG) showed steady increases after PGF treatments in treatment groups that resulted in regression of the CL, as shown in Figure 5. mRNA concentrations for all four immune-related genes were increased at the second, third, and fourth biopsies for the regressed groups (2XPGF-regressed and 4XPGF). At the fourth biopsy, there was maximal up-regulation of IL1B (25.4 ± 12.9-fold for 4XPGF, 18.4 ± 3.8-fold for 2XPGF-regressed), IL8 (36 ± 9.7-fold for 4XPGF, 28.6 ± 12.7-fold for 2XPGF-regressed), FAS (7.9 ± 2.7-fold for 4XPGF, 6.6 ± 1.5-fold for 2XPGF-regressed), and FASLG (11.9 ± 2.6-fold for 4XPGF, 7.5 ± 1.4-fold for 2XPGF-regressed). In addition, the regressed groups had a slight but significant increase in IL1B at the first biopsy (3.4 ± 0.9-fold for 4XPGF, 4.1 ± 1.1-fold for 2XPGF-regressed). In the 2XPGF-non-regressed group there was no significant increase in mRNA for IL1B or IL8 in any of the biopsy samples and only a significant increase in mRNA for FAS and FASLG in the second biopsy and not in any of the other biopsy samples.
FIG. 5.
Effect of intrauterine PGF or PBS treatment on steady-state mRNA concentrations for A) IL1B (interleukin 1, beta), B) IL8 (interleukin 8), C) FAS (Fas, TNF receptor superfamily, member 6), D) FASLG (Fas ligand). Data are shown as fold changes ± SEM. Columns with different letters (a, b, c) at each biopsy indicate differences, P < 0.05. First, second, third, and fourth indicate biopsy times.
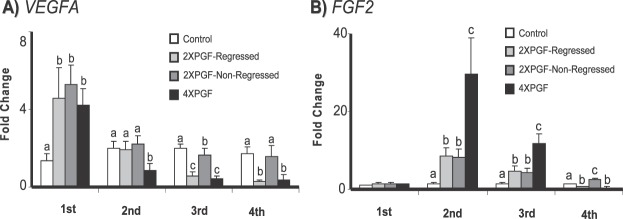
Effect of PGF on mRNA for Angiogenesis-Related Genes (VEGFA, FGF2)
The steady-state concentrations of mRNA for the two genes related to angiogenesis (VEGFA, FGF2) showed distinct patterns of changes, as shown in Figure 6. The VEGFA mRNA concentrations increased at the first biopsy for all three treatment groups (4.2 ± 0.9-fold for 4XPGF, 4.6 ± 1.6-fold for 2XPGF-regressed, 5.3 ± 1-fold for 2XPGF-non-regressed). At the second biopsy, VEGFA mRNA decreased in the 4XPGF group (48%) but not in other groups. At the third biopsy, there were decreased VEGFA mRNA concentrations for the 4XPGF (80%) and 2XPGF-regressed (72%) groups with continued decreases in VEGFA mRNA concentrations at the fourth biopsy in the regressing groups. In the 2XPGF-non-regressed group the VEGFA mRNA concentrations were similar to those of controls, except at the third biopsy when a slight but significant decrease was observed (20%).
FIG. 6.
Effect of intrauterine PGF or PBS treatment on steady-state mRNA concentrations for A) VEGFA (vascular endothelial growth factor A), B) FGF2 (fibroblast growth factor 2). Data are shown as fold changes ± SEM. Columns with different letters (a, b, c) at each biopsy indicate differences, P < 0.05. First, second, third, and fourth indicate biopsy times.
The FGF2 mRNA concentrations were not altered after PGF treatment at the first biopsy; however all three treatment groups had increased mRNA at the second biopsy, with the 4XPGF group having a greater increase (29.5 ± 9.2-fold) than the other two groups. Similarly, at the third biopsy, all three treatment groups had greater FGF2 mRNA concentrations than controls, with the 4XPGF greater than the other two groups. By the fourth biopsy, the FGF2 mRNA concentrations had decreased dramatically in the 2XPGF-regressed (53%) and the 4XPGF (65%) groups but remained elevated in the 2XPGF-non-regressed group (2.4 ± 0.5-fold).
In Situ Hybridization and Immunohistochemistry
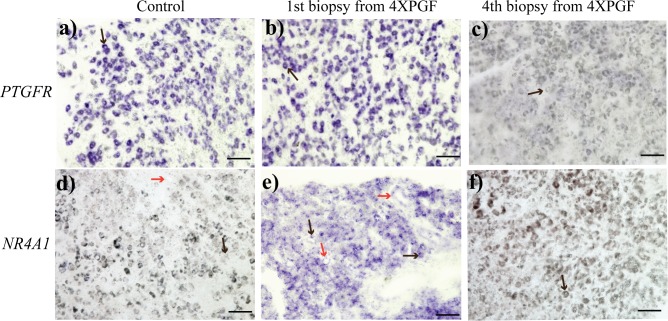
The mRNA for PTGFR was clearly localized to large luteal cells in the control biopsy sample (Fig. 7a). At 0.5 h after PGF, there was no difference in PTGFR mRNA with clear localization in the large luteal cells (Fig. 7b). After the fourth PGF treatment, there was no PTGFR mRNA detectable by in situ hybridization (Fig. 7c).
FIG. 7.
Representative images of in situ hybridization staining for PTGFR (prostaglandin F receptor, a, b, c) and NR4A1 (nuclear receptor subfamily 4, group A, member 1, d, e, f) in bovine CL in control and at first, and fourth biopsies from 4XPGF group. Large cells (black arrow) and vascular endothelial cells (red arrow) are shown. Bars = 100 μm.
