Abstract
Species of Oithona (Copepoda, Cyclopoida) are highly abundant, ecologically important, and widely distributed throughout the world oceans. Although there are valid and detailed descriptions of the species, routine species identifications remain challenging due to their small size, subtle morphological diagnostic traits, and the description of geographic forms or varieties. This study examined three species of Oithona (O. similis, O. atlantica and O. nana) occurring in the Argentine sector of the South Atlantic Ocean based on DNA sequence variation of a 575 base-pair region of 28S rDNA, with comparative analysis of these species from other North and South Atlantic regions. DNA sequence variation clearly resolved and discriminated the species, and revealed low levels of intraspecific variation among North and South Atlantic populations of each species. The 28S rDNA region was thus shown to provide an accurate and reliable means of identifying the species throughout the sampled domain. Analysis of 28S rDNA variation for additional species collected throughout the global ocean will be useful to accurately characterize biogeographical distributions of the species and to examine phylogenetic relationships among them.
Introduction
Biogeography and Ecology of the Species
Among the small size copepods, the family Oithonidae [1] is recognized as one of the most abundant groups in the ocean [2]. The abundance, biomass and ecological role of Oithona spp. have been examined in recent studies [3]–[5]. The genus has been the subject of concerted and expert taxonomic analysis and detailed descriptions of the species are in place [6]–[8]. However, routine identification of species has remained challenging due to the small body size and subtle mophological differences among species [6] and descriptions of geographic forms or varieties of widely-distributed species [9].
The Oithona species examined in this study are important components of the Argentine Sea - a region of the Southwest Atlantic Ocean -, as well as of the North Atlantic Ocean [5], [10]–[12]. Over the Argentine continental shelf, the occurrence of O. similis Claus 1866 [13] syn. O. helgolandica [14], [15], O. atlantica [16] and O. nana [17] has been extensively cited [18]–[20]. These species are abundant, ecologically-important, and geographically-widespread; their numerical dominance was recently highlighted [21], [22]. Oithona similis occurs over the Argentine continental shelf between 34° and 55° S [18], [23]–[25]. It is broadly distributed from the tropics to high latitudes of the Atlantic [10], [18], [23]–[25] and Pacific Oceans [8]; in the Indian Ocean, and Mediterranean and Red Seas [26]. Although O. similis is a widespread species, multivariate analyses of community structure in the Argentine Sea reveal that the species reaches its maximum densities in cold shelf waters [20], [27].
Oithona atlantica also has a broad biogeographical distribution throughout both the North and South Atlantic Oceans, occurring over wide ranges in salinity (24–26 ppt and 34–36 ppt) and temperature (8–19°C) [18]. Despite such wide ecological tolerances, this is the least abundant Oithona species in the Argentine Sea [24], [28], but quite common throughout the Strait of Magellan [18]. It occurs in the northern North and eastern equatorial Pacific Ocean, Bearing Sea and Sea of Japan [8]. It is also found in the Sub-Antarctic and Antarctic waters, as well as the Mediterranean Sea [8].
In Argentine waters O. nana is found throughout the year between 34° and 45°S. The species is an important component of the coastal species assemblage [27], [28], and it is potentially important as prey for fish larvae [29], [30]. It is also found in tropical and subtropical waters of the Atlantic Ocean [31], [32] as well as in the Mediterranean Sea [33], and the Pacific and Indian Oceans [8].
28S rDNA as a Taxonomic Marker
Although molecular approaches have been applied exhaustively to copepods to ensure accurate taxonomic identification of species, little information is available for cyclopoid copepods, especially for species of Oithona. DNA sequence variation of the large-subunit (28S) rRNA gene has been used extensively to examine phylogenetic relationships among marine invertebrate species, including cnidarians [34], annelids [35], nematodes [36], molluscs [37], and echinoderms [38], among others. The broad application of this gene as a character for taxonomic identification of species with subtle or ambiguous morphological characteristics makes it a useful marker to be employed for species of the cyclopoid copepod Oithona.
The relationships among Oithona species, including O. similis, O. atlantica and O. nana, have been studied for the Pacific and Indian Oceans [8]. These morphological analyses included forty five structural characters and suggested that O. atlantica and O. similis are more closely related to each other than to O. nana [8]. Here we analyze DNA sequences for a 575 base-pair (bp) region of the 28S rRNA gene and characterize patterns of variation within and among three Oithona species occurring in the South and North Atlantic Oceans.
Methods
Ethics Statement
No specific permits were required for the described field study, and no endangered or protected species were included in this study.
