Abstract
Pseudomonas aeruginosa OprF is the major porin of the organism and allows very slow, nonspecific diffusion of solutes. The low permeability of this porin channel is a major factor that enhances other types of resistance mechanisms, and that often creates strong multidrug resistance in this nosocomial pathogen. We have earlier showed that the low permeability is due to the fact that OprF folds into two conformers: a majority two-domain closed-channel conformer containing the N-terminal transmembrane β-barrel and the C-terminal periplasmic, globular domain, and a minority one-domain open-channel conformer comprising only <5 % of the protein population. Our analysis of the bifurcate folding pathway by using site-directed mutagenesis showed that slowing down of the folding of two-domain conformer increases the fraction of open, one-domain conformer. Use of mutants of the outer membrane protein assembly machinery showed that the absence of the Skp chaperone led to increased proportion of the open conformers. Since many environmental pathogens causing nosocomial infections appear to have OmpA/OprF homologs as the major porin, efforts to understand the low permeability of these “slow porins” are important in our fight against these organisms.
Keywords: outer membrane, permeability, β-barrel, disulfide bonds, periplasmic chaperone, Acinetobacter spp
A gram-negative bacterium, Pseudomonas aeruginosa, is one of the most important opportunistic human pathogens and is responsible for a large fraction of serious nosocomial infections including bacteremia [1]. Even the wild-type strains of this organism show a high level of intrinsic resistance to many commonly used antibiotics, such as common β-lactams, tetracyclines, and chloramphenicol. This is because antibiotics must penetrate into the bacterial cell usually through the water-filled porin channels in the outer membrane permeability barrier, and the OprF porin of this organism shows a permeability at least two orders of magnitude lower than the porins of other common gram-negative bacteria, such as Escherichia coli [2]. In addition, most antibiotics, even when they successfully penetrate through the porin channel, are efficiently pumped out by the RND (Resistance-Nodulation-Division) family efflux transporters [3, 4]. Thus P. aeruginosa strains can become, with a few alterations, resistant to the small number of antibiotics that normally work for this organism (fluoroquinolones, aminoglycosides, and carbapenems), producing so-called pan-resistant strains [5, 6]. In view of all this, we need to understand why the OprF porin of this organism shows such low permeability, the topic of this minireview.
Only a small fraction of OprF contains open channels
From the time of initial discovery of OprF as a porin [7], OprF was known to produce channels of wider diameter than the channels of classical E. coli porins such as OmpF or OmpC, which is barely permeable to trisaccharides [8]. OprF is abundant in the outer membrane. Yet the permeability of the P. aeruginosa outer membrane is exceedingly low. This apparent paradox produced papers offering clever “solutions,” some of which claimed that OprF is not a porin, or that it is not the major porin of this organism. However, OprF was proven to be the major porin in two ways. First, the group of R. E. W. Hancock showed, in intact cells, that OprF was responsible for the permeation of oligosaccharides [9]. At about the same time, one of us showed, by careful purification of pore-forming proteins, that OprF was indeed the major porin of this organism [10]. But it was still unclear why this abundant porin protein with its large channel diameter produced such low permeability both in vitro and in intact cells.
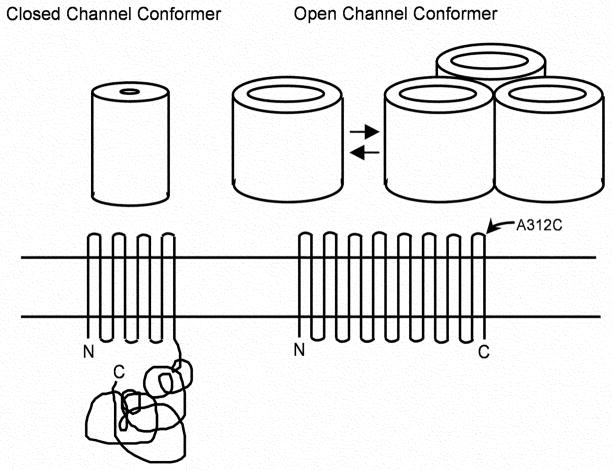
An OprF homolog, OmpA of E. coli, was always thought to fold as a two-domain protein, its N-terminal half folding into an eight-stranded β-barrel spanning the outer membrane bilayer and the C-terminal half folding into a globular domain located in the periplasm [11] (Fig. 1, left). Indeed, the crystal structures are known for the N-terminal β-barrel of OmpA [12] and for the globular domain of a homolog [13]. When OprF was isolated without the use of denaturing detergent, the CD spectrum of the protein showed the presence of both β-strands as well as α-helices [14]. Furthermore, OprF was easily cleaved by trypsin in the middle of the molecule. These data suggest the two-domain, “OmpA-like” conformation for OprF, but do not explain how large channels could exist in such a protein.