There was no detectable NR4A1 mRNA in control tissues that were not treated with PGF (Fig. 7d). At 0.5 h after PGF treatment, there was substantial NR4A1 mRNA localized throughout the luteal tissue. Although localization to large luteal cells was most dramatic, there also appeared to be localization in some capillary endothelial cells (Fig. 7e). After the fourth PGF treatment, NR4A1 mRNA was not detectable by in situ hybridization (Fig. 7f).
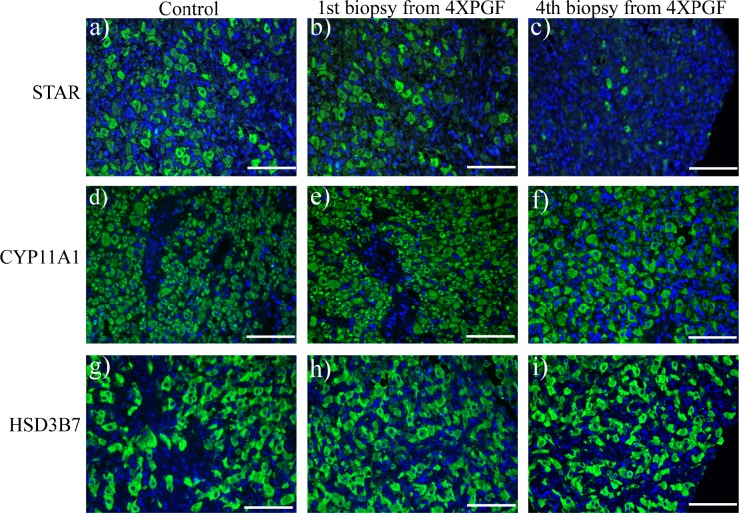
The STAR, CYP11A1, and HSD3B7 proteins could be readily visualized in cells of the CL (Fig. 8). The STAR protein was particularly apparent in large luteal cells and also appeared to be present in small luteal cells but was not visualized in endothelial cells (Fig. 8, a and b). After the fourth PGF treatment, there was very little STAR protein that could be visualized in the CL (Fig. 8c). The CYP11A1 and HSD3B7 proteins were localized in both large and small luteal cells but not in capillary endothelial cells. There were no changes in CYP11A1 or HSD3B7 protein expression levels that could be grossly observed by visual inspection of immunohistochemistry after the fourth PGF infusions (Fig. 8, f and i).
FIG. 8.
Representative images of immunofluorescent staining for STAR (steroidogenic acute regulatory protein a, b, c), CYP11A1 (cytochrome P450, family 11, subfamily A, polypeptide 1, d, e, f), and HSD3B7 (hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7, g, h, i) in bovine CL at control (30 min after saline), and at first and fourth biopsies from 4XPGF group. Labeled tissue sections were counterstained with 4′, 6-diamidino-2-phenylindole dilactate (blue staining). Bars = 100 μm.
DISCUSSION
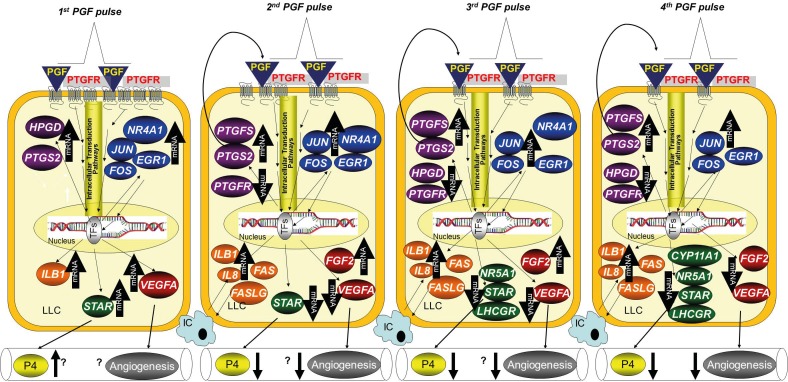
This study used intrauterine infusion of low doses of PGF to mimic the pulsatile secretion of PGF that accompanies normal luteolysis. The intrauterine infusion of 0.5 mg of PGF produced hormonal patterns similar to natural PGF pulses observed during luteolysis in previous experiments [11], although it remains to be determined whether the timing of PGF infusion used in the current experiment mimics natural luteolysis. The four luteal biopsy samples that were collected during this experiment allowed us to carefully monitor luteal gene expression following each pulse of PGF. Clearly there were distinct patterns of gene expression induced by each individual PGF pulse. The control group had the same quantity of PBS infused into the uterus and a biopsy schedule similar to that of the treatment groups. The observation that there were no substantial changes in any of the 18 mRNAs during any of the four biopsies provides some confidence that the biopsy and uterine treatments alone do not induce the observed luteal responses, consistent with previous results [11, 34]. We included a group of cows that received only two doses of PGF in order to observe responses to PGF that did not result in complete luteal regression. Some of the cows in this group reacted as expected with a partial decrease in P4 concentrations at 9 to 24 h after the first PGF treatment and a subsequent rebound in circulating P4 to near control concentrations. Unexpectedly, some of the cows treated with two doses of PGF underwent complete luteal regression of P4 concentrations from 12 to 48 h after the first PGF that were similar to those of cows receiving four doses of PGF. Thus, our experimental design allowed us to determine the temporal pattern of luteal gene expression associated with each specific PGF pulse and to differentiate PGF pulse-induced changes that were associated or not associated with complete luteal regression. We have summarized our findings using a physiological model that can guide a discussion of potential mechanisms that accompany natural luteolysis (Fig. 9).
FIG. 9.
