Abstract
Studies on ligand interaction with quadruplex DNA, and their role in stabilizing the complex at concentration prevailing under physiological condition, has attained high interest. Electrospray ionization mass spectrometry (ESI-MS) and spectroscopic studies in solution were used to evaluate the interaction of PBD and TMPyP4 ligands, stoichiometry and selectivity to G-quadruplex DNA. Two synthetic ligands from PBD family, namely pyrene-linked pyrrolo[2,1-c][1,4]benzodiazepine hybrid (PBD1), mixed imine-amide pyrrolobenzodiazepine dimer (PBD2) and 5,10,15,20-tetrakis(N-methyl-4-pyridyl)porphyrin (TMPyP4) were studied. G-rich single-stranded oligonucleotide d(5′GGGGTTGGGG3′) designated as d(T2G8), from the telomeric region of Tetrahymena Glaucoma, was considered for the interaction with ligands. ESI-MS and spectroscopic methods viz., circular dichroism (CD), UV-Visible, and fluorescence were employed to investigate the G-quadruplex structures formed by d(T2G8) sequence and its interaction with PBD and TMPyP4 ligands. From ESI-MS spectra, it is evident that the majority of quadruplexes exist as d(T2G8)2 and d(T2G8)4 forms possessing two to ten cations in the centre, thereby stabilizing the complex. CD band of PBD1 and PBD2 showed hypo and hyperchromicity, on interaction with quadruplex DNA, indicating unfolding and stabilization of quadruplex DNA complex, respectively. UV-Visible and fluorescence experiments suggest that PBD1 bind externally where as PBD2 intercalate moderately and bind externally to G-quadruplex DNA. Further, melting experiments using SYBR Green indicate that PBD1 unfolds and PBD2 stabilizes the G-quadruplex complex. ITC experiments using d(T2G8) quadruplex with PBD ligands reveal that PBD1 and PBD2 prefer external/loop binding and external/intercalative binding to quadruplex DNA, respectively. From experimental results it is clear that the interaction of PBD2 and TMPyP4 impart higher stability to the quadruplex complex.
Introduction
Ligands containing nitrogen atoms such as azido groups or triazole ring systems and other related functionalities or scaffolds were well known for their therapeutic activity [1], [2]. Number of molecules of this type have been previously reported to interact with G-quadruplex DNA and stabilizes them [3], [4]. Considering the importance of nitrogen containing ligands, couple of PBD based ligands have been selected and examined for their ability to interact and stabilize G-quadruplex DNA. Pyrrolo[2,1-c][1,4]benzodiazepine (PBD) ligands belong to the family of naturally occurring antibiotics originated from Streptomyces species [5], that exert their cytotoxic activity by binding covalently between C11-position of the PBD and C2-amino group of the guanine residues with in the minor groove of DNA and typical examples of which include DC-81, anthramycin, tomaymycin, sibiromycin and neothramycins [6]. Moreover, PBD ligands bind to DNA sequence selectively and have potential not only as antitumor agents but also as gene regulators and probes of DNA structure [7]. Recently, number of PBD dimers as well as PBD conjugates have been designed and synthesized that exhibit promising anticancer activity with remarkable DNA binding ability [8]–[10]. In addition, PBD dimers which were designed and synthesized by joining two PBD subunits via flexible linkers have been shown to form non distorting interstrand cross-links within the minor groove of DNA and increase in the chain length of the linker in PBD dimers significantly increases the efficiency as anti tumor drugs [11]–[13]. Rettig et al., has recently shown by NMR and spectroscopic studies that PBD hybrids bind strongly to the minor groove of the DNA duplex with the formation of a covalent bond between the PBD moiety and an exocyclic guanine amino group 14–16. Various porphyrin-based ligands have been extensively studied for quadruplex interaction and binding, since they can inhibit the activity of telomerase upon binding to human telomeric G-quadruplex DNA [17], [18]. The size of porphyrin ring is similar to that of guanine quartet, hence the stability of G-quadruplex DNA by porphyrin was due to π-π stacking interaction between the porphyrin ring and guanine quartet. Even though it was known that planar porphyrin ligands bind tightly to G-quadruplex structure, there is still some controversy regarding the specific site and molecular nature of their binding [19], [20].
G-quadruplex (or) G4-DNA structures were composed of stacked tetrads, each having planar association of four guanines arranged in a cyclic manner connected by Hoogsteen hydrogen bonds [21], [22]. Formation of G-quadruplex requires presence of monovalent cations like K+, NH4 + and Na+ at the centre [23], [24]. G-rich sequences with the potential to form quadruplex structure were common in genomic DNA, and these have been identified in several biologically important regions, such as the telomeric ends of chromosomes [25], oncogene promoters [26], [27], or immunoglobulin heavy chain switch regions [28]. G-quadruplexes can fold into various conformations where the strands were oriented either in parallel or antiparallel forms and can fold either intra or inter molecularly. Due to the increased interest in quadruplex structures, their interaction with unique ligands which could bind and stabilize them was tested and such ligands could be used as potential anticancer agents, diagnostic tools, and molecular probes. Small organic molecules have been proposed to interact noncovalently with G-quadruplex through tetrad stacking, groove binding, loop binding, and intercalation between the two G-tetrads. Recently, several synthetic molecules were tested for their interaction with quadruplex DNA. Most successful among them were cationic porphyrins [19], [20], acridine derivatives [29], ethidium derivatives [30], anthraquinone derivatives [31], perylene derivatives [32], and telomestatin [33], these have been explored for their ability to bind to quadruplexes and inhibit telomerase activity. Attempts were made from our laboratory in search of suitable ligands which would interact with quadruplex and stabilize [19], [20], [29], [34], [35]. In this context, the present work was designed to study the interaction between d(T2G8) and PBD ligands as well as TMPyP4. The structure of ligands and G-quadruplex complex were shown in figure 1.
Figure 1. Structure of G-quadruplex, TMPyP4, PBD1 and PBD2.
Electrospray ionization mass spectrometry (ESI-MS) has been used as a powerful technique to examine interactions between small ligands and nucleic acids as a result of its low sample consumption, sensitivity and lesser analysis time, making it adaptable to high throughput screening techniques [36]. ESI is a suitable technique which gently lifts the non-covalent complex in the gas phase while keeping the binding interactions intact. Earlier much work has been carried out to study the non-covalent interaction of different ligands with quadruplex DNA [35]–[37]. In the present study, electrospray ionization mass spectrometry and spectroscopic methods viz., UV-Visible, fluorescence, and circular dichroism (CD) have been used to examine the interaction of therapeutically potential ligands like PBD (PBD1 and PBD2) and TMPyP4 with the G-quadruplex formed by d(T2G8). Throughout the study, interaction with TMPyP4 was considered as a control, as huge data is available on its interaction with quadruplex DNA.
Materials and Methods
The telomeric region of Tetrahymena Glaucoma contains stretches of 5′ TTGGGG 3′ sequences and they were reported to form G-quadruplex complex [23], [38]. Considering this, an oligonucleotide d(5′ GGGGTTGGGG 3′) or d(T2G8), with a molecular weight of 3180.109, was synthesized using ABI 394 DNA synthesizer and purified by RP-HPLC. 1 mM stock solution of d(T2G8) was prepared in 100 mM KBPS buffer (30 mM Potassium Phosphate, pH 7.0, with 100 mM KCl, pH 7.0). The oligonucleotide solution was heated to 90°C and slowly cooled to room temperature. DNA samples were dialyzed (1000 molecular weight cutoff membrane) against two changes of buffer (1L, 24 hours each) at 4°C. The DNA solutions were diluted before each experiment to obtain a final quadruplex concentration of 5 µM for CD, UV-Visible, and fluorescence experiments and 10 nM for ESI-MS analysis. For ESI-MS experiments, as ammonium acetate buffer was volatile, analysis of G-quadruplex-ligand complexes were performed in this buffer in order to obtain clean spectra [39] and 10% methanol was used to obtain good spray.