Fig. 1.
The models of OmpA/OprF proteins, after [44]. The open (minority) conformer, which folds as a single-domain protein, has a tendency to exist as loose oligomers (see below, Fig. 3B).
On the other hand, the presence of large channels in OprF seemed to favor β-barrel structures containing many β-strands, suggesting an OmpF-like, one-domain structure (Fig. 1, right). Indeed such a structure was favored by the results showing that antigenic epitopes in the C-terminal portion of OprF were exposed on cell surface [15]. However, this hypothesis does not fit with the CD spectrum data mentioned above, nor does it explain why the permeability of OprF channel is so low.
Our 1994 study [16] on the permeability of the OmpA protein solved this longstanding paradox. We reconstituted purified OmpA protein into unilamellar liposomes containing 0.3 M urea, and then centrifuged these liposomes through an isotonic gradient containing different proportions of sucrose and urea. If any of the OmpA channel were open in the liposome, intravesicular urea was exchanged by sucrose, and the vesicles sedimented to the bottom. But if none of the channels was open, the lighter vesicles containing urea remained on top. By incorporating only a few OmpA molecules per vesicle, we could conclude, by using the Poisson statistics, that only 2–3% of OmpA contained open channels. Importantly, the open or closed nature of the channel was a stable property of the molecule, so that extraction of OmpA molecules from the vesicles floating at the top and re-insertion of them into new liposomes produced totally closed vesicles. It must be mentioned that our conclusion brought a sort of “paradigm shift” to the field. Earlier, workers were obsessed with the old paradigm that each protein folds into one final conformation which is most stable in terms of energy, but our study showed that for OmpA there are apparently two stable, final conformations.
Because OprF is a member of the OmpA family of proteins, we thought that a similar explanation may apply to the low permeability of OprF. Indeed, application of a similar procedure showed that again only a small fraction (this time estimated to be 5 % of the OprF molecules) contains large, open channels [17]. The presence of these two conformers explains not only the low permeability of unfractionated OprF but also all of the seemingly paradoxical data on the conformation of this protein, as described below.
Conformation of the Closed Conformers of OprF
The closed majority conformers appear to have the two-domain conformation that has long been assumed for the homologous OmpA protein, that is, the N-terminal half folding as an eight-stranded β-barrel located in the outer membrane, and the C-terminal half folding as a globular domain located in the periplasm. By introducing the cleavage site for a Tobbaco Etch Virus protease in the middle of the molecule, we could split this majority conformer into two halves. The N-terminal domain was indeed composed of all β-structure according to the CD spectrum, whereas the C-terminal domain contained both α- and β-structures. The N-terminal domain was also produced by deleting the promoter-distal half of the gene. This domain was completely inactive in forming pores, even when very small molecules such as glycine was used in the liposome swelling assay [18].