Collection of Samples
Zooplankton samples collected from regions across the North and South Atlantic Oceans (Figure 1, Table 1), preserved immediately and stored in 95% undenatured ethanol, as described by Bucklin [39]. A total of 150 oithonid copepods were identified to species level following [24], [25], using a Leica D1000 inverted microscope. The following specimens were removed and prepared for molecular analysis: O. similis (108 individuals), O. nana (19 individuals) and O. atlantica (23 individuals). Specimens from O. similis and O. nana type localities were also included in the molecular analysis.
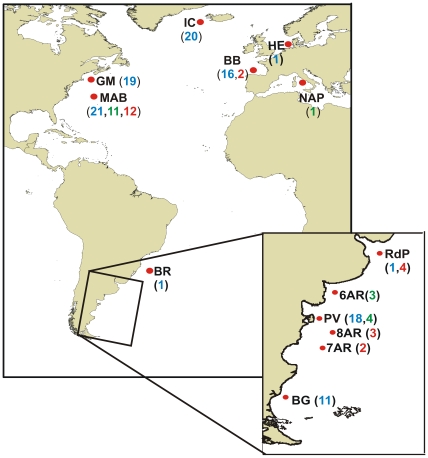
Figure 1. Collection sites and number of specimens in each site for each Oithona species.
Specimens of O. similis sampled (in blue); O. nana (green); O. atlantica (red). Explanation of abbreviations for the collection sites are given in Table I and the text.
Table 1. Sample sites, latitude, longitude, location code, sample size (N), sequence diversity (h), standarized sequence diversity (Hk) and number of kind sequences in each population of O. similis, O. nana and O. atlantica collected for this study from the Atlantic Ocean.
| Species | Sample site | Latitude | Longitude | Location Code | N | h | Hk |
| O. similis | Gulf of Maine, US | 43°10′4.8″N | 70°25′4.8″W | GM | 19 | 0.51 | 0.52 |
| Bay of Biscay, Spain | 43°42′N | 6° 9′W | BB | 16 | 0.24 | 0.24 | |
| Iceland | 64°20.15′N | 27°W | IC | 20 | 0.68 | 0.72 | |
| Mid Atlantic Bight, US | 38°16.3′N | 74°24.4′W | MAB | 21 | 0.74 | 0.72 | |
| Península Valdés, Argentina | 42°31′4.8″S | 63°12′W | PV | 18 | 0.81 | 0.82 | |
| Bahía Grande, Argentina | 51°S | 67°W | BG | 11 | 0.34 | 0.35 | |
| Río de la Plata, Argentina | 36°4′48″S | 54°32′2.4″W | RdP | 1 | N/A | N/A | |
| Helgoland Sea, Germany* | 54°10′57″N | 7°53′E | HE | 1 | N/A | N/A | |
| Torres, Brazil | 29°40′4.8″S | 49°30′W | BR | 1 | N/A | N/A | |
| 108 | |||||||
| O. nana | Mid Atlantic Bight, US | 38°16.3′N | 74°24.4′W | MAB | 11 | 0.56 | 0.56 |
| El Rincón, Argentina | 39°38′2.4″S | 61°6′3.6″W | 6AR | 3 | 0.00 | 0.00 | |
| Península Valdés, Argentina | 42°31′4.8″S | 63°12′W | PV | 4 | 0.50 | 0.50 | |
| Gulf of Naples, Italy* | 40°50′N | 14°15′E | NAP | 1 | N/A | N/A | |
| 19 | |||||||
| O. atlantica | Bay of Biscay, Spain | 43°42′N | 6° 9′W | BB | 2 | 1.00 | 1.00 |
| Mid Atlantic Bight, US | 38°16.3′N | 74°24.4′W | MAB | 12 | 0.41 | 0.41 | |
| Rio de la Plata, Argentina | 36°4′48″S | 54°32′2.4″W | RdP | 4 | 0.00 | 0.00 | |
| Argentina | 45°15′S | 62°30′3,6″W | 7AR | 2 | 0.00 | 0.00 | |
| Argentina | 43°31′4.8″S | 61°23′2.4″W | 8AR | 3 | 0.00 | 0.00 | |
| 23 |
Total sample size for each species is indicated in bold, samples from type locality are indicated by asterisk. N/A: not applicable.
Molecular Analysis
DNA was extracted from individual identified specimens using the QIAGEN Dneasy tissue Kit. The Polymerase Chain Reaction (PCR) was used to amplify a 800 bp fragment of the D1–D2 region of the large subunit (28S) ribosomal DNA (rDNA) gene using primers 28SF1: 5′-GCGGAGGAAAAGAAACTAAC-3′ and 28SR1: 5′-GCATAGTTTCACCATCTTTCGGG-3′ [34]. PCR amplifications were performed in a total volume of 25 µl including 5 µl of 5X Green GoTaq® Flexi Buffer, 2.5 µl of 25 mM MgCl2, 1 µl of dNTPs (final concentration 0.2 mM each), 1 µl of each primer (10 µM), 0.75 units of GoTaq® Flexi DNA Polymerase (Promega) and 3 µl of the DNA template solution. The PCR protocol was: 4 min initial denaturation step at 94°C; 35 cycles of 40 s denaturation step at 94°C, 40 s annealing at 50°C, and 90 s extension at 72°C; and a final extension step of 15 min at 72°C.