Schematic model shows changes in mRNAs following the four PGF pulses associated with luteolysis. Following the first PGF pulse, there were increases in mRNA for specific genes in all five pathways that were examined: immediate early genes (blue), PGF-related genes (purple), immune-related genes (orange), steroidogenic genes (green), and angiogenesis genes (red). Increases were not specific for luteal regression as similar increases were observed in CL that subsequently underwent or did not undergo regression. The second PGF pulse also increased the immediate early genes, but this pulse induced decreases in VEGFA and STAR. These changes were specific for the second PGF pulse, as they did not happen in CL that were only exposed to a single PGF pulse. After the third PGF pulse, there were dramatic changes in mRNA, consistent with a course that would result in luteolysis. Most of these changes did not happen in CL that were not destined for luteolysis, even though they also received the second PGF pulse at 30 min prior to the third biopsy. The fourth biopsy continued the luteolytic cascade with increases or decreases in essentially all genes in a luteolytic direction that was specific for CL that were undergoing the full luteolytic cascade. Names of genes are defined in the text. An up arrow (↑) indicates up-regulation; a down arrow (↓) indicates down-regulation; IC, immune cell; LLC, large luteal cells; TFs, transcription factors; P4, progesterone. The question marks indicate possible or unknown outcomes such as increased P4 following STAR or changes in angiogenesis after changes in VEGFA or FGF2.
After the first treatment with PGF (Fig. 9, first biopsy), there were similar responses in all three of the treatment groups. A single receptor for PGF has been identified, and the initial actions of PGF appear to be due to binding of PGF to this specific G-coupled receptor (PTGFR) on the plasma membrane of luteal cells [40, 41]. Use of in situ hybridization (Fig. 8) indicated that that the PTGFR was primarily, if not exclusively, localized to large luteal cells. The concentration of PTGFR mRNA and protein are extremely high in CL of all species that have been evaluated, and binding of PGF to this receptor is likely to initiate the cascade of events resulting in altered expression of multiple genes. Binding of PGF to the PTGFR increases free intracellular calcium and activity of phospholipase C, protein kinase C (PKC), and mitogen-activated protein (MAP) kinase, with subsequent activation of multiple transcription factors [42,45]. These signaling pathways, particularly activated ELK1 and ERK signaling, are involved in inducing a rapid and dramatic increase in the immediate early genes such as FOS, JUN, NR4A1, and EGR1 [46,49] that were evaluated in this study. Multiple hormones and other signaling molecules are known to rapidly induce these immediate early genes, with proposed involvement in a remarkable diversity of cellular responses [50, 51]. For example, immediate early genes have been proposed as critical mediators of apoptotic signaling cascades [52] as well as growth and mitosis pathways [53]. In the CL, it has been reported that EGR1 has an important role in expression of genes whose cognate proteins regulate luteal regression (e.g., EGR1 [54]). FOS and JUN proteins, as constituents of the activator protein-1 (AP-1) transcription factor complex, appear to be involved in programming gene expression during PGF-induced luteolysis [47]. Other studies have previously shown that these immediate early genes increase after a supraphysiological dose of PGF [54, 55], and our study confirmed the fact that these genes are also induced by low doses of PGF delivered by intrauterine infusion. After the second infusion (Fig. 9), only the group (4XPGF) that was treated with PGF had an increase in these immediate early genes. Similarly, at the third and fourth biopsies, only cows that had been treated with PGF immediately before the biopsy had an increase in immediate early gene concentrations, indicating the acute role of PGF in induction of these genes. The magnitude of the increase in immediate early genes was generally greatest at the first PGF treatment and was reduced after subsequent PGF treatments. This may be due to the clear reduction in expression of PTGFR following PGF treatment or due to other intracellular mechanisms that were down-regulated by prior PGF treatments. Reduction in PTGFR expression levels after PGF treatment is consistent with findings from other reports that used supraphysiological doses of PGF [16, 17]. However, the observation that even low doses of PGF decrease PTGFR expression is in apparent conflict with the idea that prior exposure to low doses of PGF can increase subsequent luteolytic responses to a pulse of PGF. In contrast, the CL of other species have either an increase [56, 57] or no change [58] in luteal PTGFR expression after PGF treatment. Somewhat surprisingly, in our study, the cows that did not regress after PGF treatment (2XPGF-non-regressed) had much less of a reduction in PTGFR mRNA (Fig. 4D) and generally a reduced magnitude of immediate early gene induction after PGF infusion. This may indicate that less PGF reached the CL after intrauterine PGF infusion in these cows or that these cows were less responsive to PGF that reached the CL. Although the precise role for these immediate early genes in luteolysis could not be determined in our experiment, our results are consistent with a clear induction of these genes after each PGF pulse and a role for these genes, probably due to their role as transcription factors, in PGF-induced luteolysis.
Five other genes were also found to have an increase in their mRNA at 0.5 h after PGF infusion. Three of these mRNA (STAR, HPGD, and VEGFA) were increased at the early times after PGF, with a later reduction in their mRNA following subsequent PGF pulses. This was a surprising and unexpected pattern that suggests complex regulatory mechanisms for these genes during multiple PGF pulses. Induction of these mRNA at only 0.5 h after PGF infusion suggests involvement of regulatory mechanisms similar to those found for the acute induction of the immediate early genes discussed above. Nevertheless, acute PGF-induced inhibition of degradation of these specific mRNA is another mechanism that could also produce an acute increase in these mRNA. The subsequent profound reduction in these three mRNA indicates activation of other regulatory mechanisms down-regulating these mRNA. It is noteworthy that these reductions occurred only after a second PGF treatment (second biopsy for 4XPGF group or third biopsy for 2XPGF group). Furthermore, the CL that did not undergo subsequent regression had either little or no reduction in these mRNA following the second PGF treatment, although the first PGF treatment produced similar increases in STAR, HPGD, and VEGFA mRNA in all three groups.
