Abstract
Background
Phylogenetic relationships among Asian and African colobine genera have been disputed and are not yet well established. In the present study, we revisit the contentious relationships within the Asian and African Colobinae by analyzing 44 nuclear non-coding genes (>23 kb) and mitochondrial (mt) genome sequences from 14 colobine and 4 non-colobine primates.
Principal Findings
The combined nuclear gene and the mt genome as well as the combined nuclear and mt gene analyses yielded different phylogenetic relationships among colobine genera with the exception of a monophyletic ‘odd-nosed’ group consisting of Rhinopithecus, Pygathrix and Nasalis, and a monophyletic African group consisting of Colobus and Piliocolobus. The combined nuclear data analyses supported a sister-grouping between Semnopithecus and Trachypithecus, and between Presbytis and the odd-nosed monkey group, as well as a sister-taxon association of Pygathrix and Rhinopithecus within the odd-nosed monkey group. In contrast, mt genome data analyses revealed that Semnopithecus diverged earliest among the Asian colobines and that the odd-nosed monkey group is sister to a Presbytis and Trachypithecus clade, as well as a close association of Pygathrix with Nasalis. The relationships among these genera inferred from the analyses of combined nuclear and mt genes, however, varied with the tree-building methods used. Another remarkable finding of the present study is that all of our analyses rejected the recently proposed African colobine paraphyly and hybridization hypothesis and supported reciprocal monophyly of the African and Asian groups.
Significance
The phylogenetic utility of large-scale new non-coding genes was assessed using the Colobinae as a model, We found that these markers were useful for distinguishing nodes resulting from rapid radiation episodes such as the Asian colobine radiation. None of these markers here have previously been used for colobine phylogenetic reconstruction, increasing the spectrum of molecular markers available to mammalian systematics.
Introduction
The Old World monkeys are comprised of two living subfamilies – the cheek-pouch monkeys (Cercopithecinae) and the leaf-eating monkeys (Colobinae). Although these groups are both the closest living relatives to the apes, research has historically focused on cercopithecine evolution as a model for human evolution. The systematics and evolution of the colobines, on the other hand, has been a relatively neglected topic. The colobinaes consist of 10 genera in two subtribes - the African Colobina (including the genera Colobus, Piliocolobus, and Procolobus) and the Asian Presbytina (including the genera Pygathrix, Rhinopithecus, Nasalis, Simias, Presbytis, Trachypithecus, and Semnopithecus) [1]–[4]. Currently, phylogenetic relationships among these genera remain controversial [2], [5]–[11]. The main reason is that colobines represent a typical example of an evolutionary radiation with rapid diversification events that date back to the Middle Miocene about 10–15 million years ago (MYA) [1], [4]. Close to the initial appearance of colobines in the fossil record, nearly all the extant colobine genera diversify from one another within a four million year window [12]–[14]. For this reason, attempts to clarify relationships among these colobine genera have encountered challenges. Given that they share with apes a close relatedness, historically similar distribution in the Old World, and similar timing of diversification events, elucidating colobine evolutionary history can shed light upon the evolution and dispersal of apes (and other mammals) across the Old World, including our own ancestors.
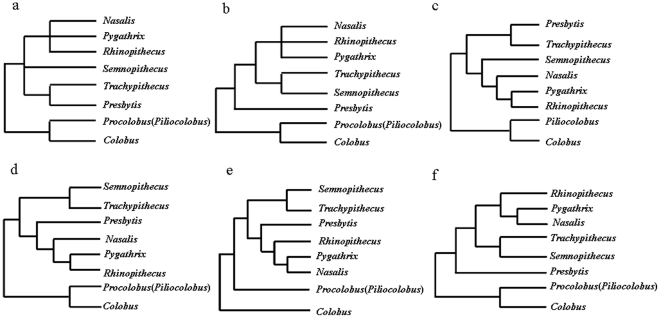
Within the Asian colobines, although the monophyly of the odd-nosed monkey group (Pygathrix, Rhinopithecus, Nasalis, and Simias) is now widely accepted [1]–[4], [15] and confirmed by genetic data [12]–[14], [16]–[18], monophyly of the langur group (Trachypithecus, Semnopithecus, and Presbytis) is disputed. In fact, recent genetic data provided contradicting relationships among langur genera and the odd-nosed monkey group (see Figure 1) [12]–[14],[16]–[18]. Also, there has been long-standing controversy over the relationships among the genera within the odd-nosed group (see Figure 1) [12]–. For example, compared with earlier investigations that mainly utilized analysis of portions of a single or a small number of mt genes [17], [19]–[21]. Sterner et al. [14] examined 12 mt protein-coding genes of six Asian colobine genera and argued for a sister-group association between Presbytis and Trachypithecus within the langur group, but failed to resolve the precise relationship among Presbytis/Trachypithecus, Semnopithecus, and the odd-nosed monkey group, as well as the relationships among Pygathrix, Rhinopithecus, and Nasalis within the odd-nosed monkeys (Figure 1a). Ting et al. [18] analyzed a 4,297 bp fragment of the X-chromosome and suggested that within the Asian colobines Presbytis diverged earliest, followed by the split between Trachypithecus/Semnopithecus and the odd-nosed monkey group. However, phylogenetic relationships among Pygathrix, Rhinopithecus, and Nasalis within the odd-nosed monkey group remained unresolved in their analyses (Figure 1b). The same results were also obtained in Perelman et al. [12], which included 54 nuclear genes of 186 primates. In contrast, Chatterjee et al [22] analyzed a 6,138 bp mt fragment and Meyer et al. [16] analyzed a 1.8 kb fragment, and both inferred an earliest divergence of Presbytis/Trachypithecus, and the close relatedness of Semnopithecus and the odd-nosed monkey group (Figure 1c), but they lacked significant support. Through an analysis of 15 mt and 43 nuclear genes, Fabre et al. [23] found support for close relationships between Trachypithecus and Semnopithecus, and between Presbytis and the odd-nosed monkeys (Figure 1d). These same relationships were also recovered Roos et al. [13] from an analysis of 83 mobile elements. However, phylogenetic relationships among Pygathrix, Rhinopithecus, and Nasalis within the odd-nosed monkey group were different between Fabre et al. [23] and Roos et al. [13] (Figure 1e). Intriguingly, in Roos et al.'s [13] nuclear sequence data (∼13 kb) analyses, they also support the former Semnopithecus-Trachypithecus clade, but suggest Presbytis as sister to the other Asian colobines (Figure 1f). The results from these studies demonstrate that the relationships among Asian colobines remain unresolved, although hybridization has been proposed as a most likely explanation for some of these incongruent relationships [13].
Figure 1. Hypotheses of phylogenetic relationships among Colobine genera.
Trees were reconstructed based on (a) 12 protein-coding mt genes [14], (b) fragment of X-chromosome [18] and 54 nuclear genes [12], (c) complete cytb gene [16] and 7 mt genes [22], (d) 15 mt genes and 43 nuclear genes [23], (e) 83 mobile elements [13], (f) nuclear genes [13].
Relationships among the African colobines at the genus-level are not as contentious as there are only 2–3 commonly recognized genera (Piliocolobus, Procolobus and Colobus). Previous studies of this group based on morphology and molecular data suggest that the African colobines represent a monophyletic group [1], [2], [7], [24], [25] that contains a sister-taxon relationship between Piliocolobus and Procolobus to the exclusion of Colobus [26]–[29]. Intriguingly, a recent study by Roos et al. [13] based on mobile elements indicated a closer association of the Piliocolobus/Procolobus clade to Asian genera than to Colobus, a relationship that was not rejected by nuclear sequence data in their study [13]. This finding led them to propose African colobine paraphyly and a hypothesis of ancient hybridization, which challenges the current well-recognized monophyly of the African colobines.