Several sets of PCR primers for various genes were tested, but most did not amplified consistently. The genes for which published primers were tested included: internal transcribed spacer [40]; mitochondrial cytochrome c oxidase subunit I [41]; cytochrome b and 12S rDNA [42]; heat shock protein 70 [43]; and AMP-activated protein kinase [44].
Approximately 5 µl of each PCR product was electrophoresed on a 1% TBE agarose gel and visualized by UV light with with Biotium GelRedTM staining. The PCR products were purified using QIAquick spin columns (Qiagen). Both strands of the template DNA were sequenced using the PCR primers and Big Dye Terminator Ver. 3.1 (Applied Biosystems Inc., ABI), and were run in an ABI 3130 Genetic Analyzer automated capillary DNA sequencer.
The 28S rDNA sequences obtained were manually edited, with comparison of aligned sequences for both strands. DNA sequences for O. similis, O. nana and O. atlantica were aligned using the default parameters by Clustal W [45], using MEGA Ver. 5.05 [46]. DNA sequences were submitted to the molecular database, GenBank (http://www.nlm.nih.ncbi.org) and were assigned a GenBank Accession Numbers: FM991727.1; JF419529-JF419547.
Genetic Distances within and between Oithona Species
Analysis was done using a final aligned length of 575 bp of the 28S rRNA gene. Numbers of kind sequence and sequence diversities (h) were calculated for each population sampled for the studied species by DnaSP Ver. 5.10 [47]. Standarized sequence diversities (Hk) were calculated considering the smallest sample size (O. similis: n = 11; O. nana: n = 3; O. atlantica: n = 2) using the software RAREFAC (http://www.pierroton.inra.fr/genetics/labo/Software/Rarefac) [48]. The appropriate best-fit substitution model of DNA evolution was determined with jModelTest Ver. 0.1.1 [49] under the Akaike information criterion (A.I.C.). Neighbor-Joining method [50] analysis implemented in MEGA Ver 5.05 [46] was used on the identified kind sequences to assess the relationships among the three Oithona species based on DNA sequence variation; relative support for the tree topology was obtained by bootstrapping [51] using 10,000 iterations.
Genetic Variation of O. similis
A total of 108 28S rDNA sequences for O. similis were aligned using MEGA Ver. 5.05 [46]. A 51-bp region showing intraspecific variation was used for this analysis; the best-fitting substitution model was determined with jModelTest [49]. The most appropriate model was found to Jukes-Cantor; the model and estimated parameters were set in Arlequin Ver. 3.5.1.2 [52] and the geographic pattern of 28S rDNA variation was assessed. ΦST genetic distances between all pairs of O. similis populations were calculated using Arlequin Ver. 3.5.1.2 [52]. Pairwise ΦST values among all conspecific populations were calculated and tested for significance through 10,000 permutations. For this analysis, all sequence types found in the populations from the Gulf of Maine (GM), Mid Atlantic Bight (MAB), Iceland (IC), Bay of Biscay (BB), Península Valdés (PV) and Bahía Grande (BG) were considered (Table 1).
An hierarchical Analysis of MOlecular VAriation [53] was performed using different groupings of populations based on the distances between sampling locations and ΦST distances. The statistical significance of the AMOVA statistics, including among groups (ΦCT), among populations within groups (ΦSC), and within populations (ΦST), was obtained after 10,000 permutations.
Results
Genetic Distances within and between Oithona Species
DNA sequences of a 575 bp region of the 28S rDNA gene for 108 O. similis individuals revealed the presence of six well-resolved kind sequences and six kind sequences with one or two ambiguous sites (H1–H12). These ambigous sites correspoded to C-T sites, and were defined by equivalents peaks of both bases (Figure S1).
Among the 19 O. nana specimens from 3 populations, five kind sequences (H13–H17) defined by ten polymorphic sites were recorded, whereas among the 23 O. atlantica individuals analyzed, distributed in 5 populations, three kind sequences (H18–H20) were found defined by thirteen polymorphic sites. For O. similis, the sequence diversity was somewhat higher at PV than at MAB or IC. An intermediate value was found at GM, while the lower ones were at BG and BB (Table 1). For O. nana, mean values of sequence diversity were found at MAB and PV, while at ER only one sequence type was recorded. In the case of O. atlantica, BB showed the highest sequence diversity value, followed by MAB, while at RdP, 7AR and 8AR, no sequence diversity was detected, since only one sequence type was found (Table 1).