The STAR, HPGD, and VEGFA genes encode proteins that are critical for maintenance of luteal function. For example, the STAR protein mediates cholesterol transport from the outer to the inner mitochondrial membrane, the key rate-limiting step for production of most steroid hormones, including P4 [59]. Similarly, it has been reported that PGF treatment causes a transient increase in circulating P4 followed by a decrease [25, 29, 60]. While it seems unlikely that the acute induction of STAR mRNA is involved in the acute increase in circulating P4 (translation of mRNA to functional protein would be expected to take a longer period), it seems possible that similar mechanisms may mediate both increases. The dramatic decrease in STAR mRNA after the second PGF pulse (second biopsy in 4XPGF or third biopsy in 2XPGF-regression) is consistent with observations from other research [61] and with the decrease in STAR-mediated cholesterol transport that is a key rate-limiting step in the decreased P4 production during luteolysis. The mRNA for other steroidogenesis-related mRNA were not decreased until the later times, with CYP11A1 lower only at the fourth biopsy and NR5A1 and LHCGR lower only at third and fourth biopsies. Nevertheless, the lack of a decrease in these mRNA in the 2XPGF-non-regressed CL suggests that decreases in these mRNA may have a role in luteolysis.
The product of the HPGD gene (formerly known as PGDH) metabolizes PGF into 13,14-dihydro-15-keto-PGF2α (PGFM), thought to be an inactive metabolite. Thus, intraluteal PGF could be inactivated by HPGD and thus protect normal luteal function. A previous study [62] reported the greatest HPGD activity in the early CL and the pregnant CL, which are relatively resistant to PGF action. Thus, the initial induction of HPGD followed by a dramatic inhibition may produce an initial luteal protection followed by an increase in luteal sensitivity to PGF due to lack of intraluteal PGF inactivation.
Finally, the third mRNA with a distinct induction followed by inhibition was VEGFA, a growth factor that is critical for induction of angiogenesis. More than 50% of the cells in a mature CL are endothelial cells [1], and inhibition of VEGF leads to inadequate CL microvasculature and inadequate luteal function [63]. Again, the initial induction of VEGFA is perplexing. Previous studies using a single, high dose of PGF have reported either an increase [64] or decrease [16] in VEGFA mRNA at earlier times after PGF. The later reduction in VEGFA mRNA is consistent with a previous report [18]. The FGF2 mRNA encodes another protein that is involved in angiogenesis, and the pattern of expression following PGF also demonstrates a pattern of initial induction followed by inhibition. Nevertheless, the initial induction of FGF2 was not observed at 0.5 h after PGF and was observed only at the second biopsy (6.5 h after initial PGF). Intriguingly, the second PGF infusion acutely stimulated a greater induction of FGF2 (Fig. 6, compare 2XPGF to 4XPGF), implying that the second PGF regulates immediate regulatory mechanisms involved in FGF2 regulation. The subsequent inhibition, while profound, was not observed until the fourth biopsy. Similar profiles for FGF2 mRNA were previously reported in sheep and cow CL during luteolysis [18, 64].
The other two mRNA that were induced at 0.5 h after PGF (PTGS2 and IL1B) continued to be induced by PGF at all biopsies following the initial PGF pulse. Both of these mRNA were also less profoundly induced in animals that did not undergo luteal regression during the treatments (2XPGF-non-regressed) than in the other two PGF treatment groups. Induction of PTGS2 mRNA in CL by higher doses of PGF has been demonstrated in every species that has been evaluated [24, 65,67]. It has been proposed that PGF-induced PTGS2 expression represents an intraluteal autoamplification pathway that increases intraluteal PGF following exposure to the low amounts of PGF that reach the CL during the PGF pulses associated with luteolysis [68]. Our results are clearly consistent with the luteal PGF autoamplification concept due to the induction PTGS2. Furthermore, the induction of PTGFS mRNA, albeit at later times after PGF treatment, would also tend to increase intraluteal PGF synthesis. Again, the CL that did not undergo luteolysis following the two doses of PGF had a greatly reduced induction of PTGFS mRNA. Thus, the induction of PTGS2 and PTGFS combined with the reduction in HPGD would all lead to increased intraluteal PGF and would amplify the PGF signal at later PGF pulses associated with natural luteolysis.
All of the immune-related mRNA increased after PGF pulses, although IL1B was the only mRNA that was increased at 0.5 h after PGF. All of the immune-related mRNAs that we examined were increased by 6.5 h after the first PGF pulse, with increasing magnitude of expression with increasing time. Most investigators have found an increase in lymphocytes and macrophages in the CL at the time of luteolysis [14, 69,71]. Thus, increases in immune-related mRNA may be the result of increased numbers of immune cells in the CL or greater induction of these mRNA in immune cells or other cells that are already present in the CL. Recent research shows findings consistent with activation of the immune system during luteolysis [14, 72] and with a functional role for the immune system in luteal regression [73]. Further studies are needed to define the precise cellular localization and physiological role of each of the immune-related molecules in luteal regression. Consistent with a role for these molecules in luteal regression, there was a reduced induction of immune-related mRNA in CL that did not undergo luteolysis. This difference is particularly stark at the later times with all four immune-related mRNA concentrations similar in CL from the 2XPGF-non-regressed and control groups (Fig. 5).