One obvious question about this majority conformer of OprF is its biological function. If we believe that OprF is the major porin of P. aeruginosa, it makes no sense to fold more than 90% of the protein into a non-functional conformation. The same problem exists, even more acutely, for E. coli OmpA, because the porin function of OmpA is not needed for the organism that produces OmpF and OmpC porins that are far more efficient. The answer was supplied by the observation that the C-terminal domain of OmpA (and also OprF) contains sequences that are predicted to interact with peptidoglycan [19, 20]. The interaction with a peptidoglycan fragment was verified by a computer docking program [13], and experimental binding to the peptidoglycan was shown with an OmpA family protein from Acidithiobacillus ferrooxidans [21]. Thus a major function for both OmpA and OprF is to serve as a linker between the outer membrane and the peptidoglycan, and this role is critical in P. aeruginosa, which lacks the Braun lipoprotein performing a similar role in E. coli. In fact, the deletion of OprF produces an unstable outer membrane and aberrant cell morphology [22], a phenotype previously observed for E. coli mutants lacking both OmpA and the Braun lipoprotein [23].
Conformation of the Open Conformers of OprF
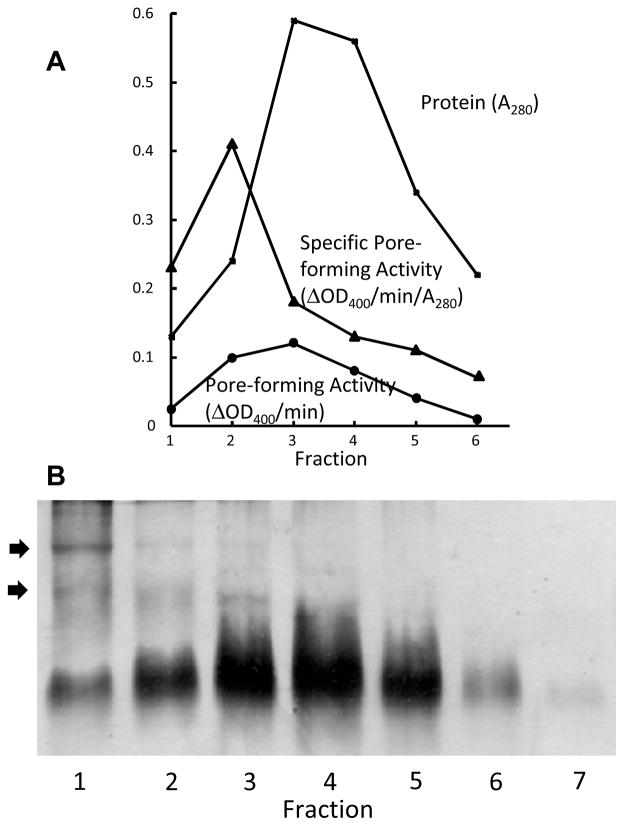
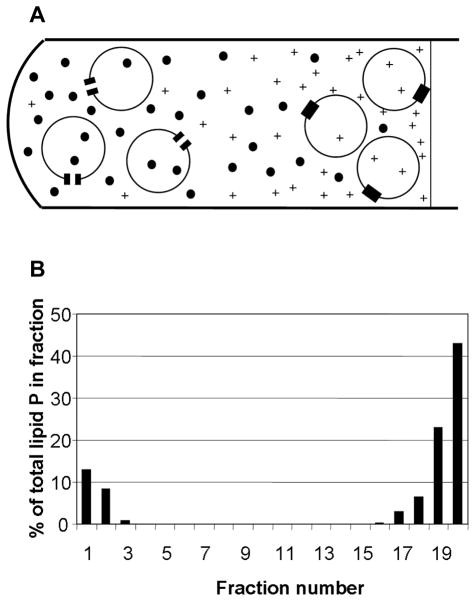
We wanted to enrich for the open conformers in the OprF population. Although the urea/sucrose gradient centrifugation of OprF-containing liposomes allowed us to get fractions highly enriched for the open conformer at the bottom, this method was not useful for the further biochemical studies of this conformer because we could insert only a few OprF molecules per vesicle in order to achieve a good separation. However, we found two ways allowing us to get open-conformer-enriched preparations of OprF in larger amounts. First, by fractionating OprF population by gel filtration on a high-resolution medium (Toyo Pearl HW-50F), we found that the pore-forming specific activity was much higher in the leading edge of the eluted OprF peak [17] (Fig. 3). SDS-PAGE showed that these fractions were enriched in oligomers of OprF (Fig. 3). Thus the open conformers have a stronger tendency to assemble into a metastable oligomer, and from the activity in liposome swelling assay we could estimate that the best fraction contained about 25 % open conformers. This fraction showed a much stronger tendency to produce open channels also in planar bilayer assay [18]. When the unfractionated OprF and this open-channel-enriched fraction were analyzed by electron spray ionization mass spectrometry, both showed main peaks corresponding to 35,241 and 35,242 m/e units. This is important because recently it was reported, for E. coli OmpA, that covalent addition of oligo-3-hydroxybutyrate strongly affects the folding of this protein. With OprF, there is no evidence of such covalent modification of the protein.