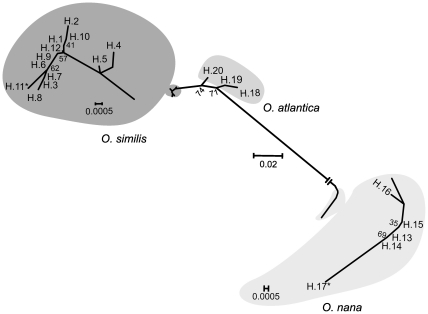
The A.I.C. selected the Jukes-Cantor [54] with alpha parameter for the gamma distribution of 0.25 as the evolutionary model that best fit the observed sequence variation. Mean Jukes-Cantor distances within species ranged from 0.001 for O. similis to 0.015 for O. atlantica (Table 2). Genetic distance between species was highest between O. nana and the other two species, with O. nana differing from O. similis by a distance of 0.224 and from O. atlantica by 0.222; the distance between O. similis and O. atlantica was much lower at 0.034 (Figure 2, Table 2).
Table 2. Relationships among the three Oithona species based on 28S rDNA.
| Species | O. atlantica | O. similis | O. nana |
| O. atlantica | 0.015 (0.008) | ||
| O. similis | 0.034 (0.009) | 0.001 (0.001) | |
| O. nana | 0.222 (0.014) | 0.244 (0.013) | 0.006 (0.005) |
Mean Jukes-Cantor distances within (diagonal) and between (below diagonal) the three Oithona species. Distances among sequence types were calculated with MEGA (Ver. 5.05 using the Jukes-Cantor model with alpha parameter of 0.25. The standard deviation about each mean is indicated in parentheses. Numbers of specimens used for the analysis are: O. similis (108), O. atlantica (23), and O. nana (19).
Figure 2. Relationships among the three Oithona species based on 28S rDNA.
Unrooted Neighbor-Joining analysis under the Jukes-Cantor model, showing relationships among the three Oithona species based on 28S rDNA sequence types of O. similis (H1–H12), O. atlantica (H13–H17) and O. nana (H18–H20). Sequence types found at each species’ type locality are indicated by asterisk (*). Numbers in the nodes indicate the percentage bootstrap recovery after 10,000 repetitions.
28S rDNA Variation of O. similis
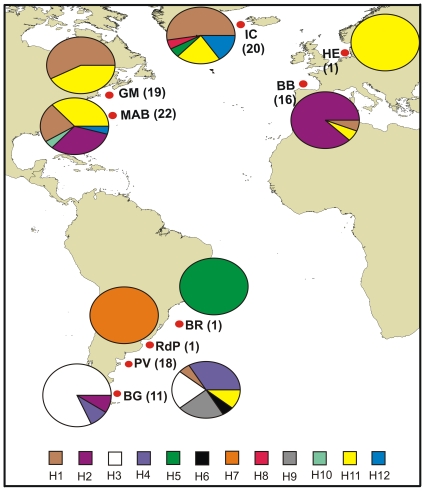
Among twelve 28S rDNA sequences detected for O. similis, H1, H2, H5 and H11 were present in both hemispheres (Figure 3). H1 was the most frequently found, distributed at GM, BB, IC, MAB and PV. H11 was found in GM, BB, IC, MAB, HE and PV, while H2 was present in BB, MAB, and BG. H5 was in IC and BR (Figure 3).
Figure 3. Distribution and frequence of Oithona similis kind sequence.
Pie diagrams depicting the kind sequence frequencies of a 51bp region of 28S for samples of O. similis collected from Gulf of Maine (GM), Iceland (IC), Middle Atlantic Bight (MAB), Bay of Biscay (BB), Península Valdés (PV) and Bahía Grande (BG). Sample size (n = number of individual copepods) in each location. The twelve O. similis sequence types (H1–H12) are represented by different colours. References in the figure.
Three sequences were exclusively found in the Northern Hemisphere. They were only present at IC (H8, H12) and MAB (H10, H12) (Figure 3). Five sequences occurred only in the Southern Hemisphere: H3, H4, H6, H7 and H9 (Figure 3). The most frequently found were H3, H4 and H9 which were present at BG and PV. H6 and H7 were only found at PV and RdP (Figure 3). ΦST values [53] derived from genetic distances were significant for all pairwise comparisons between populations except for the pairs GM-IC and PV-BG. Thus, O. similis populations were tentatively separated into four groups: GM+IC; MAB; BB; and PV+BG (Table 3). This clustering was supported by AMOVA analysis, which revealed that 53.58% of the observed genetic variation was among groups, and 37.94% was within populations (Table 4).