The cationic porphyrin, TMPyP4, was purchased from Frontier Scientific (Logan, Utah, USA) and used without further purification. The stock solution of TMPyP4 was prepared by dissolving weighed amount of ligand in sterile milli Q water. The concentration of TMPyP4 drug was calculated using the molar extinction coefficient €424 = 2.26×105 M− 1Cm− 1. Synthesis of pyrrolobenzodiazepine (PBD) ligands was carried out using the protocol mentioned in the literature [40], [41]. Stock solutions (1 mM) of these ligands were prepared by dissolving weighed amount of each ligand in methanol: water (1∶1).
Mass spectrometry
Electrospray ionization (ESI) mass spectra were recorded using Exactive Orbitrap mass spectrometer (Thermo Scientific, USA) in negative ion mode. Data was acquired using Xcalibur software (Thermo Scientific). The source conditions maintained were; sheath gas (N2) pressure, 35 psi; aux gas pressure, 5 psi; capillary temperature, 120°C; capillary voltage, −50.0 V; tube lens offset voltage, −60 V; skimmer voltage, −40 V; vaporizer temperature, 50°C. Scanning parameters were; higher energy collisional induced dissociation (HCD) gas, off; resolution, enhanced; microscans, 1; lock masses, off, AGC target, balanced and maximum injection time, 200 ms. For ESI experiments, G-quadruplex and ligands concentration was maintained at 10 nM. All the sample solutions were infused into the ESI source at a flow rate of 5 µL/min by using instrument's syringe pump.
Circular Dichroism spectroscopy
Circular dichroism (CD) experiments were performed using a JASCO 815 CD spectropolarimeter (Jasco, Tokyo, Japan). Quadruplex DNA solution was prepared in 100 mM KBPS buffer (30 mM Potassium Phosphate, pH 7.0, with 100 mM KCl, pH 7.0). G-quadruplex DNA concentration was maintained at 5.0 µM and 1.0 µM, 5.0 µM of each ligand were added. The CD spectra were recorded from 200 to 500 nm in 1 mm path length cuvette. Spectra were averaged over 3 scans, which were recorded at 100 nm/min with a response time of 1 s and a band width of 1 nm.
Fluorescence spectroscopy
Fluorescence emission spectra were measured at 25°C with Hitachi F4500 spectrofluori- meter (Maryland, USA) using a 1 cm path length quartz cuvette. Quartz cuvettes was thoroughly washed with distilled water and dilute nitric acid (approximately 0.1 N, nitric acid) to minimize nonspecific binding of the ligands to the surface of the cuvette. Throughout the fluorescence experiment, the concentration of ligands were kept constant (5 µM) and titrated with 5 µL of 5 µM G-quadruplex DNA each time. TMPyP4 was excited at 433 nm and emission spectra for each titration were collected from 600 to 800 nm. Both PBD1 and PBD2 ligands were excited at 240 nm, and emission spectra were recorded from 350 nm to 480 nm for PBD1 and 300 nm to 550 nm for PBD2.
UV-Vis absorption spectroscopy
Absorption spectra were recorded using ABI Lambda Spectrophotometer (Waltham, MA, USA) at 25°C. Experiments were carried out in polystyrene cuvettes to minimize binding of ligands to the surface of the cuvettes. 5 µM of TMPyP4 stock solution was prepared in milliQ water. 5 µM of PBDs solution was prepared in methanol and water (1∶1) and 5 µM of G-quadruplex DNA in 100 mM KBPS buffer. Each ligand (1 mL) was taken in a 1 cm path length cuvette and absorption spectra were recorded after a step wise addition of 5 µL of G-quadruplex DNA ranging from 350 nm to 500 nm. All the solutions used were freshly prepared before starting the experiment. Sample solutions were monitored until equilibrium was reached, as evidenced by constant absorbance readings.
Quadruplex DNA melting analysis using SYBR Green
To further corroborate the binding and stabilizing ability of the ligands with quadruplex DNA, melting curve analysis of G-quadruplex DNA with and without PBD ligands based on the principle of DNA-DNA relatedness assay using SYBR green (SG) was performed [42], [43]. Quadruplex DNA provides binding grooves to SYBR green molecules. Denaturation of the quadruplex DNA results in the release of SYBR green molecules into the surrounding solvent. 5 µM of quadruplex DNA with and without ligands were mixed with 10 µl of 1× SYBR green and were made to denature at a temperature gradient of 4°C to 94°C with a ramp of 0.2°C per minute in the real time PCR machine (Eppendorf Real Plex, Hamburg, Germany). Melting curve due to the dissociation of quadruplex DNA complex was analyzed and Tm was calculated.
Isothermal titration calorimetry
ITC experiments were performed using Microcal VP-ITC (Northampton, MA, USA). All the ITC experiments were done by filling the ITC cell with approximately 1.5 mL of d(T2G8) G-quadruplex DNA solution and its concentration was maintained at 20 µM and ligand concentration in the titration syringe was maintained at 5 µM. Each time 60 injections (5 µL) of PBD were added to ITC cell. Ligand addition to G-quadruplex DNA was made for every 180 seconds. Throughout the ITC experiment, the final dialysate from the appropriate oligonucleotide was used in order to maintain heat of ligand dilution constant. Each ITC experiment was performed in triplicate to avoid errors generated while performing the experiment. The integrated heat/injection data obtained in each ITC titration were fitted with algorithm developed for Mathamatica 5.0 software. In the present study, though the stoichiometry was 1∶1, the data were fitted within the experimental error using two-sites binding model to understand ligand binding affinity towards both the binding sites on quadruplex DNA. As reported earlier [44] equilibrium of macromolecules, like quadruplex DNA with multiple ligand binding sites can be illustrated by two different association constants. Thermodynamic parameters like, ΔG1, ΔG2, ΔH1, ΔH2, −TΔS1, −TΔS2, K1 and K2 were extracted directly from the fits.