These findings highlight the need to gather and analyze additional sequence data sets in order to unravel the phylogenetic relationships among colobine genera. To this end, we sequenced 44 nuclear non-coding genes comprising a total of >23 kb from 14 colobine and 4 non-colobine primates. These 44 nuclear genes are applied for the first time to study colobine phylogeny. In addition, we also undertook analyses of complete mt genomes from these 18 taxa, including 5 newly determined Asian colobine mt genomes and 13 previously published mt genomes [14]. Our objectives were to: (1) provide further insights into the relationships among the colobine genera, and (2) examine the utility of these genes in the context of colobine phylogeny, with special attention to the previously unexplored 44 nuclear non-coding genes.
Materials and Methods
Data Sets
Detailed information on the 44 nuclear non-coding genes (mainly intergenic regions and introns) used for the colobine phylogenetic reconstruction is shown in Table S1. These non-repetitive, non-coding genes were selected from Peng et al. [30], in which 280 genes were screened across primates based on bioinformatic analyses of genome sequences available for human (Homo sapiens), common chimpanzee (Pan troglodytes) and rhesus macaque (Macaca mulatta) (hg18, panTro2, rheMac2).
Our sampling includes most of the commonly recognized extant colobine genera except for Simias and Procolobus. We were unable to obtain biomaterials for these taxa, but previous studies of both morphology and genetics have established that Simias is the sister-taxon of Nasalis [13], [31], [32] and Procolobus is the sister-taxon of (and possibly congeneric to) Piliocolobus [13], [18], [25], [27] (Table 1). We also follow the classification of Brandon-Jones et al. [3] in assigning Trachypithecus johnii and Trachypithecus vetulus to the genus Semnopithecus, which morphological and molecular studies have supported [3], [17]. For each of the 14 colobine species sampled, total genomic DNA was isolated from blood or frozen tissues using a standard proteinase K or phenol/chloroform extraction [33]. To amplify these non-coding genes, a “touch-down” PCR amplification was carried out using the following parameters with the primer pairs of Peng et al. [30]: 95°C hot start (3 min), 20 cycles of 94°C denaturation (1 min), 60-40°C annealing (1 min), 72°C extension (1 min), and finally 15 cycles of 94°C denaturation (1 min), 55°C annealing (1 min), 72°C extension (1 min). The amplified DNA fragments were purified and sequenced in both directions with an ABI PRISM™ 3700 DNA or 3130xL sequencer following the manufacturer's protocol.
Table 1. Species used in this study.
| Genus | Species | Common name | Sample Source | MT genomes | Nuclear genes |
| Pygathrix | P nemaeus | Douc langur | Vietnam | NC_008220 [14] | JN103440-JN104028 |
| Rhinopithecus | R roxellana | Sichuan snub-nosed monkey | Gansu Province, China | NC_008218 [14] | JN103440-JN104028 |
| R bieti | Yunnan snub-nosed monkey | Yunnan Province, China | HM125579 [71] | JN103440-JN104028 | |
| R avunculus | Tonkin snub-nosed monkey | Vietnam | HM125578 [71] | JN103440-JN104028 | |
| Trachypithecus | T hatinhensis | Hatinh langur | Vietnam | HQ149046(this study) | JN103440-JN104028 |
| T germaini | Germain's silver langur | Vietnam | HQ149047(this study) | JN103440-JN104028 | |
| T shortridgei | Shortridge's langur | Sino-Burmese border area | HQ149048(this study) | JN103440-JN104028 | |
| Nasalis | N larvatus | Proboscis monkey | Borneo | NC_008216 [14] | JN103440-JN104028 |
| Presbytis | P melalophos | Mitered leaf monkey | Sumatra | NC_008217 [14] | JN103440-JN104028 |
| Semnopithecus | S entellus | Hanuman langur | India | NC_008215 [14] | JN103440-JN104028 |
| S johnii | Nilgiri Langur | Sri Lanka | HQ149050(this study) | JN103440-JN104028 | |
| S vetulus | Purple-faced langur | Sri Lanka | HQ149049(this study) | JN103440-JN104028 | |
| Colobus | C guereza | Eastern black and white colobus | zoo specimen | NC_006901 [14] | |
| C angolensis | Angolan black-and white colobus | zoo specimen | JN103440-JN104028 | ||
| Piliocolobus | P badius | Western red colobus | Sierra Leone (MT) | ||
| Gambia (nuclear) | NC_008219 [14] | JN103440-JN104028 | |||
| Macaca | M sylvanus | Barbary macaque | AJ309865 [30] | rheMac2 | |
| Pongo | P abelli | Sumatra orangutan | X97707 [30] | ponAbe2 | |
| Pan | P troglodytes | Chimpanzee | D38113 [30] | panTro2 | |
| Homo | H sapiens | Human | X93334 [30] | hg18 |
The complete mt genome sequences from five Asian colobine species (Semnopithecus johnii, S. vetulus, Trachypithecus hatinhensis, T. germaini, T. shortridgei) were newly determined here. The mt genome sequences were amplified using the LA PCR™ Kit from Takara Biotechnology Co., Ltd and 10 universal PCR primers (Table S2). Amplification was performed using 32 cycles of 10 sec at 97°C, 5.5 min at 58°C to 68°C, with an initial step of 1.5 min at 94°C and a final step of 10 min at 72°C. Long-Range PCR products, each with a size of ∼4000 bp, were sequenced in both directions using a primer walking strategy. Sequencing was performed in an ABI PRISM™ 3700 DNA sequencer following the manufacturer's protocols. Primer sequence information is available upon request. Where necessary, PCR products were cloned into the PMD18-T Vector and transformed into ultracompetent E. coli cells (TaKaRa Biotechnology Co., Ltd. Dalian, China) in order to resolve the difficulty of direct sequencing of control regions arising from long tandem repeats. Five positive clones per ligation reaction were sequenced. Mt sequences obtained were checked to ensure that they did not include nuclear copies of mtDNA-like pseudogenes (numts), as indicated by the fact that the amino acid sequences of protein-coding genes did not possess premature stop codons or frameshifting insertions/deletions. Also, long-range amplifications are less likely to amplify numts, and we assembled the PCR amplifications to ensure that they formed a circular molecule.
Nuclear and mt genome data from 4 non-colobine primates, i.e., human (H. sapiens), common chimpanzee (P. troglodytes), rhesus macaque (M. mulatta), and orangutan (Pongo abelii), were also included in the analyses. Their nuclear and mt genome sequences were downloaded from GenBank (for accession numbers see Table 1).
Alignments and Sequence Characterizations
Sequences were aligned using Muscle 3.8.31 [34] under the default settings. All 44 genes were analyzed separately and in a combined data set. The mt sequences were divided into five data sets: (1) all 13 protein-coding genes combined, (2) 12S and 16S rRNA genes combined, (3) all 22 tRNA genes combined, (4) control region (CR), (5) tRNAs, rRNAs, CR, and protein-coding genes combined. In the analyses of rRNAs (alignment 2) and tRNAs (alignment 3), the data were also partitioned into single-strand stem and base-paired loops based on the models of Gutell et al. [35] and Springer and Douzery [36]. In addition, the 44 nuclear genes and mt genomes were combined into one alignment. Although arguments can be made against combining genomic regions that possibly have different histories, a combined approach (of all nuclear regions and of nuclear and mitochondrial regions) is thought to detect the phylogenetic signal that is most prevalent across the genome, which is also most likely to represent the species tree [37]. Respective alignments are available upon the authors' request.
Pairwise comparisons and sequence characterizations were estimated using MEGA 4.0 [38].