Table 3. Pairwise ΦST distances between all O. similis populations with n>1.
| GM | IC | PV | BG | MAB | BB | |
| GM | - | |||||
| IC | 0.139 | - | ||||
| PV | 0.603** | 0.528** | - | |||
| BG | 0.907** | 0.856** | 0.216 | - | ||
| MAB | 0.419** | 0.248* | 0.286** | 0.586** | - | |
| BB | 0.886** | 0.817** | 0.504** | 0.687** | 0.405* | - |
Asterisks indicate the significance level (p) for each comparison calculated from 10,000 permutations: p<0.001 (*); p<0.0001 (**). Numbers of specimens used for the analysis are: GM (19), BB (16), IC (20), MAB (21), PV (18) and BG (11).
Table 4. Analysis of MOlecular VAriance (AMOVA) based on 28S rDNA sequence data for Oithona similis.
| Observed partition | |||||
| Source of variation | Variance | % | Φ-statistics | P-value | d.f. |
| Among groups | 0.646 | 53.58 | ΦST = 0.535 | 0.02 | 3 |
| Among populations within groups | 0.102 | 8.49 | ΦSC = 0.182 | 0.01 | 2 |
| Within populations | 0.457 | 37.94 | ΦST = 0.620 | < 0.01 | 102 |
Variance and percentage of variance explained (%), fixation indexes (Φ-statistics), P-value indicates probability of obtaining a higher Φ value by chance estimated by 10,000 permutations, d.f.: degrees of freedom. (Refer to Method text for the definition of group and population).
Discussion
Accurate and reliable identification of species is a necessary foundation for assessment of biodiversity, especially for important but lesser-known regions of the world ocean, such as the Argentine Sea [55]. DNA sequence variation of target genes provides invaluable tools for such analyses.
This study examined variation of a portion of 28S (large subunit) rDNA as a marker to identify and discriminate species of the ecologically-important but understudied cyclopoid copepod genus Oithona, found in the Argentine Sea - Southwest Atlantic Ocean - and North Atlantic Ocean. The species analyzed here, O. similis, O. nana and O. atlantica, were confirmed by molecular analysis to be distinct species, as previously characterized by morphological taxonomic analysis [7], [8], [24], [25]. Inclusion in our analysis of O. similis and O. nana from the type localities was particularly useful to allow determination of reference sequences for these species for future comparisons.
The genetic distances observed within and between species of Oithona agreed somewhat with those reported by Ueda et al. [56]. Our distance values were higher than those registered by these authors; which could be related to the fact that they analyzed two size forms of O. dissimilis. Our interspecific genetic distances may reflect the relationships registered by Nishida [8].
In addition to characterizing differences between species, the present work provided preliminary analysis of the levels and patterns of 28S rDNA sequence variation within each of the studied species based on samples collected from a broad latitudinal range of the Atlantic Ocean. Shared kind sequences were detected between North and South Atlantic collections for each of the Oithona species analyzed, despite the large distances between sampling locations. This finding confirms that 28S rDNA serves as a useful genetic marker for identification of these – and likely all – Oithona species, even those with global distributions.
Levels of intraspecific variation differed among the species: DNA sequence variation (measured as the percentages of bases) was higher for O. atlantica (1.5%) than either O. nana (0.6%) or O. similis (0.1%). The lower values recorded for O. nana and O. similis, which are both found commonly in coastal and shelf waters, might be due in part to their introduction by ballast water. For the Argentine Sea, [57] reported the presence of O. nana and O. similis in ballast water from commercial vessels from several origins (e.g., Indian and Pacific Ocean, Mediterranean and Baltic Seas and Atlantic ports north of 20°S). At the Russian port of Novorossiysk, high abundances (10,000 individuals/m3) of live individuals of O. nana were found in samples taken from ships’ ballast water [58]. Interestingly, O. similis exhibited significantly different genetic differences among populations sampled for this study, although these differences were the lowest of the three species examined were not correlated with geographic distances, since some samples differed markedly despite their geographic proximity (e.g., GM and MAB).
Based on 28S rDNA, O. similis is a single, genetically-cohesive species throughout the studied distributional range. Even for this conserved genetic marker, the species showed significant genetic differentiation among regions of the North and South Atlantic Oceans. It seems likely that geographic populations of O. similis might be primarily isolated by large-scale patterns of ocean circulation, as has been suggested by other genetic analysis of zooplankton in the Atlantic Ocean basin [44], [59], [60].
Our analysis of intraspecific and interspecific patterns of variation for three species of Oithona in selected regions of the North and South Atlantic Oceans demonstrated the usefulness of the 28S rDNA as an accurate and reliable means of identifying and discriminating the species. The 28S rDNA fragment we focused on is included the D1–D2 region, and has been suggested by Sonnenberg et al. [61] as a taxonomic marker due to its variability. Previous studies have used this marker for analysis of copepods [62] and other taxa [63]. Additional analysis of intraspecific variation, including studies using more highly variable molecular markers, will be needed to addresss questions of population connectivity, barriers to genetic cohesion, and discovery of cryptic species among such globally-distributed taxa.