Results and Discussion
Mass spectrometry
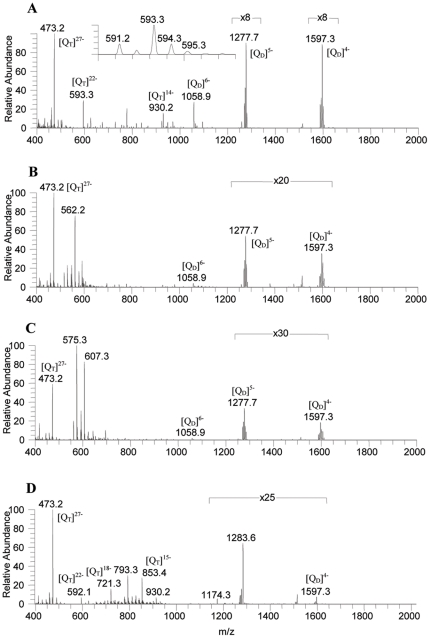
Electrospray ionization (ESI) is a soft ionization technique which allows detecting intact non-covalent complexes at very low concentration [36]. It is essential to study G-quadruplex-ligand interaction at low concentrations (close to in vivo condition), stoichiometry, affinity of each ligand towards quadruplex, and number of cations in each complex. To investigate the interaction of PBDs and TMPyP4 with G-quadruplex DNA, ESI mass spectra of each ligand with G-quadruplex were recorded and the details of mass spectral studies were given in table 1. The ESI-MS spectrum of G-rich single strand d(5′GGGGTTGGGG 3′) or d(T2G8) in 50 mM ammonium acetate solution shows deprotonated G-quadruplex ions at m/z 473 (base peak), 593, 930, 1060, 1278 and m/z 1597 with charge states of −27, −22, −14, −6, −5, and −4, respectively (Figure 2a). The ions at m/z 473, 593, and 930 correspond to the four-stranded structure d(T2G8)4 with presumably two, nine, and eight potassium cations, respectively ([QT+2K−H]27−, [QT+9K−H]22−, and [QT+8K−H]14−). We speculate that eight potassium cations stabilize eight G-quartets with one more potassium ion placed in the central region of quadruplex DNA, associate with a quartet formed by thymidines. It was shown that like guanines, thymidines also form clusters stabilized by central cations [45]. Formation of m/z 1060, 1278 and 1597 ions can be attributed to the two hairpin structures formed by d(T2G8)2 with specifically one and two ammonium adducts, at charge states of −6, −5 and −4, respectively ([QD+NH4−3H]6−, [QD+2NH4−H]5− and [QD+2NH4−2H]4−). QD and QT corresponds to quadruplex in the dimeric (two hairpin structures) and tetrameric forms respectively. Since cations were known to sit between two stacks or in the centre of quadruplex [46], complex formed by d(T2G8)2 and d(T2G8)4 was stabilized by ammonium and potassium cations, respectively, indicate that these ions were placed in the centre of the quadruplex. The ammonium or potassium ions are expected to be readily eliminated if they are non-specifically attached to the DNA phosphate groups.
Table 1. ESI-MS spectra of PBDs (PBD1 and PBD2) and TMPyP4 with G-quadruplex DNA with m/z (Relative abundances) and stoichiometry.
| G-quadruplex | Ligand | m/z (Relative abundance) | Charge of the complex | Quadruplex∶Ligand |
| d(T2G8) | No ligand | 473 (100) | [QT+2K−H]27− | 1∶0 |
| 593 (30) | [QT+9K−H]22− | 1∶0 | ||
| 930 (15) | [QT+8K−H]14− | 1∶0 | ||
| 1060 (27) | [QD+NH4−3H]6− | 1∶0 | ||
| 1278 (12) | [QD+2NH4−H]5− | 1∶0 | ||
| 1597 (11) | [QD+2NH4−2H]4− | 1∶0 | ||
| d(T2G8) | PBD1 | 473 (100) | [QT+2K−H]27− | 1∶0 |
| 516 (15) | [QT+PBD1+5K−H]26− | 1∶1 | ||
| 530 (23) | [QT+PBD1+K−H]25− | 1∶1 | ||
| 545 (23) | [QT+10K−H]24− | 1∶0 | ||
| 562 (75) | [QT+PBD1+7K−H]24− | 1∶1 | ||
| 578 (15) | [QT+PBD1+2K−H]23− | 1∶1 | ||
| 592 (28) | [QT+8K+NH4−H]22− | 1∶0 | ||
| 608 (3) | [QT+PBD1+4K−H]22− | 1∶1 | ||
| 622 (3) | [QT+9K+NH4−H]21− | 1∶0 | ||
| 638 (3) | [QT+PBD1+4K−H]21− | 1∶1 | ||
| 652 (2) | [QT+9K−H]20− | 1∶0 | ||
| 1060 (4) | [QD+NH4−3H]6− | 1∶0 | ||
| 1277 (2.6) | [QD+2NH4−2H]5− | 1∶0 | ||
| 1597 (2) | [QD+2NH4−2H]4− | 1∶0 | ||
| d(T2G8) | PBD2 | 473 (60) | [QT+2K−H]27− | 1∶0 |
| 561 (17) | [QT+PBD2+5K−H]24− | 1∶1 | ||
| 575 (100) | [QT+PBD2+NH4−2H]23− | 1∶1 | ||
| 593 (30) | [QT+9K−H]22− | 1∶0 | ||
| 607 (82) | [QT+PBD2+2K−H]22− | 1∶1 | ||
| 653 (4) | [QT+9K−H]20− | 1∶0 | ||
| 667 (4) | [QT+PBD2+2K−H]20− | 1∶1 | ||
| 1060 (3) | [QD+NH4−4H]6− | 1∶0 | ||
| 1278 (1.0) | [QD+2NH4−H]5− | 1∶0 | ||
| 1597 (0.5) | [QD+2NH4−2H]4− | 1∶0 | ||
| d(T2G8) | TMPyP4 | 473 (100) | [QT+2K−H]27− | 1∶0 |
| 592 (5) | [QT+8K+NH4−H]22− | 1∶0 | ||
| 721 (15) | [QT+7K−H]18− | 1∶0 | ||
| 793 (30) | [QT+TMPyP4+6K−2H]18− | 1∶1 | ||
| 853 (27) | [QT+2K+NH4−H]15− | 1∶0 | ||
| 930 (7) | [QT+8K−H]14− | 1∶0 | ||
| 1174 (0.3) | [QT+TMPyP4+NH4−H]12− | 1∶1 | ||
| 1284 (2.6) | [QT+TMPyP4+3NH4−H]11− | 1∶1 | ||
| 1597 (0.4) | [QD+2NH4−2H]4− | 1∶0 |
Figure 2. ESI-MS spectra of A d(T2G8) G-quadruplex, B G-quadruplex with PBD1, C G- quadruplex with PBD2, and D G-quadruplex with TMPyP4 drug.