Thus, our results are consistent with changes in four key regulatory pathways in the CL being increasingly shunted into a luteolytic direction with increasing numbers of low dose PGF pulses. This is illustrated in Figure 9. For example, the P4 synthesis pathway is increasingly inhibited at later times after the initial PGF treatments, probably primarily due to changes in STAR and cholesterol transport. The lack of substantial decreases in the two key proteins involved in P4 production, CYP11A and HSD3B7, as determined by immunohistochemistry, indicates the CL still has functional capacity for P4 synthesis, even at the time of the fourth luteal biopsy (18.5 h after initial PGF) when circulating P4 is low. The dramatic decrease in the two key angiogenic mRNA by the fourth biopsy is consistent with multiple PGF pulses causing regression of the luteal vasculature. Although the decrease in PTGFR would tend to reduce luteal PGF sensitivity, all of the other genes involved in PG pathways that were examined would tend to increase intraluteal PGF concentrations and move this pathway in a luteolytic direction. Similarly the increase in immune-related mRNA is consistent with multiple PGF pulses activating this luteolytic pathway. For almost all of the mRNA, PGF responses were absent or less profound after the second PGF treatment in CL that recovered normal function after PGF pulses compared to CL that underwent regression after two or four PGF pulses. Generally, most of these intracellular mechanisms would tend to prevent luteal regression following a single exposure to a low dose PGF pulse with the surprising finding of acute and dramatic increases in a multitude of mRNA after a single PGF pulse even in CL that are not destined for regression. However, this initial PGF pulse appeared to amplify the luteolytic effects of subsequent low amplitude PGF pulses in CL that subsequently underwent regression. These results are likely to have important implications for the mechanisms involved in normal luteolysis.
Supplementary Material
ACKNOWLEDGMENT
We thank Jerry N. Guenther, Dr. Kishore K. Baruah, Paulo D. Carvalho, Murillo A.P. Meschiatti, Wolff C. Filho, and Katie S. Hackbart for technical assistance with luteal biopsies and blood collection. In addition, we appreciate the assistance of Josie A. Lewandowski for laboratory analysis of P4.
Footnotes
Supported by National Institutes of Health grant R01 HD050616, the Wisconsin Experiment Station, and The Council of Higher Education, Turkey.
REFERENCES
- O'Shea JD, Rodgers RJ, D'Occhio MJ. Cellular composition of the cyclic corpus luteum of the cow. J Reprod Fertil 1989; 85: 483 487 [DOI] [PubMed] [Google Scholar]
- Wiltbank MC. Cell types and hormonal mechanisms associated with mid-cycle corpus luteum function. J Anim Sci 1994; 72: 1873 1883 [DOI] [PubMed] [Google Scholar]
- Niswender GD, Juengel JL, McGuire WJ, Belfiore CJ, Wiltbank MC. Luteal function: the estrous cycle and early pregnancy. Biol Reprod 1994; 50: 239 247 [DOI] [PubMed] [Google Scholar]
- Niswender GD, Juengel JL, Silva PJ, Rollyson MK, McIntush EW. Mechanisms controlling the function and life span of the corpus luteum. Physiol Rev 2000; 80: 1 29 [DOI] [PubMed] [Google Scholar]
- Schams D, Berisha B. Regulation of corpus luteum function in cattle—an overview. Reprod Domest Anim 2004; 39: 241 251 [DOI] [PubMed] [Google Scholar]
- McCracken JA, Custer EE, Lamsa JC. Luteolysis: a neuroendocrine-mediated event. Physiol Rev 1999; 79: 263 323 [DOI] [PubMed] [Google Scholar]
- Kindahl H, Edqvist LE, Granstrom E, Bane A. Release of prostaglandin-F2alpha as reflected by 15-keto-13,14-dihydroprostaglandin F2alpha in peripheral-circulation during normal luteolysis in heifers. Prostaglandins 1976; 11: 871 878 [DOI] [PubMed] [Google Scholar]
- Mann GE, Lamming GE. Timing of prostaglandin F-2 alpha release episodes and oxytocin receptor development during luteolysis in the cow. Anim Reprod Sci 2006; 93: 328 336 [DOI] [PubMed] [Google Scholar]
- Silvia WJ, Lewis GS, McCracken JA, Thatcher WW, Wilson L., Jr Hormonal regulation of uterine secretion of prostaglandin F2 alpha during luteolysis in ruminants. Biol Reprod 1991; 45: 655 663 [DOI] [PubMed] [Google Scholar]
- Ginther OJ, Silva LA, Araujo RR, Beg MA. Temporal associations among pulses of 13,14-dihydro-15-keto-PGF2alpha, luteal blood flow, and luteolysis in cattle. Biol Reprod 2007; 76: 506 513 [DOI] [PubMed] [Google Scholar]
- Ginther OJ, Araujo RR, Palhao MP, Rodrigues BL, Beg MA. Necessity of sequential pulses of prostaglandin F2alpha for complete physiologic luteolysis in cattle. Biol Reprod 2009; 80: 641 648 [DOI] [PubMed] [Google Scholar]
- Penny LA, Armstrong DG, Baxter G, Hogg C, Kindahl H, Bramley T, Watson ED, Webb R. Expression of monocyte chemoattractant protein-1 in the bovine corpus luteum around the time of natural luteolysis. Biol Reprod 1998; 59: 1464 1469 [DOI] [PubMed] [Google Scholar]