Supporting Information
Alignment of the twelve 28S rDNA kind sequences of Oithona species.
(FASTA)
Acknowledgments
We appreciate and acknowledge captains and crews of the different cruises for collecting samples. We are grateful to G. Veit-Köhler, Y. Carotenuto and M. Mazzocchi for collection of specimens from types localities, and to M. Sabatini for donation of specimens from southern Argentina.
Footnotes
Competing Interests: Dirk Steinke is Campaign Coordinator for the MarBOL initiative and a co-organiser of the PLoS ONE MarBOL Collection.
Funding: This work was partially supported by Instituto Nacional de Investigación y Desarrollo Pesquero, Fundación para Investigaciones Biológicas Aplicadas; Agencia Nacional de Promoción Científica y Tecnológica [Grant No. 15227/03 to M.D.V.]; Universidad Nacional de Mar del Plata [Grant No. 15/E269 to M.D.V.]; and a PhD. fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) to G.C. Additional support was provided by the Department of Marine Sciences at the University of Connecticut, and by the Alfred P. Sloan Foundation (Marine Barcode of Life project, MarBOL). This study is a contribution from the Census of Marine Zooplankton (CMarZ, see www.CMarZ.org), an ocean realm field project of the Census of Marine Life. This is INIDEP contribution N 1736. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dana JD. United States Exploring Expedition during the years 1838 1839 1840 1841 1842 under the command of Charles Wilkes U.S.N. Atlas Philadelphia; 1853. Crustacea. [PMC free article] [PubMed] [Google Scholar]
- 2.Paffenhöfer GA. On the ecology of marine cyclopoid copepods (Crustacea Copepoda Cyclopoida). J Plankton Res. 1993;15:37–55. [Google Scholar]
- 3.Gallienne CP, Robins DB. Is Oithona the most important copepod in the world’s oceans? J Plankton Res. 2001;23:1421–1432. [Google Scholar]
- 4.Nielsen TG, Møller EF, Satapoomin S, Ringuette M, Hopcroft RR. Egg hatching rate of the cyclopoid copepod Oithona similis in arctic and temperate waters. Mar Ecol Prog Ser. 2002;236:301–306. [Google Scholar]
- 5.Castellani C, Irigoien X, Harris RP, Holliday NP. Regional and temporal variation of Oithona spp. biomass, stage structure and productivity in the Irminger Sea, North Atlantic. J Plankton Res. 2007;29:1051–1070. [Google Scholar]
- 6.Nishida S, Tanaka O, Omori M. Cyclopoid copepods of the family Oithonidae in Suruga Bay and adjacent waters. Bull Plankton Soc Japan. 1977;24:119–158. [Google Scholar]
- 7.Shuvalov VS. Nauka Press, Leningrad (in Russian); 1980. Copepod cyclopoids of the family Oithonidae of the World Ocean. [Google Scholar]
- 8.Nishida S. Taxonomy and distribution of the family Oithonidae (Copepoda: Cyclopoida) in the Pacific and Indian Oceans. Bull Ocean Res Inst Univ Tokyo. 1985;20:1–167. [Google Scholar]
- 9.Dvoretsky VG, Dvoretsky AG. Morphological plasticity in the small copepod Oithona similis in the Barents and White Seas. Mar Ecol Prog Ser. 2009;385:165–178. [Google Scholar]
- 10.Fish CJ. The biology of Oithona similis in the Gulf of Maine and Bay of Fundy. Biological Bulletin, Marine Biological Laboratory, Woods Hole. 1936;71:168–187. [Google Scholar]
- 11.Nielsen TG, Sabatini ME. Role of cyclopoid copepods Oithona spp. in North Sea plankton communities. Mar Ecol Prog Ser. 1996;139:79–93. [Google Scholar]
- 12.Williams JA, Muxagata E. The seasonal abundance and production of Oithona nana (Copepoda: Cyclopoida) in Southampton Water. J Plankton Res. 2006;28:1055–1065. [Google Scholar]
- 13.Claus C. Die Copepoden-Fauna von Nizza. Ein Beitrag zur Charakteristik der Formen und deren Abänderungen “im Sinne Darwin’s”. Schr Ges Beförd ges Naturw. 1866;Marburg,(suppl. 1):1–34. [Google Scholar]
- 14.Claus C. Die freilebenden Copepoden mit besonderer Berücksichtigung der Fauna Deutslands, der Nordsee und des Mittelmeeres, Leipzig. 1863. pp. 1–230.