The ESI-MS spectrum of G-quadruplex with PBD1 was shown in figure 2b. The spectrum exhibits the ions at m/z 473, 516, 530, 545, 562, 578, 592, 608, 622, 638, 652, 1060, 1277, and 1597. It has been observed that the intensities of lower charged species at m/z 1060 ([QD+NH4−3H]6−), 1277 ([QD+2NH4−2H]5−) and m/z 1597 ([QD+2NH4−2H]4−) decreased after the addition of PBD1. The peaks at m/z 473, 545, 592, 622, and 652 can be attributed to ([QT+2K−H]27−), ([QT+10K−H]24−), ([QT+8K+NH4−H]22−), ([QT+9K+NH4−2H]21−) and ([QT+9K−H]20−), respectively. Whereas, the peaks at m/z 516, 530, 562, 578, 608, and 638 correspond to ([QT+PBD1+5K−H]26−), ([QT+PBD1+K−H]25−), ([QT+PBD1+7K−H]24−), ([QT+PBD1+2K−H]23−), ([QT+PBD1+4K−H]22−) and ([QT+PBD1+4K−H]21−), respectively formed with the interaction of PBD1. Figure 2c shows the ESI-MS spectrum of G-quadruplex with PBD2. The spectrum exhibits the peaks at m/z 473, 561, 575, 593, 607, 653, 667, 1060, 1278, and 1597. It has been observed that after the addition of PBD2, the intensities of m/z 1060 ([QD+NH4−4H]6−), 1277 ([QD+2NH4−2H]5−) and 1597 ([QD+2NH4−2H]4−) decreased. The ions at m/z 473, 593 and 653 can be attributed to ([QT+2K−H]27−), ([QT+9K−H]22−) and ([QT+9K−H]20−), respectively. The formation of ions at m/z 561, 575, 607 and 667 corresponding to ([QT+PBD2+5K−H]24−), ([QT+PBD2+NH4−3H]23−), ([QT+PBD2+2K−H]22−) and ([QT+PBD2+2K−H]20−), respectively indicate that the interaction of PBD2 with the G-quadruplex DNA. Figure 2d shows the ESI-MS spectrum of the G-quadruplex with TMPyP4. The spectrum shows the ions at m/z 473, 592, 721, 793, 853, 930, 1174, 1284, and 1597. The ions at m/z 721, 793, 853, 930, 1174, and 1284 can be attributed to ([QT+7K−H]18−), ([QT+TMPyP4+6K−2H]18−), ([QT+2K+NH4−H]15−), ([QT+8K−H]14−), ([QT+TMPyP4+NH4−H]12−) and ([QT+TMPyP4+3NH4−H]11−), respectively. Occurrence of peaks with m/z 793, 1174, and 1284 ions clearly indicate an interaction between TMPyP4 and G-quadruplex DNA. From the present study, it was understood that when TMPyP4 was added to G-quadruplex DNA in 1∶1 molar ratio (10 nM), the relative abundance of peaks, corresponding to the G-quadruplex DNA decreased significantly. This indicates an effective interaction of TMPyP4 and stabilization of d(T2G8) G-quadruplex. Probably due to lower ammonium ion concentration and formation of secondary structures, the peaks in the lower m/z region (m/z 400–1000) were sharp and intense with low abundance cationic adducts around them. In order to view these peaks clearly with their cationic adducts, the lower m/z region was expanded and shown in (Figure S1, Figure S2, Figure S3, Figure S4).
A parameter, IRa [47], that denotes the relative binding affinity of PBD ligands (PBD1 and PBD2) and TMPyP4 to the d(T2G8) G-quadruplex DNA, was calculated by using the following equation.
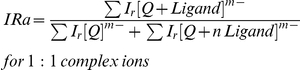 |
Where ∑ Ir[Q]m− and ∑ Ir[Q+Ligand]m− were the total intensities of G-quadruplex and 1∶1 quadruplex complex: ligand, respectively. Table 2 shows the relative binding affinity (IRa) of the ligands with the G-quadruplex. These ligands have different binding affinities for 1∶1 complexes. The IRa values show that the binding affinity of PBD1 was comparatively less than PBD2. This indicates that the interaction of PBD2 with the G-quadruplex DNA is more as compared to the interaction of PBD1 with the G-quadruplex. Intercalation between quadruplex and ligand depends on certain factors like, planarity and orientation of the interacting ligand. PBDs were previously reported to possess planar structure and intercalate to DNA [12], [48]. The relative binding affinity values for PBD1 and PBD2 were found to be 0.45 and 0.67 respectively. This indicates that the binding affinity of PBD ligands was in the order, PBD2>PBD1. TMPyP4 ligand was reported to bind to the G-quadruplex DNA through intercalation, because of its planarity [49].
Table 2. IRa values of ligands (PBD1, PBD2, and TMPyP4) with the G-quadruplex DNA.
| Ratio of Quadruplex∶Ligand | IRa (Relative binding affinity) |
| Q:PBD1 (1∶1) | 0.45 |
| Q:PBD2 (1∶1) | 0.67 |
| Q:TMPyP4 (1∶1) | 0.18 |
CD spectroscopy
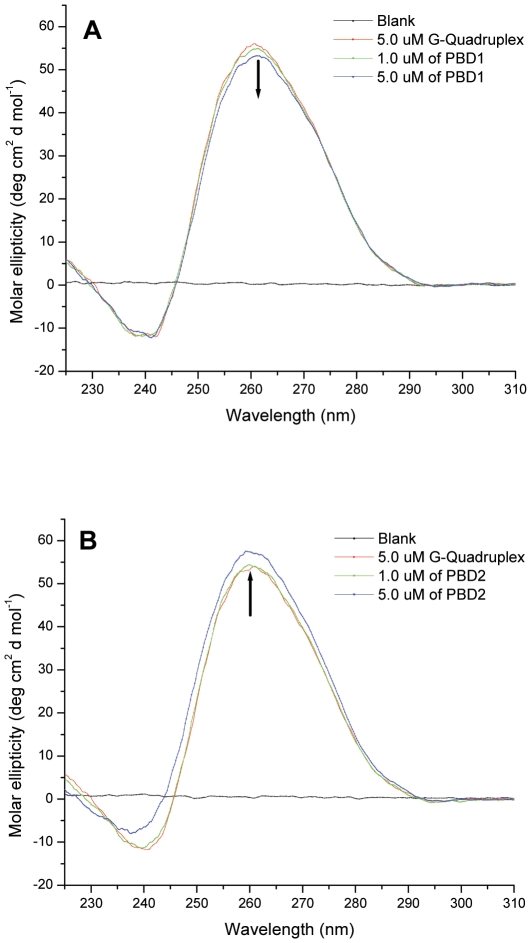
Circular dichroism has been used to understand the changes in the conformation of G-quadruplex on interaction with ligand. Parallel quadruplexes display a positive CD peak around 265 nm and a negative peak at 240 nm, while antiparallel ones exhibit a positive peak at around 295 nm, and a negative peak around 260 nm [50]. CD spectrum of 5.0 µM G-quadruplex with d(T2G8) showed a prominent positive peak at around 265 nm and a negative peak at 240 nm, indicating the presence of parallel inter-molecular quadruplex conformation in 100 mM KBPS buffer. Figure 3a demonstrates the effect of PBD1 on interaction with d(T2G8) quadruplex complex. On addition of 1.0 µM and 5.0 µM of PBD1 to G-quadruplex DNA, the peak at 265 nm exhibits hypochromicity and very low negative cotton effect. Hypochromicity of CD band was low and no shift of soret band was observed. The hypochromicity of CD band can be attributed to the partial unfolding of G-quadruplex DNA [51]. Figure 3b shows the CD spectrum of G-quadruplex with PBD2. On addition of 1.0 µM and 5.0 µM of PBD2 to G-quadruplex DNA, hyperchromicity and lesser positive cotton effect was observed. This indicates that PBD2 interacts with the parallel inter-molecular G-quadruplex DNA and stabilizes the complex [52].
Figure 3. CD spectra of A PBD1on interaction with G-quadruplex and B PBD2 interaction with G-quadruplex in 100 mM KBPS buffer (pH 7.0).
The CD spectrum of G-quadruplex DNA with 1.0 µM and 5.0 µM of TMPyP4 exhibits a more hypochromicity, induced CD (ICD) at 463 nm and negative cotton effect probably due to higher intercalation and end loop binding of TMPyP4 to G-quadruplex DNA (spectrum is not shown). Among the porphyrins, TMPyP4 was known to have better interaction with G-quadruplex DNA [19], [20]. From circular dichroism experiments, it was evident that the interaction of the PBD ligands with G-quadruplex DNA was in the order; PBD2>PBD1.