Phylogenetic Analyses
Phylogenetic analyses of the individual nuclear non-coding genes and mt alignments 1–4, were performed using PAUP* 4.0b10 [39] for maximum-parsimony [MP] and maximum-likelihood [ML] analyses. MrBayes 3.1.2 [40] was used for the Bayesian inference. We used three hominoid species (Homo, Pan, Pongo) for outgroup rooting in all analyses. In MP analyses, a heuristic search was performed with tree-bisection-reconnection (TBR) branch swapping, random addition of taxa, and 1000 replicates per search. Only one of the best trees found during branch swapping was saved. In ML analyses, the best-fit models of sequence evolution were selected using the Akaike Information Criterion (AIC) [41], [42] with Modeltest 3.7 [43]. The chosen models (see Table 2) and their parameters were used to infer ML trees with the heuristic algorithm, 10 random-addition sequence replicates, and TBR branch swapping. The tree reliability under ML analysis was assessed using a bootstrap analysis of 100 replicates [44]. In Bayesian inference, each Metropolis-coupled Markov chain Monte Carlo (MCMC) run for all individual genes employed the model selected by Modeltest for that gene, or the nearest model to that model that could be implemented in MrBayes. Three heated chains and a single cold chain were used in all MCMC analyses and run for 2 million generations. Three simultaneous independent runs were performed. Trees were sampled every 100 generations. The average standard deviation of split frequencies was close to 0.001 when the runs were finished. The first 25% of the trees were discarded as burn-in. A 50% majority-rule consensus of post burn-in trees was constructed to summarize the posterior probability (PP) for each branch.
Table 2. Characterization of nuclear non-coding and mt genes examined in the present study.
| Sequence type | Fragment name | Aligned length | Parsimony-informative sites | Best fit model | Among-site Rate Variation | Pairwise Distance(%) | |
| I | α | ||||||
| Nuclear genes | chr1-4 | 462 | 30 | K81+G | 0 | 0.6033 | 3.10 |
| chr1-6 | 567 | 36 | GTR | 0 | 0 | 3.40 | |
| chr2-1 | 504 | 32 | K80 | 0 | 0 | 3.10 | |
| chr2-8 | 413 | 28 | HKY+G | 0 | 0.6264 | 3.10 | |
| chr3-2 | 533 | 28 | HKY+G | 0 | 0.4657 | 2.50 | |
| chr3-5 | 337 | 33 | TVM+I | 0.4589 | 0 | 5.10 | |
| chr4-2 | 486 | 26 | HKY+G | 0 | 1.3216 | 5.40 | |
| chr4-7 | 492 | 34 | HKY | 0 | 0 | 2.90 | |
| chr5-6 | 534 | 41 | HKY | 0 | 0.9808 | 3.60 | |
| chr5-8 | 480 | 39 | TIM | 0 | 0 | 3.80 | |
| chr6-5 | 456 | 43 | TIM+G | 0 | 0.7324 | 4.10 | |
| chr6-6 | 367 | 23 | HKY+I | 0.5102 | 0 | 3.30 | |
| chr7-6 | 514 | 49 | K81uf | 0 | 0 | 5.20 | |
| chr8-1 | 577 | 51 | HKY+G | 0 | 0.7446 | 4.50 | |
| chr8-2 | 526 | 29 | HKY | 0 | 0 | 2.80 | |
| chr9-5 | 522 | 26 | GTR+G | 0 | 1.1437 | 3.20 | |
| chr10-1 | 503 | 46 | TrN+I | 0.4591 | 0 | 4.20 | |
| chr10-5 | 498 | 9 | TVMef+I | 0.6203 | 0 | 1.40 | |
| chr11-2 | 522 | 30 | TIM+G | 0 | 0.5405 | 4.00 | |
| chr12-1 | 586 | 44 | TVMef+I | 0.4245 | 0 | 3.50 | |
| chr12-2 | 439 | 30 | HKY | 0 | 0 | 3.20 | |
| chr13-3 | 401 | 23 | HKY | 0 | 0 | 3.00 | |
| chr13-6 | 472 | 31 | K81uf | 0 | 0 | 3.30 | |
| chr15-1 | 862 | 302 | HKY | 0 | 0 | 22.60 | |
| chr15-3 | 398 | 21 | TrN | 0 | 0 | 2.50 | |
| chr17-4 | 788 | 141 | TVMef+G | 0 | 1.1752 | 8.60 | |
| chr17-8 | 497 | 44 | TrN+G | 0 | 0.9342 | 4.70 | |
| chr18-4 | 504 | 30 | HKY+I | 0.6339 | 0 | 2.60 | |
| chr19-1 | 550 | 55 | HKY+I | 0.5819 | 0 | 4.50 | |
| chr19-5 | 458 | 45 | HKY+I | 0.5243 | 0 | 3.70 | |
| chr20-4 | 588 | 58 | K81uf+I | 0.5311 | 0 | 4.00 | |
| chr20-5 | 457 | 32 | HKY+G | 0 | 0.8146 | 3.70 | |
| ENC5 | 641 | 39 | HKY+I | 0.4338 | 0 | 3.00 | |
| ENC14 | 539 | 30 | GTR | 0 | 0 | 2.50 | |
| ENC15 | 868 | 49 | HKY | 0 | 0 | 3.00 | |
| ENC19 | 530 | 29 | TVM | 0 | 0 | 2.90 | |
| ENC25 | 401 | 32 | HKY+G | 0 | 0.7782 | 3.90 | |
| ENC35 | 548 | 34 | TVM+I | 0.4981 | 0 | 2.60 | |
| X2 | 565 | 37 | HKY+G | 0 | 0.457 | 3.60 | |
| X5 | 510 | 38 | HKY+G | 0 | 1.04 | 3.50 | |
| X37 | 598 | 56 | TVM+G | 0 | 0.4556 | 4.20 | |
| X45 | 490 | 24 | GTR+G | 0 | 1.2507 | 3.00 | |
| X61 | 602 | 39 | TVM+G | 0 | 0.8788 | 2.90 | |
| X65 | 549 | 32 | K81uf+G | 0 | 0.603 | 2.70 | |
| Combined | 23134 | 1951 | TVM+G | 0 | 0.7034 | 3.90 | |
| Mt genes | ND1 | 957 | 337 | GTR+I+G | 0.4389 | 1.2434 | 19.00 |
| ND2 | 1044 | 426 | TrN+I+G | 0.3765 | 1.3323 | 22.80 | |
| COX1 | 1545 | 501 | TVM+I+G | 0.5698 | 1.9262 | 16.90 | |
| COX2 | 684 | 227 | HKY+I+G | 0.5132 | 1.2176 | 17.10 | |
| ATP8 | 211 | 104 | TrN+I+G | 0.2533 | 1.1544 | 28.80 | |
| ATP6 | 681 | 281 | TIM+I+G | 0.3344 | 0.964 | 23.60 | |
| COX3 | 784 | 274 | HKY+I+G | 0.5063 | 1.237 | 18.60 | |
| ND3 | 346 | 144 | K81uf+I+G | 0.4127 | 2.2742 | 23.40 | |
| ND4L | 297 | 108 | K81uf+I+G | 0.4101 | 1.204 | 19.80 | |
| ND4 | 1378 | 532 | TrN+I+G | 0.4155 | 1.2687 | 21.00 | |
| ND5 | 1806 | 709 | TIM+I+G | 0.3871 | 1.2705 | 22.40 | |
| ND6 | 528 | 181 | TrN+I+G | 0.3847 | 0.7419 | 18.90 | |
| CYTB | 1135 | 430 | HKY+I+G | 0.4267 | 1.1196 | 20.70 | |
| 12SrRNA | 961 | 223 | GTR+I+G | 0.4386 | 0.6382 | 10.90 | |
| 16SrRNA | 1582 | 375 | GTR+I+G | 0.4575 | 0.7409 | 12.70 | |
| tRNA | 1573 | 377 | GTR+I+G | 0.3032 | 0.4305 | 12.10 | |
| D-loop | 1015 | 415 | TVM+I+G | 0.2362 | 0.81 | 24.80 | |
| Combined | 16527 | 5644 | GTR+I+G | 0.4328 | 1.0995 | 18.50 | |
Note: Ti = Transition; Tv = Transversion; I = Proportion of invariable sites; α = Gamma distribution shape parameter.