- 15.Razouls C, de Bovée F, Kouwenberg J, Desreumaux N. Diversité et répartition géographique chez les Copépodes planctoniques marins. 2005-2010;6 Available: http://copepodes.obs-banyuls.fr. Accessed 2011 Dec. [Google Scholar]
- 16.Farran GP. Second report on the Copepoda of the Irish Atlantic slope. Fish Ireland Sci Inv. 1908;2:1–104. [Google Scholar]
- 17.Giesbrecht W. Systematik und Faunistik der pelagischen Copepoden des Golfes von Neapel. Fauna Und Flora des Golfes von Neapel, 1892;19:1–831. [Google Scholar]
- 18.Mazzocchi MG, Zagami G, Ianora A. Systematic Account. In: Guglielmo L, Ianora A, editors. Atlas of Marine Zooplancton Straits of Magellan. Springer Verlag; 1995. pp. 23–246. [Google Scholar]
- 19.Berasategui A, Menu Marque S, Gómez Erache M, Ramírez F, Mianzan H, et al. Copepod assemblages in a highly complex hydrographic region. Estuar Coast Shelf Sci. 2006;66:483–492. [Google Scholar]
- 20.Sabatini ME. El ecosistema de la plataforma patagónica austral, Marzo-Abril 2000. Composición, abundancia y distribución del zooplancton. Rev Invest Desarr Pesq. 2008;19:5–21. [Google Scholar]
- 21.Di Mauro R, Capitanio F, Viñas M. Capture efficiency for small dominant mesozooplankters (Copepoda, Appendicularia) off Buenos Aires province (34°S–41°S), Argentina, using two plankton mesh sizes. Braz J Oceanogr. 2009;57:205–214. [Google Scholar]
- 22.Antacli JC, Hernández D, Sabatini ME. Estimating copepods' abundance with paired nets: Implications of mesh size for population studies. J Sea Res. 2010;63:71–77. [Google Scholar]
- 23.Marques E. Copépodes des eaux de Boma et de l'embouchure du fleuve Congo. Revue Zool Bot Afr. 1966;73:1–16. [Google Scholar]
- 24.Ramírez FC. Copépodos ciclopoideos y harpacticoideos del plancton de Mar del Plata. Physis. 1966;26:285–292. [Google Scholar]
- 25.Ramírez FC. Copépodos planctónicos de los sectores bonaerense y norpatagónico. Resultados de la Campaña Pesquería III. Revista del Museo de La Plata, n. s Zoología. 1971;11:73–94. [Google Scholar]
- 26.Mori T. Soyo. Tokyo; 1964. The pelagic copepods from the neighbouring waters of Japan. [Google Scholar]
- 27.Viñas MD, Negri RM, Ramírez FC, Hernández D. Zooplankton assemblages and hydrography in the spawning area of anchovy (Engraulis anchoita) off Rio de la Plata estuary (Argentina–Uruguay). Mar Freshwater Res. 2002;53:1031–1043. [Google Scholar]
- 28.Marrari M, Viñas MD, Martos P, Hernández D. Spatial patterns of mesozooplankton distribution in the Southwestern Atlantic Ocean (34°S–41°S) during austral spring: relationship with the hydrographic conditions. ICES J Mar Sci. 2004;61:667–679. [Google Scholar]
- 29.Viñas MD, Ramírez FC. Gut analysis of first-feeding anchovy larvae from the Patagonian spawning areas in relation to food availability. Arch Fish Mar Res. 1996;43:231–256. [Google Scholar]
- 30.Viñas MD, Santos BA. First-feeding of hake (Merluccius hubbsi) larvae and prey availability in the North Patagonian spawning area. Comparison with anchovy. Arch Fish Mar Res. 2000;48:242–254. [Google Scholar]
- 31.Ferrari FD, Bowman TE. Pelagic copepods of the family Oithonidae (Cyclopoida) from the east coasts of Central and South America. Smithsonian Contributions to Zoology. 1980;312:1–111. [Google Scholar]
- 32.Bradford-Grieve JM, Markhaseva EL, Rocha CEF, Abiahy B. Copepoda. In Boltovskoy D, editors. South Atlantic Zooplakton. Vol. 2. Backhuys Publishers, Leiden, 1999;869–1706 [Google Scholar]
- 33.Früchtl F. Plankton Copepoden aus der nordlichen Adria. Sber Akad Wiss Wien Math Nat Kl. 1920;129:463–509. [Google Scholar]
- 34.Ortman BD. Ph.D. Dissertation, University of Connecticut, Storrs, CT; 2008. DNA barcoding the medusozoa and ctenophora. [Google Scholar]
- 35.Struck TH, Schult N, Kusen T, Hickman E, Bleidorn C, et al. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evolutionary Biology 7:57. 2007;6 doi: 10.1186/1471-2148-7-57. Available: http://www.biomedcentral.com/1471-2148/7/57. Accessed 2011 Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bik HM, Lambshead PJD, Thomas WK, Lunt DH. 2010. Moving towards a complete molecular framework of the Nematoda: a focus on the Enoplida and early-branching clades. BMC Evolutionary Biology 10:353. Available: http://www.biomedcentral.com/1471-2148/10/353. [DOI] [PMC free article] [PubMed]