UV-Visible spectroscopy
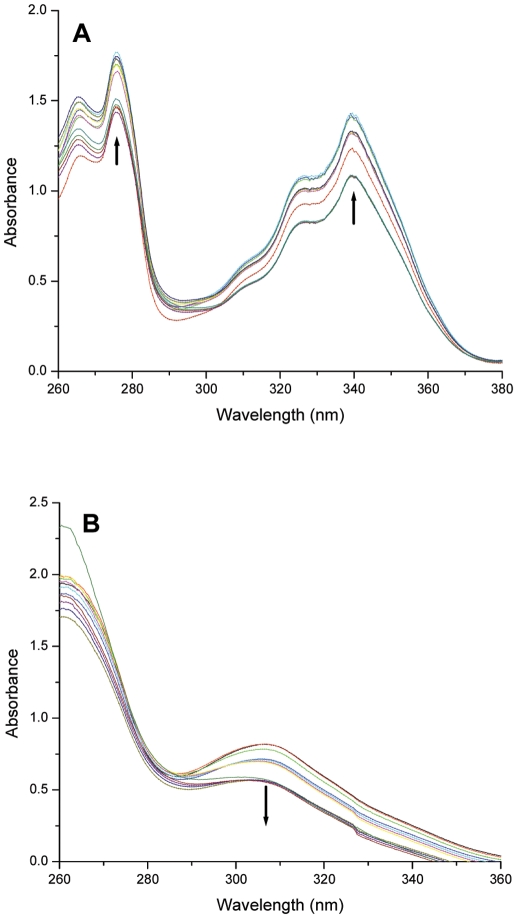
UV-Visible spectroscopy is an excellent technique to comprehend DNA-ligand binding. To gain insight into the interaction between ligands and the G-quadruplex, the UV-Visible spectra of PBD ligands (PBD1 and PBD2) and TMPyP4 were recorded in the absence and presence of G-quadruplex DNA. The titration was continued until the wavelength and intensity of the absorption band of ligand did not change any more upon three successive additions of G-quadruplex. UV-Vis spectrum of PBD1 was remarkably different, consisting of two absorption peaks at 276 and 339 nm (Figure 4a). The soret band exhibited less hyperchromicity and does not show any shift either from 276 or from 339 nm. No shift of the soret UV-Visible peak and lesser hyperchromicity indicate that interaction of PBD1 with quadruplex DNA takes place through external binding. The isobestic point was observed at around 280 nm. The isobestic point was not a sharp, tight single point. The slight deviation of isobestic point from a single point indicates that the interaction of PBD1 with G-quadruplex DNA occurs through multiple steps.
Figure 4. UV-Visible spectra of A G-quadruplex (5 µM) on interaction with PBD1 ligand and B G-quadruplex (5 µM) on interaction with PBD2 ligand.
The UV-Visible spectrum of PBD2 exhibits the soret band at 306 nm (Figure 4b). The soret band shows hypochromicity and does not show any shift. Presence of less intense hypochromic band indicates a mild intercalation of PBD2 with the G-quadruplex DNA. The isobestic point was observed at around 276 nm and the deviation of isobestic point from a single point shows that the interaction of PBD2 with the G-quadruplex DNA takes place through multiple steps. On interaction of d(T2G8) quadruplex complex to TMPyP4, the porphyrin soret band has shown bathochromic shift of 10 nm and higher percentage of hypochromicity compared to PBD2. This indicates that TMPyP4 has a higher level of interaction (mostly by intercalation) with quadruplex DNA. Higher level of TMPyP4 interaction with quadruplex DNA may be due to planar structure of TMPyP4 molecule compared to PBD2. The UV-Visible data obtained on ligand-quadruplex DNA interaction was shown in table 3. Percentage of hypochromicity for each ligand was calculated by following the procedure described by Keating and Szalai [53]. From the results obtained from the spectroscopic and mass experiments, we speculate that PBD1 bind externally and PBD2 exhibit both modes of interaction (external binding/intercalation) with quadruplex DNA. TMPyP4 interacts more efficiently to the G-quadruplex DNA than PBD ligands.
Table 3. UV-Visible spectral titration results for binding of PBD1, PBD2, and TMPyP4 to d(T2G8) G-quadruplex.
| G-quadruplex | Ligand | Soret band shift (nm) | Isobestic point (nm) | % hypochromicity | % hyperchromicity |
| d(T2G8) | PBD1 | No shift | 280 | - | 16.2 |
| d(T2G8) | PBD2 | No shift | 276 | 29.5 | - |
| d(T2G8) | TMPyP4 | 424–434 | 435 | 38.2 | - |
Fluorescence spectroscopy
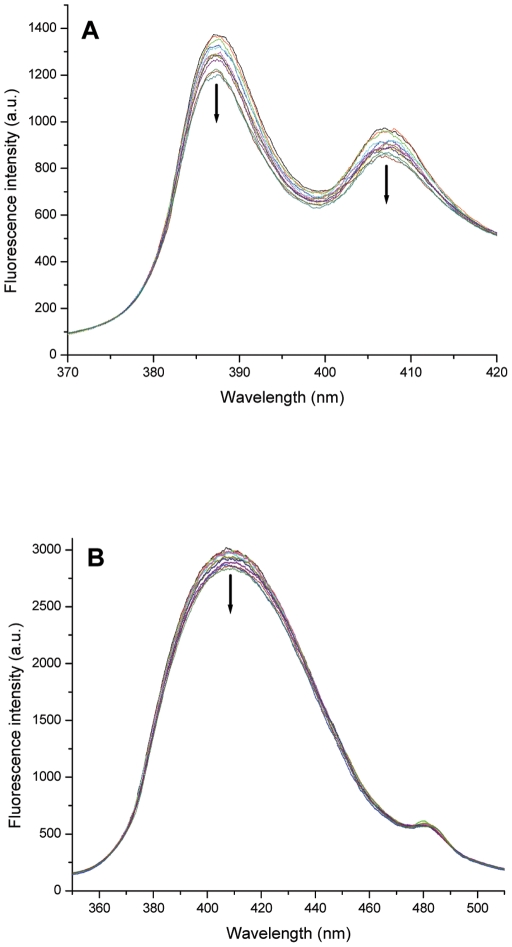
In order to understand the mode of ligand interaction with G-quadruplex DNA fluorescence emission spectra for PBD ligands (PBD1 and PBD2) and TMPyP4 were recorded in the absence and presence of different amounts of G-quadruplex DNA. Figure 5a shows the representative fluorescence spectrum of PBD1 titrated with different amounts of G-quadruplex DNA. Fluorescence emission spectrum with PBD1 shows two peaks corresponding to 387 and 407 nm, which gradually decrease on addition of G-quadruplex. The fluorescence quenching observed for two peaks at 387 nm and 407 nm were found to be 13.13% and 11.03%, respectively. The emission spectrum of PBD2 shows two emission peaks at 410 nm and 482 nm. The fluorescence emission peak at 410 nm slightly decreased on addition of G-quadruplex DNA and its quenching was found to be 4.9%. From figure 5a and 5b it was evident that the fluorescence quantum yield of PBD2 was much higher than PBD1. This may be due to π-π electronic transitions occurring between the ligand and the guanine bases when PBD2 intercalates between two quartets. We speculate that on addition of G-quadruplex DNA, fluorescence quenching of PBD1 and PBD2 occurs due to two reasons, firstly, the PBDs prefers to bind externally to the G-quadruplex and expose to the solvent environment, resulting in the quenching of fluorescence. Secondly, binding of PBD ligands takes place externally due to the smaller size of the quadruplex complex formed by d(T2G8). Quenching of PBD1 (13.13% and 11.03% for peaks at 387 nm and 407 nm, respectively) was more compared to PBD2 (4.9% for 410 nm peak) on addition of G-quadruplex DNA, indicating the preferential binding of PBD1 to quadruplex DNA, externally. From figure 5b, it was evident that PBD2 fluorescence quantum yield was high and quenching was low. This may be due to moderate intercalation of PBD2 with quadruplex DNA. This is in agreement with UV-Visible experimental results.