In addition to individual analyses, phylogenetic reconstructions were performed on the combined nuclear dataset and the combined mt genome dataset (mt alignment 5) as well as the combined nuclear and mt genome dataset. We used PAUP* for the MP analysis, the RAxML online web server [45] for a partitioned ML analysis with a GTR model, and MrBayes and PhyloBayes for Bayesian analyses [46]. For each combined dataset, we identified model partitions based on partitioning matrices by gene. That is, in the analysis of the combined nuclear data set, each nuclear non-coding gene was considered a different partition, and in the combined mt data set, each of the 13 individual protein-coding genes, all tRNAs, and each of the two rRNA genes were considered to be different partitions. Based on the selected models using the AIC [41], [42] as mentioned above for individual analyses (see Table 2), we assigned a separate substitution model for each of the data partitions in the MrBayes analysis. Three heated chains and a single cold chain were used in all MCMC analyses and run for 5 million generations, sampling trees every 100 generations. The average standard deviation of split frequencies was close to 0.001 when the run ended. The first 25% of trees were discarded as burn-in. A 50% majority-rule consensus of post burn-in trees was constructed to summarize PPs for each branch. In addition, the site-heterogeneous mixture model CAT-GTR was used for the above three combined datasets in PhyloBayes analysis [47] with two independent (MCMC) chains. Compared to other phylogenetic MCMC samplers, the main distinguishing feature of PhyloBayes is the underlying probabilistic model, CAT [48]. CAT is a mixture model especially devised to account for site-specific features of sequence evolution. It is particularly well suited for large multigene alignments. To check for convergence, the program bpcomp [49] was used to compare the bipartitions between the two runs. With a burn-in of 1000 and taking every two trees, the largest discrepancy (maxdiff) between the bipartitions was less than 0.1.
Testing Potential Tree Incongruence
The incongruence among different tree topologies was evaluated using the Shimodaira-Hasegawa (SH) test [50] and the approximately unbiased (AU) test [51], as implemented in the CONSELV0.1i program [52] with default scaling and replicate values. The site-wise log-likelihood values were estimated by PAUP*.
Results
Characteristics of the Nuclear Non-Coding Data and Mt Genomes
The general characteristics of the nuclear non-coding data and mt genomes are summarized in Table 2. The 44 nuclear non-coding genes of 18 species varied in length from 337 bp (chr3-5) to 862 bp (ENC15) aligned positions. The numbers of parsimony-informative sites range from 9 (1.81%) (chr10-5) to 302 (35.03%) (chr15-1). The combined alignment of the 44 non-coding genes was comprised of 23,134 bp, 1,951 bp (8.43%) of which are parsimony-informative sites. The nuclear sequence divergence ranged from 1.40% (chr10-5) to 22.60% (chr15-1), and averaged 4.01%.
The complete mt genomes range from 16,499–16,648 bp in size. Length differences were largely due to the variation in copy number of tandem repeat sequences in the conserved sequence block (CSB) domains of the mt control region. All genomes shared not only 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a control region, but also the same gene order. The mt genome sequence divergence ranged from 16.90% (COX1) to 28.80% (ATP8) for the protein-coding dataset (average 21.00%), from 10.90% (12S rRNA) to 12.70% (16S rRNA) for the rRNA dataset (average 11.80%), 12.10% for the 22 tRNA dataset, 24.80% for the control region, and 18.50% for the complete dataset.
Phylogenetic Inference
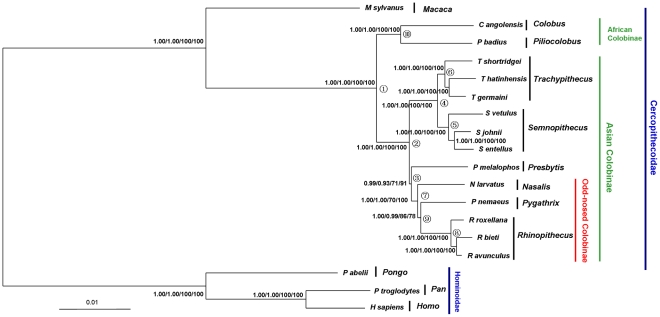
Although individual nuclear gene analyses produced incongruent topologies with low levels of nodal support (Figure S1), possibly due to limited phylogenetic information harbored in a single marker, the analyses of the combined nuclear data set using four different tree-building methods (MP, ML, Bayesian and PhyloBayes) yielded an identical, well-resolved tree topology with strong support for most nodes (Figure 2). All analyses divided colobines into reciprocally monophyletic Asian and African clades (MP BS = 100%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00). African paraphyly as suggested by Roos et al. [13] was rejected by our nuclear data (P<0.05). The Asian colobines were grouped into two clades. Clade 1 included Presbytis and the odd-nosed monkey group (MP BS = 91%; ML BS = 71%; Bayesian PP = 0.99; PhyloBayes PP = 0.93), and Clade 2 included Semnopithecus and Trachypithecus (MP BS = 100%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00). Within the monophyletic odd-nosed monkey group (MP BS = 100%; ML BS = 70%; Bayesian PP = 1.00; PhyloBayes PP = 1.00), Rhinopithecus and Pygathrix clustered together (MP BS = 78%; ML BS = 86%; Bayesian PP = 1.00; PhyloBayes PP = 0.99) to the exclusion of Nasalis. The other two alternative phylogenetic relationships among Rhinopithecus, Nasalis and Pygathrix, however, were not rejected by our nuclear data (P>0.05). In addition, the alternative placement of Presbytis as either the sister taxon to all other Asian colobines or to the Semnopithecus/Trachypithecus clade was not rejected (P>0.05).
Figure 2. Phylogenetic tree inferred from the combined 44 nuclear non-coding genes.
The nodal supports (Bayesian PP/PhyloBayes PP/ML BS/MP BS) are shown above the nodes. Node numbers in the tree indicate the nodes that were used in divergence time estimations and phylogenetic performance evaluation.
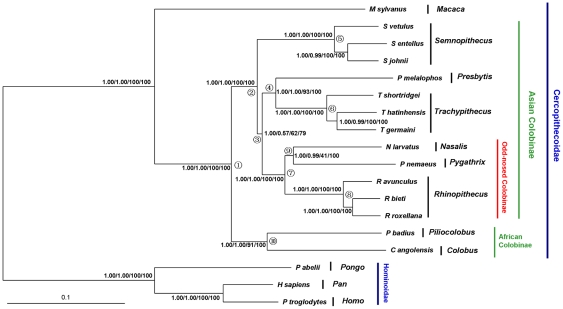
For mt gene analyses, the combined rRNA and combined tRNA data demonstrated low resolving power for phylogenetic inference compared to the combined protein-coding gene analyses and the control region analysis (see Figure S2). In comparison, the complete mt genome-based analyses, irrespective of the tree-building method that was used, produced a well-resolved and well-supported tree (Figure 3), with the tree topology being identical to those of Bayesian, PhyloBayes and ML analyses of the combined protein-coding gene analysis (Figure S2). The analyses divided colobines into reciprocally monophyletic Asian and African clades (MP BS = 100%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00). African paraphyly was rejected by our mt data (P<0.05). Within the Asian Colobinae, Semnopithecus diverged first (MP BS = 79%; ML BS = 62%; Bayesian PP = 1.00; PhyloBayes PP = 0.57), and the odd-nosed monkey group was sister to Presbytis and Trachypithecus. Monophyly of these respective clades was strongly supported (MP BS = 100%; ML BS = 93%; Bayesian PP = 1.00; PhyloBayes PP = 1.00). Within the odd-nosed monkey group, Pygathrix and Nasalis clustered together to the exclusion of Rhinopithecus (MP BS = 100%; ML BS = 41%; Bayesian PP = 1.00; PhyloBayes PP = 0.99), but alternative relationships were not rejected (P>0.05). Although Semnopithecus is suggested as the first lineage to diverge, a clade together with the odd-nosed monkey group, which was indicated in previous mt studies, is not rejected (P>0.05).
Figure 3. Phylogenetic tree inferred from the mt genome sequences.