- 37.Holznagel WE, Colgan DJ, Lydeard C. Pulmonate phylogeny based on 28S rRNA gene sequences: A framework for discussing habitat transitions and character transformation. Mol Phyl Evol. 2010;57:1017–1025. doi: 10.1016/j.ympev.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Borchiellini C, Chombard C, Manuel M, Alivon E, Vacelet J. Molecular phylogeny of Demospongiae: implications for classification and scenarios of character evolution. Mol Phyl Evol. 2004;32:823–837. doi: 10.1016/j.ympev.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Bucklin A. Methods for population genetic analysis of zooplankton. In: Harris RP, Weibe PH, Lenz J, Skjoldal HR, Huntley M, editors. ICES Zooplankton Methodology Manual. Academic Press, London; 2000. pp. 533–570. [Google Scholar]
- 40.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: MA InInnis, DH Gelfand, JJ Sninsky, TJ White., editors. editors. PCR Protocols: A Guide to Methods and Applications, Academic Press, New York; 1990. pp. 315–322. [Google Scholar]
- 41.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 42.Machida RJ, Miya MU, Nishida M, Nishida S. Complete mitochondrial DNA sequence of Tigriopus japonicus (Crustacea: Copepoda). Mar Biotech. 2002;4:406–417. doi: 10.1007/s10126-002-0033-x. [DOI] [PubMed] [Google Scholar]
- 43.Voznesenskya M, Lenz PH, Spanings-Pierrot C, Towle DW. Genomic approaches to detecting thermal stress in Calanus finmarchicus (Copepoda: Calanoida). J Exp Mar Biol Ecol. 2004;311:37–46. [Google Scholar]
- 44.Unal E, Bucklin A. Basin-scale population genetic structure of the planktonic copepod Calanus finmarchicus in the North Atlantic Ocean. Progr Oceanogr. 2010;87:175–186. [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson JJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 48.Petit JR, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Cons Biol. 1998;12:844–855. [Google Scholar]
- 49.Posada D. jModelTest: Phylogenetic Model Averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Excoffier L, Lischer HL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 53.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jukes TH, Cantor CR. In Munro, HN. (ed), Mammalian Protein Metabolism. Academic Press, New York; 1969. Evolution of Protein Molecules. [Google Scholar]
- 56.Ueda H, Yamaguchi A, Saito S, Sakaguchi S, Tachihara K. Speciation of two salinity-associated size forms of Oithona dissimilis (Copepoda: Cyclopoida) in estuaries. J Nat Hist. 2011;45:2069–2079. [Google Scholar]
- 57.Boltovskoy D, Almada P, Correa N. Biological invasions: assessment of threat from ballast-water discharge in Patagonian (Argentina) ports. Environ Sci Pol. 2011;14:578–583. [Google Scholar]
- 58.Selifonova, ZhP Oithona brevicornis Giesbrecht (Copepoda, Cyclopoida) in harborages of the northeastern part of the Black Sea shelf. Inland Water Biol. 2009;2:30–32. [Google Scholar]
- 59.Bucklin A, Astthorsson OS, Gislason A, Allen LD, Smolenack SB, et al. Population genetic variation of Calanus finmarchicus in Icelandic waters: preliminary evidence of genetic differences between Atlantic and Polar populations. ICES J Mar Sci. 2000;57:1592–1604. [Google Scholar]
- 60.Goetze E. Cryptic speciation on the high seas; global phylogenetics of the copepod family Eucalanidae. Proc R Soc Lond B. 2003;270:2321–2331. doi: 10.1098/rspb.2003.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonnenberg R, Nolte A, Tautz D. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front Zool 4, 6. 2007;6 doi: 10.1186/1742-9994-4-6. Available: http://www.frontiersinzoology.com/content/4/1/6. Accessed 2011 Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayward CJ, Svane I, Lachimpadi SK, Itoh N, Bott N, J, et al. Sea lice infections of wild fishes near ranched southern bluefin tuna (Thunnus maccoyii) in South Australia. Aquaculture. 2011;320:178–182. [Google Scholar]
- 63.Brown L, Bresnan E, Graham J, Lacaze JP, Turrell E, et al. Distribution, diversity and toxin composition of the genus Alexandrium (Dinophyceae) in Scottish waters. Eur. J Phycol. 2010;45:375–393. doi: 10.1111/j.1529-8817.2009.00678.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the twelve 28S rDNA kind sequences of Oithona species.
(FASTA)