Figure 5. Fluorescence emission spectra of A G-quadruplex on interaction with PBD1 ligand and B G-quadruplex on interaction with PBD2 ligand.
The fluorescence emission spectrum of TMPyP4 shows a peak at 780 nm. On addition of G-quadruplex DNA, the fluorescence intensity of TMPyP4 was enhanced to 18.8%. From this, it was evident that fluorescence enhancement of TMPyP4 may be attributed to intercalative and end loop binding [54]. On intercalation, TMPyP4 was sandwiched between two quartets, which shield the ligand from surrounding solvent molecules, resulting in the fluorescence intensity enhancement. From the experimental results, we speculate that PBD1 binds externally and PBD2 moderately intercalates to d(T2G8) quadruplex DNA. The experimental details obtained from fluorescence titrations were given in table 4. Studies on the interaction of PBD ligands with quadruplex DNA structures formed by various promoter regions of oncogenes are in progress.
Table 4. Fluorescence emission titration results for binding of PBD1, PBD2, and TMPyP4 to d(T2G8) G-quadruplex.
| G-quadruplex | Ligand | Soret band (nm) | % of fluorescence quenching | % of fluorescence enhancement |
| d(T2G8) | PBD1 | 387 | 13.13 | - |
| d(T2G8) | PBD1 | 407 | 11.03 | |
| d(T2G8) | PBD2 | 410 | 4.9 | - |
| d(T2G8) | TMPyP4 | 780 | - | 18.79 |
Quadruplex DNA melting analysis using SYBR Green
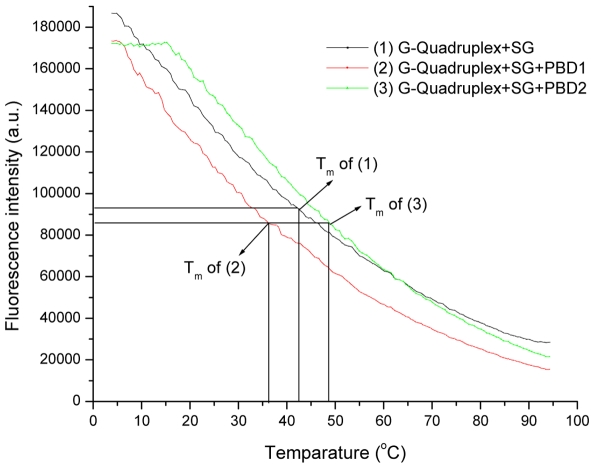
From earlier studies, it was known that dsDNA specific dyes like SYBR Green I (SG) exhibits fluorescence via surface or groove binding, when the ratio of base pair to SG molecule complexes exceeds 0.15 [55]. In the present study, stabilization and binding affinity of PBD ligands (PBD1 and PBD2) to G-quadruplex DNA was studied by exploiting the specificity and higher fluorescence of SG on binding to G-quadruplex DNA. On increase of temperature, quadruplex DNA melts to single stranded form releasing the SG molecules into the surrounding environment. This will decrease SG fluorescence. Figure 6 demonstrates the melting profiles of quadruplex DNA alone, quadruplex with PBD1 and PBD2 with SG which were marked 1, 2, and 3 respectively. From the melting profile, it was evident that Tm of quadruplex alone, with PBD1 and PBD2 was 43°C, 36°C and 49°C respectively. Lowering of Tm after the addition of PBD1 indicates unfolding of quadruplex DNA to single stranded form. About 6°C increase of Tm with PBD2 confirms stabilization of G-quadruplex DNA. The results obtained from DNA melting experiments using SG were in agreement with the spectroscopic and ESI-MS studies. The present study was aimed to understand the affinity and the interaction of ligands from a PBD family with quadruplex DNA, a non-canonical DNA structure, presumably involved in transcription regulation; indicate that PBD2 and similar structured ligands might function as an efficient anti-cancer agents.
Figure 6. Melting of (1) G-quadruplex using SYBR Green alone, (2) with PBD1, and (3) with PBD2.
Isothermal titration calorimetry
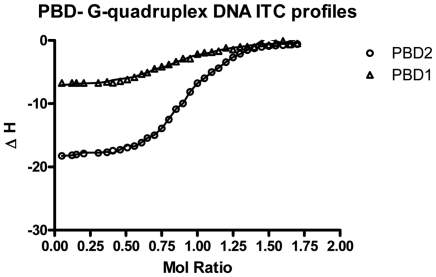
Isothermal calorimetry was used as a sensitive and direct micro calorimetric technique with precession for determining the binding affinity, stoichiometry and thermodynamic parameters [56]. Interaction between d(T2G8) G-quadruplex DNA and PBD ligands (PBD1 and PBD2) was analyzed at 25°C using 100 mM KBPS (pH 7.0) buffer was documented in figure 7 and the results obtained were shown in table 5. From the results, it was clear that PBD2 interaction with quadruplex occurs in two ways. One is a most favorable interaction process (mode-1), which takes place in combination of contribution from enthalpy (−2 to −4 kcal/mol) and significant favorable entropy (−3.0 to −6.0 kcal/mol). The second process (mode-2), which was relatively unfavorable process, proceeds with more significant enthalpy (−5.0 to −7.0 kcal/mol) contri- bution and a smaller entropy range (−1 to −3 kcal/mol). Analyzing the ITC results, the energy profiles show that interaction of PBD1 takes place with one binding mode closer to higher affinity binding (mode-1) of PBD2, considering the relative entropy and enthalpy contribution to the overall binding free energy change.
Figure 7. ITC binding isotherms for titration of PBD ligands (PBD1 and PBD2) with G- quadruplex DNA in 100 mM KBPES (pH 7.0) buffer.
Table 5. Thermodynamic parameters for the interaction of PBD ligands (PBD1 and PBD2) with G-quadruplex determined by ITC at 25°C and pH 7.0.
| ITC Derived Thermodynamic Parameters | PBD1-Quadruplex DNA | PBD2-Quadruplex DNA |
| K1×10−8 | 0.0597 (±0.01) | 0.0875 (±0.03) |
| ΔG1 (kcal/mol) | −8.73 (±0.02) | −6.35 (±0.01) |
| ΔH1 (kcal/mol) | −2.57(±0.03) | −3.46 (±0.02) |
| −TΔS1 (kcal/mol) | −6.09 (±0.01) | −3.47 (±0.01) |
| K2×10−6 | 0.222(±0.15) | 0.483 (±0.11) |
| ΔG2 (kcal/mol) | −4.65 (±0.01) | −3.23 (±0.03) |
| ΔH2 (kcal/mol) | −6.54 (±0.01) | −5.86 (±0.01) |
| −TΔS2 (kcal/mol) | −2.87 (±0.02) | −1.82 (±0.01) |
Parameters mentioned above are for a two-sites binding model. The uncertainties mentioned for the two fitting parameters, Ki and ΔHi, were determined from Monte Carlo analysis.
From the ITC results, the swift and higher binding process, which is more entropy driven resembles the binding of ligands to the exterior of DNA. It may be the binding of PBD ligands to the exterior or the two terminal end loops (end staking) of the quadruplex DNA. It may be due to binding of non planar (like PBD1) ligands externally, which cannot interact with quadruplex DNA by staking between the two orderly placed tetrads. The second process is enthalpy driven as the binding of a planar molecule between the two orderly placed tetrads occurs with a typical exothermic enthalpy change due to increased π-π staking interaction between the interacting ligand and the DNA bases [57]–[61]. We speculate that the second process which is significantly enthalpy driven is a more likely an “intercalation process”.