- Arosh JA, Banu SK, Chapdelaine P, Madore E, Sirois J, Fortier MA. Prostaglandin biosynthesis, transport, and signaling in corpus luteum: a basis for autoregulation of luteal function. Endocrinology 2004; 145: 2551 2560 [DOI] [PubMed] [Google Scholar]
- Penny LA, Armstrong D, Bramley TA, Webb R, Collins RA, Watson ED. Immune cells and cytokine production in the bovine corpus luteum throughout the oestrous cycle and after induced luteolysis. J Reprod Fertil 1999; 115: 87 96 [DOI] [PubMed] [Google Scholar]
- Berisha B, Meyer HH, Schams D. Effect of prostaglandin F2 alpha on local luteotropic and angiogenic factors during induced functional luteolysis in the bovine corpus luteum. Biol Reprod 2010; 82: 940 947 [DOI] [PubMed] [Google Scholar]
- Shirasuna K, Sasahara K, Matsui M, Shimizu T, Miyamoto A. Prostaglandin F2alpha differentially affects mRNA expression relating to angiogenesis, vasoactivation and prostaglandins in the early and mid corpus luteum in the cow. J Reprod Dev 2010; 56: 428 436 [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Wiltbank MC. Prostaglandin F2alpha regulates distinct physiological changes in early and mid-cycle bovine corpora lutea. Biol Reprod 1998; 58: 346 352 [DOI] [PubMed] [Google Scholar]
- Neuvians TP, Berisha B, Schams D. Vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) expression during induced luteolysis in the bovine corpus luteum. Mol Reprod Dev 2004; 67: 389 395 [DOI] [PubMed] [Google Scholar]
- Neuvians TP, Pfaffl MW, Berisha B, Schams D. The mRNA expression of the members of the IGF-system in bovine corpus luteum during induced luteolysis. Domest Anim Endocrinol 2003; 25: 359 372 [DOI] [PubMed] [Google Scholar]
- Kliem H, Berisha B, Meyer HH, Schams D. Regulatory changes of apoptotic factors in the bovine corpus luteum after induced luteolysis. Mol Reprod Dev 2009; 76: 220 230 [DOI] [PubMed] [Google Scholar]
- Kliem H, Welter H, Kraetzl WD, Steffl M, Meyer HH, Schams D, Berisha B. Expression and localisation of extracellular matrix degrading proteases and their inhibitors during the oestrous cycle and after induced luteolysis in the bovine corpus luteum. Reproduction 2007; 134: 535 547 [DOI] [PubMed] [Google Scholar]
- Neuvians TP, Schams D, Berisha B, Pfaffl MW. Involvement of pro-inflammatory cytokines, mediators of inflammation, and basic fibroblast growth factor in prostaglandin F2alpha-induced luteolysis in bovine corpus luteum. Biol Reprod 2004; 70: 473 480 [DOI] [PubMed] [Google Scholar]
- Shirasuna K, Watanabe S, Nagai K, Sasahara K, Shimizu T, Ricken AM, Spanel-Borowski K, Miyamoto A. Expression of mRNA for cell adhesion molecules in the bovine corpus luteum during the estrous cycle and PGF2alpha-induced luteolysis. J Reprod Dev 2007; 53: 1319 1328 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Acosta TJ, Berisha B, Kobayashi S, Ohtani M, Schams D, Miyamoto A. Changes in prostaglandin secretion by the regressing bovine corpus luteum. Prostaglandins Other Lipid Mediat 2003; 70: 339 349 [DOI] [PubMed] [Google Scholar]
- Ginther OJ, Shrestha HK, Fuenzalida MJ, Imam S, Beg MA. Stimulation of pulses of 13,14-dihydro-15-keto-PGF2alpha (PGFM) with estradiol-17beta and changes in circulating progesterone concentrations within a PGFM pulse in heifers. Theriogenology 2010; 74: 384 392 [DOI] [PubMed] [Google Scholar]
- Ginther OJ, Shrestha HK, Fuenzalida MJ, Shahiduzzaman AK, Beg MA. Characteristics of pulses of 13,14-dihydro-15-keto-prostaglandin f2alpha before, during, and after spontaneous luteolysis and temporal intrapulse relationships with progesterone concentrations in cattle. Biol Reprod 2010; 82: 1049 1056 [DOI] [PubMed] [Google Scholar]
- Ginther OJ, Shrestha HK, Fuenzalida MJ, Shahiduzzaman AK, Hannan MA, Beg MA. Intrapulse temporality between pulses of a metabolite of prostaglandin F 2alpha and circulating concentrations of progesterone before, during, and after spontaneous luteolysis in heifers. Theriogenology 2010; 74: 1179 1186 [DOI] [PubMed] [Google Scholar]
- Imam S, Shrestha HK, Beg MA, Ginther OJ. Intrapulse changes in progesterone and LH concentrations and luteal blood flow during an estradiol-induced pulse of a metabolite of prostaglandin F2alpha in heifers. Anim Reprod Sci 2010; 121: 34 38 [DOI] [PubMed] [Google Scholar]
- Shrestha HK, Beg MA, Imam S, Ginther OJ. Luteal blood flow and concentrations of circulating progesterone and other hormones associated with a simulated pulse of 13,14-dihydro-15-keto-prostaglandin F2alpha in heifers. Reproduction 2010; 139: 673 683 [DOI] [PubMed] [Google Scholar]
- Shrestha HK, Beg MA, Siddiqui MA, Ginther OJ. Dynamic progesterone responses to simulation of a natural pulse of a metabolite of prostaglandin F(2alpha) in heifers. Anim Reprod Sci 2010; 118: 118 123 [DOI] [PubMed] [Google Scholar]