The nodal supports (Bayesian PP/PhyloBayes PP/ML BS/MP BS) are shown above the nodes. Node numbers in the tree indicate the nodes that were used in divergence time estimations and phylogenetic performance evaluation.
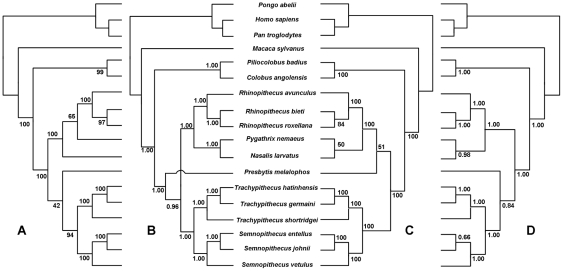
For the combined nuclear genes and mt genome dataset analyses, four different tree topologies were produced using four tree-building methods (MP, ML, Bayesian, PhyloBayes) (Figure 4). These tree topologies all support the monophyly of the Asian and African clades (MP BS = 100%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00) and the monophyly of the odd-nosed monkey group (MP BS = 100%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00), as well as the sister-group relationship between Semnopithecus and Trachypithecus (MP BS = 94%; ML BS = 100%; Bayesian PP = 1.00; PhyloBayes PP = 1.00). In comparison, the relationships among Pygathrix, Nasalis and Rhinopithecus within the odd-nosed monkey group and the placement of Presbytis within the Asian clade varied with the analytic methods used.
Figure 4. Phylogenetic trees inferred from the combined nuclear and mt dataset.
Trees are reconstructed using MP (A), Bayesian Inference (B), ML (C) and PhyloBayes (D). The nodal support values are shown above the nodes, respectively.
Assessing the Performance of Individual Nuclear and Mt Genes
The use of such a large nuclear DNA dataset and mt genome sequences from Asian colobines provides the opportunity to not only infer a colobine phylogeny but to evaluate the phylogenetic performance of the individual nuclear and mt genes as well.
The same four tree-building methods as described above were also performed on the 44 nuclear non-coding genes, 13 mt protein-coding and 2 rRNA genes individually (see Figure S1 and Figure S3; only Bayesian analyses are shown). As can be seen from the resulting phylogeny of these single-gene analyses, including those from tRNA genes and the control region (Figure S2), the relationships among Asian colobine genera were either not recovered at all or varied considerably with little or no nodal support.
We also assessed the phylogenetic utility of individual non-coding genes and mt genes in their ability to resolve the inter-generic relationships of the Colobinae by counting the number of congruent nodes between the individual phylogenies and the combined gene trees (Table 3). In the individual nuclear gene analyses, the chr12-1 gene recovered the highest number of nodes (8) congruent with the combined nuclear gene tree. The chr1-4, chr6-6, chr12-2, chr15-1 chr19-1, ENC25, chr15-3, ENC14, and X37 genes showed the fewest congruent nodes with the combined nuclear gene tree and thus had the lowest phylogenetic performance. In regard to the mt gene analyses, we observed that the COX1 gene recovered all 10 nodes of the mt genome tree. Ranking the single mt gene shows that the COX1, ND2 and ND4 genes are better indicators of colobine phylogeny at the genus level than are other genes, such as the ATP8, ND3 and ND4L genes. This result agrees broadly with previous conclusions about the rough classification of mt genes into good, medium, and poor performance categories [53]–[59] (Table S3). In summary, the assessment of the phylogenetic utility and limits of these individual nuclear and mt genes make it possible to preselect subsets of genes for future molecular studies of vertebrate phylogeny.
Table 3. Phylogenetic performance of nuclear and mt genes. Node numbers correspond to those indicated in Figure 2 and 3.
| Genea | no. congruent branches (BP>0.95) | no. congruent branches (BP<0.95) | total no. congruent branches | Nodeb | ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||||
| Nuclear genes | chr12-1 | 7 | 1 | 8 | * | * | * | * | # | * | * | * | ||
| X61 | 6 | 2 | 8 | # | * | * | * | # | * | * | * | |||
| chr17-4 | 6 | 1 | 7 | * | * | * | * | * | # | * | ||||
| chr20-4 | 4 | 3 | 7 | * | * | # | # | # | * | * | ||||
| chr5-6 | 5 | 1 | 6 | * | * | # | * | * | * | |||||
| chr20-5 | 4 | 2 | 6 | # | * | * | * | # | * | |||||
| ENC35 | 5 | 1 | 6 | # | * | * | * | * | * | |||||
| ENC15 | 0 | 5 | 5 | # | # | # | # | # | ||||||
| chr1-6 | 1 | 3 | 4 | # | # | * | # | |||||||
| chr2-8 | 1 | 3 | 4 | # | * | # | # | |||||||
| chr3-5 | 2 | 2 | 4 | # | # | * | * | |||||||
| chr6-5 | 2 | 2 | 4 | * | * | # | # | |||||||
| chr17-8 | 2 | 2 | 4 | * | * | # | # | |||||||
| X45 | 2 | 2 | 4 | * | * | # | # | |||||||
| chr4-2 | 1 | 2 | 3 | # | * | # | ||||||||
| chr7-6 | 1 | 2 | 3 | # | * | # | ||||||||
| chr8-1 | 2 | 1 | 3 | * | * | # | ||||||||
| chr9-5 | 0 | 3 | 3 | # | # | # | ||||||||
| chr13-3 | 2 | 1 | 3 | * | * | # | ||||||||
| chr18-4 | 1 | 2 | 3 | * | # | # | ||||||||
| ENC5 | 2 | 1 | 3 | # | * | * | ||||||||
| ENC19 | 0 | 3 | 3 | # | # | # | ||||||||
| X5 | 2 | 1 | 3 | # | * | * | ||||||||
| X65 | 1 | 2 | 3 | # | * | # | ||||||||
| chr2-1 | 0 | 2 | 2 | # | # | |||||||||
| chr3-2 | 1 | 1 | 2 | # | * | |||||||||
| chr4-7 | 2 | 0 | 2 | * | * | |||||||||
| chr5-8 | 0 | 2 | 2 | # | # | |||||||||
| chr8-2 | 1 | 1 | 2 | # | * | |||||||||
| chr10-1 | 2 | 0 | 2 | * | * | |||||||||
| chr10-5 | 0 | 2 | 2 | # | # | |||||||||
| chr11-2 | 1 | 1 | 2 | # | * | |||||||||
| chr13-6 | 0 | 2 | 2 | # | # | |||||||||
| chr19-5 | 2 | 0 | 2 | * | * | |||||||||
| X2 | 0 | 2 | 2 | # | # | |||||||||
| chr1-4 | 1 | 0 | 1 | * | ||||||||||
| chr6-6 | 0 | 1 | 1 | # | ||||||||||
| chr12-2 | 0 | 1 | 1 | # | ||||||||||
| chr15-1 | 1 | 0 | 1 | * | ||||||||||
| chr19-1 | 1 | 0 | 1 | * | ||||||||||
| ENC25 | 0 | 1 | 1 | # | ||||||||||
| chr15-3 | 0 | 0 | 0 | |||||||||||
| ENC14 | 0 | 0 | 0 | |||||||||||
| X37 | 0 | 0 | 0 | |||||||||||
| Mt genes | COX1 | 9 | 1 | 10 | * | * | * | * | * | * | * | * | # | * |
| ND2 | 8 | 1 | 9 | * | * | * | * | * | * | * | # | * | ||
| ND4 | 7 | 2 | 9 | * | * | # | # | * | * | * | * | * | ||
| ATP6 | 6 | 2 | 8 | * | * | * | * | # | * | # | * | |||
| ND5 | 8 | 0 | 8 | * | * | * | * | * | * | * | * | |||
| 16SrRNA | 7 | 1 | 8 | * | * | * | * | * | * | # | * | |||
| tRNA | 7 | 1 | 8 | * | * | * | * | * | * | * | # | |||
| D-loop | 8 | 0 | 8 | * | * | * | * | * | * | * | * | |||
| ND1 | 6 | 1 | 7 | * | * | * | * | # | * | * | ||||
| COX2 | 3 | 4 | 7 | * | # | # | * | * | # | # | ||||
| COX3 | 6 | 1 | 7 | * | * | * | * | * | # | * | ||||
| ND6 | 6 | 1 | 7 | # | * | * | * | * | * | * | ||||
| CYTB | 6 | 0 | 6 | * | * | * | * | * | * | |||||
| 12SrRNA | 6 | 0 | 6 | * | * | * | * | * | * | |||||
| ND4L | 3 | 2 | 5 | * | * | # | # | * | ||||||
| ND3 | 2 | 2 | 4 | # | * | * | # | |||||||
| ATP8 | 1 | 1 | 2 | * | # | |||||||||
genes are ranked by the total number of congruent branches in the combined topologies.
there are 10 nodes in total indicated in the combined nuclear gene tree (Figure 2) and the mt genome tree (Figure 3).
branches with PP>0.95 congruent in the combined topology.
branches with PP<0.95 congruent in the combined topology.