Conclusions
In this study, electrospray ionization mass spectrometry (ESI-MS), microcalorimetry and spectroscopy have been used to evaluate the stoichiometry, stability, ligand-DNA interaction, and selectivity of PBDs (PBD1 and PBD2) and TMPyP4 to quadruplex DNA. ESI-MS, ITC, UV-Visible and fluorescence experiments indicate that PBD1 exhibits external/end-loop binding whereas PBD2 moderately intercalates/bind externally to quadruplex DNA. CD experimental data demonstrate the existence of parallel inter-molecular G-quadruplex in solution. CD and melting studies with SYBR green reveals that PBD1 unfolds and PBD2 stabilizes the quadruplex DNA.
Supporting Information
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with PBD1. The interaction peaks were marked with “•”.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with PBD2. The interaction peaks were marked with “♦”.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with TMPyP4. The interaction peaks were marked with “▪”.
(TIF)
Acknowledgments
The authors thank Dr. J. S. Yadav, Director, IICT, Hyderabad and Dr.Ch Mohan Rao, Director, CCMB for their encouragement and support. G. R. and V.S.R thank UGC and CSIR, New Delhi for the award of Senior Research Fellowships.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no funding or support to report.
References
- 1.Reyes S, Huigens RW, Su Z, Simon ML, Melander C. Synthesis and biological activity of 2-aminoimidazole triazoles accessed by Suzuki–Miyaura cross-coupling. Org Biomol Chem. 2011;9:3041–3049. doi: 10.1039/c0ob00925c. [DOI] [PubMed] [Google Scholar]
- 2.Lavie A, Su Y, Ghassemi M, Novak RM, Caffrey M, et al. Restoration of the antiviral activity of 3′-azido-3′-deoxythymidine (AZT) against AZT-resistant human immunodeficiency virus by delivery of engineered thymidylate kinase to T cells. Journal of General Virology. 2008;89:1672–1679. doi: 10.1099/vir.0.2008/000273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakali JE, Klupsch F, Guédin A, Brassart B, Fontaine G, et al. 2,6-Diphenylthiazolo[3,2-b][1,2,4]triazoles as telomeric G-quadruplex stabilizers. Bioorg Med Chem Lett. 2009;19:3434–3438. doi: 10.1016/j.bmcl.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 4.Tajmir-Riahi HA. AZT Binding to DNA and RNA: Molecular Modeling and Biological Significance. Journal of the Iranian Chemical Society. 2005;2:78–84. [Google Scholar]
- 5.Thurston DE. In Molecular Aspects of Anticancer Drug-DNA Interactions: Neidle S, Waring MJ Eds. 1993. pp. 54–88. The Macmillan Press Ltd. London, UK.
- 6.Hadjivassileva T, Thurston DE, Taylor PW. Pyrrolobenzodiazepine dimers: novel sequence-selective, DNA-interactive, cross-linking agents with activity against Gram-positive bacteria. Journal of Antimicrobial Chemotherapy. 2005;56:513–518. doi: 10.1093/jac/dki256. [DOI] [PubMed] [Google Scholar]
- 7.Hurley LH. DNA and associated targets for drug design. J Med Chem. 1989;32:2027–2033. doi: 10.1021/jm00129a001. [DOI] [PubMed] [Google Scholar]
- 8.Kamal A, Kashi Reddy M, Janaki Ramaiah M, Srikanth YVV, Rajender, et al. Synthesis of Aryl-Substituted Naphthalene-Linked Pyrrolobenzodiazepine Conjugates as Potential Anticancer Agents with Apoptosis-Inducing Ability. ChemMedChem. 2011;6:1665–1679. doi: 10.1002/cmdc.201100207. [DOI] [PubMed] [Google Scholar]
- 9.Kamal A, Kashi Reddy M, Janaki Ramaiah M, Rajender, Surendranadha Reddy J, et al. Synthesis and biological evaluation of estradiol linked pyrrolo[2,1-c]-[1,4]benzodiazepine (PBD) conjugates as potential anticancer agents. Bioorg Med Chem. 2011;19:2565–2581. doi: 10.1016/j.bmc.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Kamal A, Prabhakar S, Janaki Ramaiah M, Venkat Reddy P, Ratna Reddy Ch, et al. Synthesis and anticancer activity of chalcone-pyrrolobenzodiazepine conjugates linked via 1,2,3-triazole ring side-armed with alkane spacers. European Journal of Medicinal Chemistry. 2011;46:3820–3831. doi: 10.1016/j.ejmech.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 11.Cipolla L, Araújo AC, Airoldi C, Bini D. Pyrrolo[2,1-c][1,4]benzodiazepine as a Scaffold for the Design and Synthesis of Anti Tumour Drugs. Anti-Cancer Agents in Medicinal Chemistry. 2009;9:1–31. doi: 10.2174/187152009787047743. [DOI] [PubMed] [Google Scholar]
- 12.Hopton SR, Thompson AS. Nuclear Magnetic Resonance Solution Structures of Inter- and Intrastrand Adducts of DNA Cross-Linker SJG-136. Biochemistry. 2011;50:4720–4732. doi: 10.1021/bi102017e. [DOI] [PubMed] [Google Scholar]
- 13.Reddy BS, Damayanthi Y, Reddy BS, Lown JW. Design, synthesis and in vitro cytotoxicity studies of novel pyrrolo[2,1-c][1,4]benzodiazepine (PBD)-polymade conjugates and 2,2′-PBD dimers. Anticancer Drug Des. 2000;15:225–238. [PubMed] [Google Scholar]
- 14.Rettig M, Langel W, Kamal A, Weisz K. NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate. Org Biomol Chem. 2010;8:3179–3187. doi: 10.1039/c001893g. [DOI] [PubMed] [Google Scholar]
- 15.Rettig M, Weingarth M, Langel W, Kamal A, Praveen Kumar P, et al. Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine Benzimidazole Hybrid to a 10mer DNA Duplex. Biochemistry. 2009;48:12223–12232. doi: 10.1021/bi901655t. [DOI] [PubMed] [Google Scholar]
- 16.Rettig M, Kamal A, Ramu R, Mikolajczak J, Weisz K. Spectroscopic and calorimetric studies on the DNA recognition of pyrrolo[2,1-c][1,4]benzodiazepine hybrids. Bioorg Med Chem. 2009;17:919–928. doi: 10.1016/j.bmc.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Han H, Langley DR, Rangan A, Hurley LH. Selective Interactions of Cationic Porphyrins with G-Quadruplex Structures. J Am Chem Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 18.Dixon IM, Lopez F, Estève JP, Tejera AM, Blasco MA, et al. Porphyrin Derivatives for Telomere Binding and Telomerase Inhibition. ChemBioChem. 2005;6:123–132. doi: 10.1002/cbic.200400113. [DOI] [PubMed] [Google Scholar]
- 19.Nagesh N, Sharma VK, Ganesh Kumar A, Lewis EA. Effect of Ionic Strength on Porphyrin Drugs Interaction with Quadruplex DNA Formed by the Promoter Region of C-myc and Bcl2 Oncogenes. Journal of Nucleic Acids. 2010 doi: 10.4061/2010/146418. doi: 10.4061/2010/146418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagesh N, Buscaglia R, Dettler JM, Lewis EA. Studies on the Site and Mode of TMPyP4 Interactions with Bcl-2 Promoter Sequence G-Quadruplexes. Biophys J. 2010;98:2628–2633. doi: 10.1016/j.bpj.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, et al. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 22.Williamson JR. G-quartet structures in telomeric DNA. Ann Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 23.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis JT. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew Chem Int Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 25.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J Bio Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 27.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annu Rev Pharmacol Toxicol. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 29.Nagesh N, Krishnaiah AA. A comparative study on the interaction of acridine and synthetic bis-acridine with G-quadruplex structure. J Biochem Biophys Methods. 2003;57:65–74. doi: 10.1016/s0165-022x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 30.Koeppel F, Riou JF, Laoui A, Mailliet P, Arimondo PB, et al. Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res. 2001;29:1087–1096. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the First Parallel DNA Quadruplex-Drug Complex. J Am Chem Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 32.Tuntiwechapikul W, Lee JT, Salazar M. Design and synthesis of the G-quadruplex-specific cleaving reagent perylene-EDTA.iron(II). J Am Chem Soc. 2001;123:5606–5607. doi: 10.1021/ja0156439. [DOI] [PubMed] [Google Scholar]