- Callahan N, Tsang P, Milvae R, Schreiber D, Keator C, McCracken J. In vivo temporal expression of protein levels of tissue inhibitor of metalloproteinase-3 (TIMP-3) during prostaglandin F2alpha-induced luteolysis in sheep. Biol Reprod 2009; 179 179
- Ferguson D, Tsang P, Schreiber D, Keator CT, Milvae R. In vivo changes in protein expression of the gelatinases, matrix metalloproteinases-2 and-9 (MMP-2 and MMP-9), and tissue inhibitors of metalloproteinases-1 (TIMP-1) in the ovine corpus luteum (CL) after multiple pulses of prostaglandin F2alpha (PGF2A). Biol Reprod 2007; 99 100
- Ferguson DE, Tsang PCW, Kneebone J, Milvae RA, Schreiber DT, Keator CS, McCracken JA. In vivo temporal expression of proteins associated with luteolysis following multiple pulses of prostaglandin F (2)alpha in sheep. Biol Reprod 2009; 177 177 19339708
- Tsai SJ, Kot K, Ginther OJ, Wiltbank MC. Temporal gene expression in bovine corpora lutea after treatment with PGF2alpha based on serial biopsies in vivo. Reproduction 2001; 121: 905 913 [PubMed] [Google Scholar]
- Beindorff N, Nagai K, Shirasuna K, Herzog K, Hoeffmann K, Sasaki M, Bollwein H, Miyamoto A. Vascular changes in the corpus luteum during early pregnancy in the cow. J Reprod Dev 2010; 56: 263 270 [DOI] [PubMed] [Google Scholar]
- Goravanahally MP, Salem M, Yao J, Inskeep EK, Flores JA. Differential gene expression in the bovine corpus luteum during transition from early phase to midphase and its potential role in acquisition of luteolytic sensitivity to prostaglandin F2 alpha. Biol Reprod 2009; 80: 980 988 [DOI] [PubMed] [Google Scholar]
- Murayama C, Kaji A, Miyauchi K, Matsui M, Miyamoto A, Shimizu T. Effect of VEGF (vascular endothelial growth factor) on expression of IL-8 (interleukin-8), IL-1beta and their receptors in bovine theca cells. Cell Biol Int 2010; 34: 531 536 [DOI] [PubMed] [Google Scholar]
- Abler LL, Mehta V, Keil KP, Joshi PS, Flucus CL, Hardin HA, Schmitz CT, Vezina CM. A high throughput in situ hybridization method to characterize mRNA expression patterns in the fetal mouse lower urogenital tract. J Vis Exp 2011; 54: e2912, DOI 10.3791/2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402 408 [DOI] [PubMed] [Google Scholar]
- Chegini N, Lei ZM, Rao CV, Hansel W. Cellular distribution and cycle phase dependency of gonadotropin and eicosanoid binding sites in bovine corpora lutea. Biol Reprod 1991; 45: 506 513 [DOI] [PubMed] [Google Scholar]
- Mamluk R, Chen DB, Greber Y, Davis JS, Meidan R. Characterization of messenger ribonucleic acid expression for prostaglandin F-2 alpha and luteinizing hormone receptors in various bovine luteal cell types. Biol Reprod 1998; 58: 849 856 [DOI] [PubMed] [Google Scholar]
- Davis JS, Rueda BR. The corpus luteum: an ovarian structure with maternal instincts and suicidal tendencies. Front Biosci 2002; 7: d1949 d1978 [DOI] [PubMed] [Google Scholar]
- Boiti C, Zampini D, Zerani M, Guelfi G, Gobbetti A. Prostaglandin receptors and role of G protein-activated pathways on corpora lutea of pseudopregnant rabbit in vitro. J Endocrinol 2001; 168: 141 151 [DOI] [PubMed] [Google Scholar]
- Duncan RA, Davis JS. Prostaglandin F2 alpha stimulates inositol 1,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate formation in bovine luteal cells. Endocrinology 1991; 128: 1519 1526 [DOI] [PubMed] [Google Scholar]
- Stocco CO, Zhong LP, Sugimoto Y, Ichikawa A, Lau LF, Gibori G. Prostaglandin F-2 alpha-induced expression of 20 alpha-hydroxysteroid dehydrogenase involves the transcription factor NUR77. J Biol Chem 2000; 275: 37202 37211 [DOI] [PubMed] [Google Scholar]
- Chen DB, Westfall SD, Fong HW, Roberson MS, Davis JS. Prostaglandin F2alpha stimulates the Raf/MEK1/mitogen-activated protein kinase signaling cascade in bovine luteal cells. Endocrinology 1998; 139: 3876 3885 [DOI] [PubMed] [Google Scholar]
- Chen DB, Fong HW, Davis JS. Induction of c-fos and c-jun messenger ribonucleic acid expression by prostaglandin F2 alpha is mediated by a protein kinase C-dependent extracellular signal-regulated kinase mitogen-activated protein kinase pathway in bovine luteal cells. Endocrinology 2001; 142: 887 895 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Bach K, Thiel G. The extracellular signal-regulated protein kinases Erk1/Erk2 stimulate expression and biological activity of the transcriptional regulator Egr-1. Biol Chem 2001; 382: 1077 1081 [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Shore P, Sharrocks AD, Davis RJ. Integration of map kinase signal-transduction pathways at the serum response element. Science 1995; 269: 403 407 [DOI] [PubMed] [Google Scholar]
- Carter DA. Rhythms of cellular immediate-early gene expression: more than just an early response. Exp Physiol 1997; 82: 237 244 [DOI] [PubMed] [Google Scholar]
- Hughes P, Dragunow M. Induction of immediate-early genes and the control of neurotransmitter-regulated gene expression within the nervous system. Pharmacol Rev 1995; 47: 133 178 [PubMed] [Google Scholar]