Discussion
Among mammalian phylogenies, those characterized by rapid species radiations have long been a challenging problem in species tree reconstruction [60]. This is among the first studies to utilize data from such large-scale nuclear non-coding genes in the Colobinae, which provides new insights into the phylogenetic resolution of the colobines.
Phylogeny of the Asian Colobinae
In our study, different phylogenetic relationships among Asian colobine genera were recovered by analyzing combined nuclear data, combined mt genome data, as well as combined nuclear and mt data, except for the consensus of clustering Rhinopithecus, Pygathrix and Nasalis together. This corroborates the prevailing definition of a monophyletic ‘odd-nosed’ group within the Asian Colobinae composed of Rhinopithecus, Pygathrix, Nasalis and Simias [61]–[64] and supports previous findings of mitochondrial and nuclear gene tree discordance due to ancient hybridization among the langurs [13], [18].
All analyses of combined nuclear non-coding data (Figure 2), and the ML analyses of combined nuclear and mt data (Figure 4C) supported the sister-grouping between Semnopithecus and Trachypithecus (BS = 100%; PP = 1.00), and that between Presbytis and the odd-nosed monkey group (BS = 51–91%; PP = 0.93–0.99). The recovery of a close affinity between Semnopithecus and Trachypithecus here is in accordance with previous nuclear analyses [12], [13], [18] and retroposon integration analyses [13], [17] as well as previous combined mt and nuclear dataset analyses [19]. The close relatedness of Presbytis and the odd-nosed monkey group, however, is interesting, because this finding disagrees with the Bayesian analyses of combined nuclear and mt data (Figure 4B) (PP = 0.96), as well as those based on previous and recent nuclear sequence analyses where Presbytis was placed as the earliest diverging genus among the Asian Colobinae [12], [13], [18], but it is consistent with that from mobile elements analysis [13] and previous combined mt and nuclear dataset analyses [23]. Interestingly, the MP and PhyloBayes analysis of the combined nuclear and mt dataset clusters Presbytis with the Semnopithecus/Trachypithecus clade, supporting the monophyly of the langur group (Figure 4A and D). But this relationship receives low support values. Despite our findings on the placement of Presbytis, tree topology tests do not reject alternative placements of this taxon within the Asian colobine tree. More nuclear data need to be collected to elucidate this issue.
The mt genome data analysis yielded a different tree topology (Figure 3). Semnopithecus diverged earliest within the Asian colobines and the odd-nosed monkey group was the sister-taxon to a clade uniting Presbytis and Trachypithecus (BS = 62–79%; PP = 1.00). In previous mt analyses that were based on partial genes and/or less taxonomic sampling, Semnopithecus either formed an unresolved polytomy with the other Asian colobine genera [13], [14], [17], [18] or was more closely related to the odd-nosed monkey group [16], [21], [22]. Thus, our mt genome data analysis provides support for a new phylogenetic hypothesis. However, alternative positions of Semnopithecus among Asian colobines are not rejected by our mt dataset (P>0.05). Therefore, evidence from additional data is necessary to further elucidate the placement of Semnopithecus within the Asian colobine mt gene tree. The association of Presbytis with Trachypithecus revealed here is consistent with most previous mt studies [14], [16], [18], [22].
In addition, an overview of our phylogenetic results revealed a topological discrepancy for the relationships among Rhinopithecus, Pygathrix and Nasalis within the odd-nosed monkey group. All analyses of combined nuclear data (Figure 2) and the MP analysis of the combined nuclear and mt dataset (Figure 4A) indicated a sister-taxon association between Pygathrix and Rhinopithecus to the exclusion of Nasalis (BS = 65–86%; PP = 0.99–1.00), whereas all analyses of mt genome data and the ML, Bayesian and PhyloBayes analyses of combined nuclear and mt dataset datasets (Figure 3, Figure 4B, C, D) supported a close association of Pygathrix with Nasalis (BS = 41–100%; PP = 0.98–1.00). The former result is consistent with those inferred from morphological and previous mt gene fragment analyses [5], [16],[19],[20],[31], and the latter result is in agreement with Roos et al. [13], who inferred relationships from analyses of mobile elements, nuclear sequence data, and mt genome sequences. Tree topology tests indicated that the sister-taxon association of Pygathrix and Rhinopithecus revealed by the nuclear data analyses was not rejected by both our mt dataset and the combined nuclear and mt dataset (P>0.05), and the association of Pygathrix with Nasalis revealed by the mt dataset was not rejected by both our nuclear data and the combined nuclear and mt dataset (P>0.05). The alternative hypothesis grouping Nasalis and Rhinopithecus inferred from the mt cytb analysis [21] was not recovered here in any of the analyses
Monophyly of the African Colobinae
Our combined nuclear data and mt genome data as well as the combined nuclear and mt datasets all clustered the two African colobine genera, i.e., Colobus and Piliocolobus, together with robust support (BS = 91–100%; PP = 1.00) (Figure 2, 3, 4). In the individual analyses of nuclear genes, six genes favored the paraphyly of the African colobines, but only one of them grouped Piliocolobus with the Asian colobines with high support (sensu Roos et al. [13]; Figure S1). In addition, the three combined datasets all significantly rejected the grouping of Piliocolobus with the Asian colobines (P<0.05), as well as the six individual alternative gene tree topologies (P<0.05). Our study thus supports the traditional view of African colobine monophyly and disagrees with the African colobinae paraphyly hypothesis proposed by Roos et al. [13], in which mitochondrial and nuclear gene tree discordance was explained by female introgression from Procolobus/Piliocolobus into Colobus. It is thus possible that the nuclear genes (three transposable element insertion events) that supported African colobine paraphyly in Roos et al. failed to sort into lineages that represent the species phylogeny. An alternative explanation is that female introgression from Procolobus/Piliocolobus into Colobus did indeed occur, but it was so extensive that very little evidence remains in the nuclear genome. Such minor signal would not be detected in a combined gene analysis.
Utility of the nuclear non-coding genes and mt genes in phylogenetic analysis of the Colobinae
Several recent studies have indicated that nuclear non-coding genes hold considerable signals for resolution of difficult phylogenies at both shallow and deeper species level hierarchies [59], [65]–[70]. We are among the first to use large-scale nuclear non-coding genes in inferring colobine phylogeny. Our analysis not only brings new perspectives on the phylogenetic relationships among colobine genera, but provides another example demonstrating that nuclear non-coding genes can be an effective data source for reconstructing evolutionary histories in a group that has undergone rapid bursts of speciation.