- 33.Kim MY, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. ) Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular G-Quadruplex. J Am Chem Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 34.Nagesh N, Chatterji D. Ammonium ion at low concentration stabilizes the G-quadruplex formation by telomeric sequence. J Biochem Biophy Methods. 1995;30:1–8. doi: 10.1016/0165-022x(94)00057-k. [DOI] [PubMed] [Google Scholar]
- 35.Nagesh N, Krishnaiah A, Dhople VM, Sundaram CS, Jagannadham MV. Noncovalent interaction of G-quadruplex DNA with acridine at low concentration monitored by MALDI-TOF mass spectrometry. Nucleosides Nucleotides and Nucleic Acids. 2007;26:303–315. doi: 10.1080/15257770701257590. [DOI] [PubMed] [Google Scholar]
- 36.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nature Rev. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 37.Yuan G, Zhang Q, Zhou J, Li H. Mass spectrometry of G-quadruplex DNA: formation, recognition, property, conversion, and conformation. Mass Spec Rev. 2011;30:1121. doi: 10.1002/mas.20315. [DOI] [PubMed] [Google Scholar]
- 38.Viglasky V, Bauer L, Tluckova K. Structural Features of Intra- and Intermolecular G-Quadruplexes Derived from Telomeric Repeats. Biochemistry. 2010;49:2110–2120. doi: 10.1021/bi902099u. [DOI] [PubMed] [Google Scholar]
- 39.Rosu F, Gabelica V, Poncelet H, De Pauw E. Tetramolecular G-quadruplex formation pathways studied by electrospray mass spectrometry. Nucleic Acids Res. 2010;38:5217–5225. doi: 10.1093/nar/gkq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamal A, Ramesh G, Srinivas O, Ramulu P. Synthesis and antitumour activity of pyrene-linked pyrrolo[2,1-c][1,4]benzodiazepine hybrids. Bioorg Med Chem Lett. 2004;14:471–474. doi: 10.1016/j.bmcl.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 41.Kamal A, Ramesh G, Laxman N, Ramulu P, Srinivas O, et al. Design, Synthesis, and Evaluation of New Noncross-Linking Pyrrolobenzodiazepine Dimers with Efficient DNA Binding Ability and Potent Antitumor Activity. J Med Chem. 2002;45:4679–4688. doi: 10.1021/jm020124h. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez JM, Saiz-Jimenez C. A simple fluorimetric method for the estimation of DNA-DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles. 2005;9:75–79. doi: 10.1007/s00792-004-0417-0. [DOI] [PubMed] [Google Scholar]
- 43.Reddy TVK, Mahmood S, Paris L, Reddy YH, Wellington EM, et al. Streptomyces hyderabadensis sp. nov., an actinomycete isolated from soil. Int J Syst Evol Microbiol. 2011;61:76–80. doi: 10.1099/ijs.0.020446-0. [DOI] [PubMed] [Google Scholar]
- 44.Brown A. Analysis of cooperativity by isothermal titration calorimetry. Int J Mol Sci. 2009;10:3457–3477. doi: 10.3390/ijms10083457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu B, Qin Z, Liu J, Luo H. Thymine quintets and their higher order assemblies studied by electrospray ionization mass spectrometry and theoretical calculation. J Mass Spectrom. 2011;46:587–594. doi: 10.1002/jms.1928. [DOI] [PubMed] [Google Scholar]
- 46.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw ET. Triplex and quadruplex DNA structures studied by electrospray mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 47.Cui X, Yuan G. Formation and recognition of G-quadruplex in promoter of c-myb oncogene by electrospray ionization mass spectrometry. J Mass Spectrom. 2011;46:849–855. doi: 10.1002/jms.1959. [DOI] [PubMed] [Google Scholar]
- 48.Kraus GA, Selvakumar N. A novel Fremy's salt-mediated oxidation and rearrangement of anilines into amino ortho-diketones. Applications to the synthesis of pyrrolobenzodiazepines. Tetrahedron Letters. 1999;40:2039. [Google Scholar]
- 49.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J Am Chem Soc. 1999;121:3561–3570. [Google Scholar]
- 50.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukuda H, Katahira M, Tsuchiya N, Enokizono Y, Sugimura T, et al. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. PNAS. 2002;99:12685–12690. doi: 10.1073/pnas.152456899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Wen Y, Liu J, Zhou J, Li C, et al. Promoting the formation and stabilization of human telomeric G-quadruplex DNA, inhibition of telomerase and cytotoxicity by phenanthroline derivatives. Org Biomol Chem. 2011;9:2648–2653. doi: 10.1039/c0ob00961j. [DOI] [PubMed] [Google Scholar]
- 53.Keating LR, Szalai VA. Parallel-Stranded Guanine Quadruplex Interactions with a Copper Cationic Porphyrin. Biochemistry. 2004;43:15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 54.Lubitz I, Borovok N, Kotlyar A. Interaction of monomolecular G4-DNA nanowires with TMPyP: evidence for intercalation. Biochemistry. 2007;46:12925–12929. doi: 10.1021/bi701301u. [DOI] [PubMed] [Google Scholar]
- 55.Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:1–5. doi: 10.1093/nar/gng135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 57.Chaires JB. Analysis and interpretation of ligand-DNA binding isotherms. Methods Enzymol. 2001;340:3–22. doi: 10.1016/s0076-6879(01)40415-0. [DOI] [PubMed] [Google Scholar]
- 58.Suh D, Chaires JB. Criteria for the mode of binding of DNA binding agents. Bioorg Med Chem. 1995;3:723–728. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]
- 59.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 60.Hyun KM, Choi SD, Lee S, Kim SK. Can energy transfer be an indicator for DNA intercalation? Biochim Biophys Acta. 1997;1334:312–316. doi: 10.1016/s0304-4165(96)00111-0. [DOI] [PubMed] [Google Scholar]
- 61.Chaires JB. A thermodynamic signature for drug-DNA binding mode. Arch Biochem Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with PBD1. The interaction peaks were marked with “•”.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with PBD2. The interaction peaks were marked with “♦”.
(TIF)
Expanded ESI-MS spectrum (from m/z 400–1000) of d(T2G8) G-quadruplex with TMPyP4. The interaction peaks were marked with “▪”.
(TIF)