- Liu CT, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther 1998; 5: 3 28 [PubMed] [Google Scholar]
- Sukhatme VP. Early transcriptional events in cell growth: the Egr family. J Am Soc Nephrol 1990; 1: 859 866 [DOI] [PubMed] [Google Scholar]
- Hou XY, Arvisais EW, Jiang C, Chen DB, Roy SK, Pate JL, Hansen TR, Rueda BR, Davis JS. Prostaglandin F2 alpha stimulates the expression and secretion of transforming growth factor B1 via induction of the early growth response 1 gene (EGR1) in the bovine corpus luteum. Mol Endocrinol 2008; 22: 403 414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JE, Stormshak F. In vivo and in vitro responses of the bovine corpus luteum after exposure to exogenous gonadotropin-releasing hormone and prostaglandin F2alpha. Endocrine 1996; 4: 165 173 [DOI] [PubMed] [Google Scholar]
- Olofsson JI, Leung CH, Bjurulf E, Ohno T, Selstam G, Peng C, Leung PC. Characterization and regulation of a mRNA encoding the prostaglandin F2alpha receptor in the rat ovary. Mol Cell Endocrinol 1996; 123: 45 52 [DOI] [PubMed] [Google Scholar]
- Ottander U, Leung CH, Olofsson JI. Functional evidence for divergent receptor activation mechanisms of luteotrophic and luteolytic events in the human corpus luteum. Mol Hum Reprod 1999; 5: 391 395 [DOI] [PubMed] [Google Scholar]
- Beg MA, Gastal EL, Gastal MO, Ji S, Wiltbank MC, Ginther OJ. Changes in steady-state concentrations of messenger ribonucleic acids in luteal tissue during prostaglandin F2 alpha induced luteolysis in mares. Anim Reprod Sci 2005; 90: 273 285 [DOI] [PubMed] [Google Scholar]
- Stocco DM. StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol 2001; 63: 193 213 [DOI] [PubMed] [Google Scholar]
- Skarzynski DJ, Jaroszewski JJ, Bah MM, Deptula KM, Barszczewska B, Gawronska B, Hansel W. Administration of a nitric oxide synthase inhibitor counteracts prostaglandin F2-induced luteolysis in cattle. Biol Reprod 2003; 68: 1674 1681 [DOI] [PubMed] [Google Scholar]
- Pescador N, Soumano K, Stocco DM, Price CA, Murphy BD. Steroidogenic acute regulatory protein in bovine corpora lutea. Biol Reprod 1996; 55: 485 491 [DOI] [PubMed] [Google Scholar]
- Silva PJ, Juengel JL, Rollyson MK, Niswender GD. Prostaglandin metabolism in the ovine corpus luteum: catabolism of prostaglandin F(2alpha) (PGF(2alpha)) coincides with resistance of the corpus luteum to PGF(2alpha). Biol Reprod 2000; 63: 1229 1236 [DOI] [PubMed] [Google Scholar]
- Fraser HM, Dickson SE, Lunn SF, Wulff C, Morris KD, Carroll VA, Bicknell R. Suppression of luteal angiogenesis in the primate after neutralization of vascular endothelial growth factor. Endocrinology 2000; 141: 995 1000 [DOI] [PubMed] [Google Scholar]
- Vonnahme KA, Redmer DA, Borowczyk E, Bilski JJ, Luther JS, Johnson ML, Reynolds LP, Grazul-Bilska AT. Vascular composition, apoptosis, and expression of angiogenic factors in the corpus luteum during prostaglandin F2alpha-induced regression in sheep. Reproduction 2006; 131: 1115 1126 [DOI] [PubMed] [Google Scholar]
- Zerani M, Dall'Aglio C, Maranesi M, Gobbetti A, Brecchia G, Mercati F, Boiti C. Intraluteal regulation of prostaglandin F2 alpha-induced prostaglandin biosynthesis in pseudopregnant rabbits. Reproduction 2007; 133: 1005 1016 [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Crenshaw TD, Wiltbank MC. Prostaglandin f(2alpha) induces distinct physiological responses in porcine corpora lutea after acquisition of luteolytic capacity. Biol Reprod 2000; 63: 1504 1512 [DOI] [PubMed] [Google Scholar]
- Narayansingh RM, Senchyna M, Carlson JC. Treatment with prostaglandin F2alpha increases expression of prostaglandin synthase-2 in the rat corpus luteum. Prostaglandins Other Lipid Mediat 2002; 70: 145 160 [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Anderson LE, Wu YL, Rabot A, Tsai SJ, Wiltbank MC. Regulation of progesterone and prostaglandin F2alpha production in the CL. Mol Cell Endocrinol 2002; 191: 65 80 [DOI] [PubMed] [Google Scholar]
- Bagavandoss P, Kunkel SL, Wiggins RC, Keyes PL. Tumor necrosis factor-a (TNF-a) production and localization of macrophages and T lymphocytes in the rabbit corpus luteum. Endocrinology 1988; 122: 1185 1187 [DOI] [PubMed] [Google Scholar]
- Takaya R, Fukaya T, Sasano H, Suzuki T, Tamura M, Yajima A. Macrophages in normal cycling human ovaries; immunohistochemical localization and characterization. Hum Reprod 1997; 12: 1508 1512 [DOI] [PubMed] [Google Scholar]
- Bauer M, Reibiger I, Spanel-Borowski K. Leucocyte proliferation in the bovine corpus luteum. Reproduction 2001; 121: 297 305 [PubMed] [Google Scholar]
- Pate JL. Cellular components involved in luteolysis. J Anim Sci 1994; 72: 1884 1890 [DOI] [PubMed] [Google Scholar]
- Pate JL. Landis Keyes P. Immune cells in the corpus luteum: friends or foes? Reproduction 2001; 122: 665 676 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.