As can be seen from the results of individual gene analyses, we found that among the 44 non-coding nuclear genes, chr17-4, chr12-1, X61 and chr20-4 provide a higher level of phylogenetic resolution, while chr1-4, chr6-6, chr12-2, chr15-1 chr19-1, ENC25, chr15-3, ENC14, and X37 genes contribute the lowest levels of phylogenetic signal. When ranking single mt genes by their respective contribution to the mt genome tree, we found that some genes, such as COX1, ND2 and ND4 genes are better indicators of colobine evolutionary relationships than are other genes, such as ATP8, ND3 and ND4L genes (Table 3). Our results agree globally with those from previous mt studies of other mammalian groups regarding the rough classification of mt individual genes into good, medium, and poor categories [53]–[59] (Table S3). In all of these studies, ND2 and COX1 genes were always included in the good category, whereas ND4L and ATP8 were included in the poor category. In contrast to previous conclusions, the present work indicates that the CYTB and 12SrRNA genes are poor genetic markers for reconstructing the genus-level relationships within the Colobinae. The assessment of phylogenetic values of these nuclear and mt genes makes it possible to preselect subsets of nuclear and mt genes for phylogenetic questions at different taxonomic levels in the case of unavailable genome sequences.
Reasons for gene tree incongruence
This study raises questions regarding why the gene trees inferred here from different markers sometimes differ from one another and also from those inferred in other studies. The main areas of gene tree incongruence seem to be 1) the interrelations of the langurs between mt and nuclear DNA data (Presbytis, Semnopithecus, Trachypithecus), 2) the relationships among the African colobines in the transposable element tree versus trees inferred from mitochondrial and nuclear sequence data, 3) the placement of Presbytis in the nuclear DNA tree, 4) the interrelations of the odd-nosed monkeys. This research agrees with previous work [13], [18] in showing that the mitochondrial and nuclear gene tree discordance in the langurs is likely the result of ancient hybridization. Multiple independent nuclear markers now show that Trachypithecus and Semnopithecus are sister taxa. It is unlikely that these are all the result of incomplete lineage sorting, and it is likely that the mitochondrial lineages sorted prior to the nuclear lineages to represent the original tree (due to the smaller effective population size of the mitochondrial genome). We also believe that the African colobine paraphyly is likely due to incomplete lineage sorting of the genomic regions from where the transposable elements were sampled in Roos et al. [13]. However, it is also possible that these areas of the genome are actually representative of an ancient hybridization event, but the vast majority of the genome no longer carries that signal because hybridization was extensive. These two scenarios of incomplete lineage sorting and extensive hybridization are very difficult to disentangle. The remaining issues of gene tree discord among the colobines found here are likely due to the presence of very short internodes that preclude the capture of sufficient variation that is required to produce well-supported and well-resolved phylogenetic relationships. This is the reason why different methods generated different arrangements and why alternative arrangements are not rejected despite apparent high support. The combination of nuclear and mitochondrial datasets did not overcome this issue, possibly because of drastically different rates of evolution and different population histories. However, it is important to point out that the use of 44 non-coding regions (>23 kb) provided as much resolution, if not more, than the much faster evolving mitochondrial genomes. Thus, we believe that the collection of even more nuclear sequence data, particularly from non-coding regions that are not under purifying selection, will provide even greater resolution to the phylogenetic relationships among the colobines. Regardless, it is now even more apparent that the colobines underwent a very rapid radiation, especially among the Asian taxa, which might have required rapid and successive biogeographic vicariance events that would have affected other taxa in the Late Miocene as well.
Supporting Information
Bayesian trees of the individual nuclear non-coding genes. PPs are presented above nodes.
(TIF)
Bayesian trees of the mt datasets (1)–(4). PPs are shown above nodes. Since the resulting tree topologies for the 13 combined protein-coding gene data set differ among the MP/Bayesian/PhyloBayes/ML analyses, trees from all four reconstructions are shown. BS values are shown above nodes.
(TIF)
Bayesian trees of the individual mt genes. PPs are shown above nodes.
(TIF)
Detailed information of the 44 non-coding genes used in the present study.
(DOC)
The universal long-range PCR primer information used for mt genome amplification.
(DOC)
Comparison of phylogenetic performance of mt genes between our study and previous studies.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the State Key Basic Research and Development Plan (2007CB411600) and the National Natural Science Foundation of China (U0836603). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davies AG, Oates JF. Colobine monkeys: their ecology, behavior, and evolution. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- 2.Groves CP. Primate Taxonomy. Washington: Smithsonian Institution Press; 2001. [Google Scholar]
- 3.Brandon-Jones D, Eudey AA, Geissmann T, Groves CP, Melnick DJ, et al. Asian primate classification. Int J Primatol. 2004;25:97–164. [Google Scholar]
- 4.Groves CP. A Theory of Human and Primate Evolution. Oxford: Oxford University Press; 1989. [Google Scholar]
- 5.Jablonski NG. Quaternary environments and the evolution of primates in East Asia, with notes on two new specimens of fossil Cercopithecidae from China. Folia Primatol. 1993;60:118–132. doi: 10.1159/000156681. [DOI] [PubMed] [Google Scholar]
- 6.Napier JR, Napier PH. Old World Monkeys. New York: Academic Press; 1970. [Google Scholar]
- 7.Szalay FS, Delson E. Evolutionary History of the Primates. New York: Academic Press; 1979. [Google Scholar]
- 8.Peng Y, Pan R, Ye Z, Wang H. Comparative study on cranioface and brain case in Asian colobines. Acta Anthropol Sin. 1991;10:346–356. [Google Scholar]
- 9.Pan R. Dental variation among Asian colobines, with specific reference to the macaques on the same continent. Zool Res. 2007;28:569–579. [Google Scholar]
- 10.Bigoni F, Stanyon R, Wimmer R, Schempp W. Chromosome painting shows that the proboscis monkey (Nasalis larvatus) has a derived karyotype and is phylogenetically nested within Asian colobines. Am J Primatol. 2003;60:85–93. doi: 10.1002/ajp.10095. [DOI] [PubMed] [Google Scholar]
- 11.Bigoni F, Houck M, Ryder O, Wienberg J, Stanyon R. Chromosome painting shows that Pygathrix nemaeus has the most basal karyotype among Asian Colobinae. Int J Primatol. 2004;25:679–688. [Google Scholar]
- 12.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roos C, Zinner D, Kubatko LS, Schwarz C, Yang M, et al. Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys. BMC Evol Biol. 2011;11:77. doi: 10.1186/1471-2148-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterner KN, Raaum RL, Zhang YP, Stewart CB, Disotell TR. Mitochondrial data support an odd-nosed colobine clade. Mol Phylogenet Evol. 2006;40:1–7. doi: 10.1016/j.ympev.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 15.Weitzel V, Yang CM, Groves CP. A catalogue of primate in the Singapore Zoological Reference Collection. Raffles Bull Zool. 1988;36:1–166. [Google Scholar]
- 16.Meyer D, Rinaldi ID, Ramlee H, Perwitasari-Farajallah D, Hodges JK, et al. Mitochondrial phylogeny of leaf monkeys (genus Presbytis, Eschscholtz, 1821) with implications for taxonomy and conservation. Mol Phylogenet Evol. 2011;59:311–319. doi: 10.1016/j.ympev.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Osterholz M, Walter L, Roos C. Phylogenetic position of the langur genera Semnopithecus and Trachypithecus among Asian colobines, and genus affiliations of their species groups. BMC Evol Biol. 2008;8:58. doi: 10.1186/1471-2148-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ting N, Tosi AJ, Li Y, Zhang YP, Disotell TR. Phylogenetic incongruence between nuclear and mitochondrial markers in the Asian colobines and the evolution of the langurs and leaf monkeys. Mol Phylogenet Evol. 2008;46:466–474. doi: 10.1016/j.ympev.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Wei F, Huang C, Pan R, Ruiter JD. Phylogeny of snub-nosed monkeys inferred from Mitochondrial DNA, Cytochrome B, and 12S rRNA sequences. Int J Primatol. 2004;25:861–873. [Google Scholar]
- 20.Wang W, Forstner MRJ, Zhang YP, Liu ZM, Wei Y, et al. A phylogeny of Chinese leaf monkeys using mitochondrial ND3-ND4 gene sequences. Int J Primatol. 1997;18:305–320. [Google Scholar]
- 21.Zhang YP, Ryder OA. Mitochondrial cytochrome b gene sequences of Old World monkeys: with special reference on evolution of Asian colobines. Primates. 1998;39:39–49. [Google Scholar]
- 22.Chatterjee HJ, Ho SY, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabre PH, Rodrigues A, Douzery EJ. Patterns of macroevolution among primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 2009;53:808–825. doi: 10.1016/j.ympev.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Napier JR, Napier JR. A Handbook of Living Primates. London: Academic Press; 1967. [Google Scholar]
- 25.Ting N. Mitochondrial relationships and divergence dates of the African colobines: evidence of Miocene origins for the living colobus monkeys. J Hum Evol. 2008;55:312–325. doi: 10.1016/j.jhevol.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Groves C, Angst R, Westwood C. The status of Colobus polykomos dollmani Schwarz. Int J Primatol. 1993;14:573–586. [Google Scholar]
- 27.Grubb P, Butynski TM, Oates JF, Bearder SK, Disotell TR, et al. Assessment of the diversity of African primates. Int J Primatol. 2003;24:1301–1357. [Google Scholar]
- 28.Oates JF, Trocco TF. Taxonomy and phylogeny of black-and-white colobus monkeys: inferences from an analysis of loud call variation. Folia Primatol. 1983;40:83–113. doi: 10.1159/000156092. [DOI] [PubMed] [Google Scholar]
- 29.Struhsaker TT. Vocalizations, phylogeny and palaeogeography of red colobus monkeys (Colobus badius). Afr J Ecol. 1981;19:265–283. [Google Scholar]
- 30.Peng ZG, Elango N, Wildman DE, Yi SV. Primate phylogenomics: developing numerous nuclear non-coding, non-repetitive markers for ecological and phylogenetic applications and analysis of evolutionary rate variation. BMC Genomics. 2009;10:247. doi: 10.1186/1471-2164-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delson E. Evolutionary history of the Cercopithecidae. Contrib Primatol. 1975;5:167–217. [PubMed] [Google Scholar]
- 32.Whittaker DJ, Ting N, Melnick DJ. Molecular phylogenetic affinities of the simakobu monkey (Simias concolor). Mol Phylogenet Evol. 2006;39:887–892. doi: 10.1016/j.ympev.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook E, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutell RR, Gray MW, Schnare MN. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Springer MS, Douzery E. Secondary structure and patterns of evolution among mammalian mitochondrial 12S rRNA molecules. J Mol Evol. 1996;43:357–373. doi: 10.1007/BF02339010. [DOI] [PubMed] [Google Scholar]
- 37.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4. Sunderland MA: Sinauer Associates; 2002. [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- 42.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 43.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 46.Larget B, Simon DL. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol. 1999;16:750–759. [Google Scholar]
- 47.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 48.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 49.Dutheil J, Boussau B. Non-homogeneous models of sequence evolution in the Bio++ suite of libraries and programs. BMC Evol Biol. 2008;8:255. doi: 10.1186/1471-2148-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimodaira H, Hasegawa M. Multiple comparisons of loglikelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 51.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 52.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 53.Zardoya R, Meyer A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol. 1996;13:933–942. doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
- 54.Russo CA, Takezaki N, Nei M. Efficiencies of different genes and different tree-building methods in recovering a known vertebrate phylogeny. Mol Biol Evol. 1996;13:525–536. doi: 10.1093/oxfordjournals.molbev.a025613. [DOI] [PubMed] [Google Scholar]
- 55.Miya M, Nishida M. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 2000;17:437–455. doi: 10.1006/mpev.2000.0839. [DOI] [PubMed] [Google Scholar]
- 56.Mueller RL. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst Biol. 2006;55:289–300. doi: 10.1080/10635150500541672. [DOI] [PubMed] [Google Scholar]
- 57.Yu L, Li YW, Ryder OA, Zhang YP. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007;7:198. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunha RL, Grande C, Zardoya R. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol Biol. 2009;9:210. doi: 10.1186/1471-2148-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L, Peng D, Liu J, Luan PT, Liang L, et al. On the phylogeny of Mustelidae subfamilies: analysis of seventeen nuclear non-coding loci and mitochondrial complete genomes. BMC Evol Biol. 2011;11:92. doi: 10.1186/1471-2148-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strasser E, Delson E. Cladistic analysis of cercopithecid relationships. J Hum Evol. 1987;16:81–99. [Google Scholar]
- 61.Groves CP. The forgotten leaf-eaters and the phylogeny of Colobinae. In: Napier JR, Napier PH, editors. Old World Monkeys. New York: Academic Press; 1970. pp. 555–586. [Google Scholar]
- 62.Jablonski NG. The evolution of the doucs and snub-nosed monkeys and the question of the phyletic unity of the odd-nosed colobines. In: Jablonski NG, editor. The Natural History of the Doucs and Snub-Nosed Monkeys. Singapore: World Scientific Press; 1998. pp. 13–52. [Google Scholar]
- 63.Jablonski NG, Peng YZ. The phylogenetic relationships and classification of the doucs and snub-nosed langurs of China and Vietnam. Folia Primatol. 1993;60:36–55. doi: 10.1159/000156674. [DOI] [PubMed] [Google Scholar]
- 64.Oates JF, Davies AG, Delson E. The diversity of living colobines. In: Davies AG, Oates JF, editors. Colobine Monkeys: Their Ecology, Behavior, and Evolution. Cambridge: Cambridge University Press; 1994. pp. 45–74. [Google Scholar]
- 66.Dalebout ML, Steel D, Baker CS. Phylogeny of the beaked whale genus Mesoplodon (Ziphiidae: Cetacea) revealed by nuclear introns: implications for the evolution of male tusks. Syst Biol. 2008;57:857–875. doi: 10.1080/10635150802559257. [DOI] [PubMed] [Google Scholar]
- 67.Matthee CA, Eick G, Willows-Munro S, Montgelard C, Pardini AT, et al. Indel evolution of mammalian introns and the utility of non-coding nuclear markers in eutherian phylogenetics. Mol Phylogenet Evol. 2007;42:827–837. doi: 10.1016/j.ympev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Moller-Krull M, Delsuc F, Churakov G, Marker C, Superina M, et al. Retroposed elements and their flanking regions resolve the evolutionary history of xenarthran mammals (armadillos, anteaters, and sloths). Mol Biol Evol. 2007;24:2573–2582. doi: 10.1093/molbev/msm201. [DOI] [PubMed] [Google Scholar]
- 69.Yu L, Luan PT, Jin W, Ryder OA, Chemnick LG, et al. Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora). Syst Biol. 2011;60:175–187. doi: 10.1093/sysbio/syq090. [DOI] [PubMed] [Google Scholar]
- 70.Schroder C, Bleidorn C, Hartmann S, Tiedemann R. Occurrence of Can-SINEs and intron sequence evolution supports robust phylogeny of pinniped carnivores and their terrestrial relatives. Gene. 2009;448:221–226. doi: 10.1016/j.gene.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 71.Yu L, Wang XP, Ting N, Zhang YP. Mitogenomic analysis of Chinese snub-nosed monkeys: Evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion. 2011;11:497–503. doi: 10.1016/j.mito.2011.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian trees of the individual nuclear non-coding genes. PPs are presented above nodes.
(TIF)
Bayesian trees of the mt datasets (1)–(4). PPs are shown above nodes. Since the resulting tree topologies for the 13 combined protein-coding gene data set differ among the MP/Bayesian/PhyloBayes/ML analyses, trees from all four reconstructions are shown. BS values are shown above nodes.
(TIF)
Bayesian trees of the individual mt genes. PPs are shown above nodes.
(TIF)
Detailed information of the 44 non-coding genes used in the present study.
(DOC)
The universal long-range PCR primer information used for mt genome amplification.
(DOC)
Comparison of phylogenetic performance of mt genes between our study and previous studies.
(DOC)