Fig. 3.
Enrichment for Open Conformers of OprF by High Resolution Gel Filtration. A. The OprF protein extracted by using a non-ionic detergent was fractionated on a 1 × 90 cm column of Toyo-Pearl HW-50F in the presence of 0.1 % dodecyl β-D-maltoside. Protein concentrations (squares) are in A280. The specific pore-forming activity was determined by proteoliposome swelling rate in isotonic L-arabinose and is shown in triangles (in units of ΔOD400/min/10 μg protein). Closed circles show the total pore-forming activity (ΔOD400/min/10 μl fraction). Note that in these latter two curves, the values of ΔOD400 are expressed in milliabsorbance units. B. Portions of each fraction were separated by SDS-PAGE. When the samples were not heat-denatured at 100 °C, fraction 1 is seen to contain oligomers of OprF (arrows), which collapse to monomers upon heat denaturation (not shown). From [17].
The other method for enriching for open conformer is based on the predicted folding model. The eight-stranded β-barrel structure of the N-terminal domain in the closed, two-domain conformer contains an essentially closed central channel, which cannot allow the permeation of organic compounds. In order to allow the passage of fairly large solutes, the open conformer must contain many more β-strands, if it folds as a β-barrel as in most outer membrane proteins. Thus the open conformer is expected to fold as a single domain, containing perhaps more than 14 β-strands. According to this folding model (Fig. 1, right), the residue A312 will be exposed on cell surface in the open conformer, but in the closed two-domain conformer will be buried in the periplasm as a part of the C-terminal globular domain. Thus the A312 residue was mutated into cysteine, and it was labeled in intact cells briefly with a cleavable biotinylation agent biotin-HPDP. The outer membrane was then solubilized, and the biotinylated OprF was isolated by using an avidin affinity column. This fraction was shown to have ten times higher pore-forming activity in comparison with the bulk, unfractionated OprF [17], suggesting that our folding model is essentially correct.
Interestingly, OprF protein was often thought to fold as a single domain conformer before these studies of ours. For example, some monoclonal anti-OprF antibodies were shown to react with sequences in the C-terminal domain [24], and even more importantly, a 4-residue malarial epitope inserted in the C-terminal half (e.g. at position 310) was shown to react with the specific monoclonal antibody in intact cells [15]. Such findings seemed to prove the surface exposure of C-terminal half residues, expected for a one-domain folding model of OprF. However, our knowledge that a minority of OprF folds as a one-domain conformer makes these results understandable. In fact, because such residues are exposed on cell surface only in a minority fraction of OprF populations, their reactivity with antibody should be rather weak. Indeed, they were shown to react much more weakly than the residues in the N-terminal domain, which are exposed in both the majority and minority conformers [15].
How are the divergent folding pathways regulated? Studies relying on spontaneous or random mutations
Because the open-channel conformer of OprF is stable and is apparently produced by a divergent folding pathway, we thought it important to find ways to influence this pathway so that the fraction of this conformer becomes increased, making this difficult-to-treat pathogen more susceptible to antimicrobial agents. As we know little about the folding pathway, we thought that the best approach would be to do a random mutagenesis and select for mutants producing more permeable OprF porin.
To select for such mutants, we chose raffinose, a trisaccharide, as the nutrient because it was shown to diffuse through OprF channel [9], and because other nutrients could be transported by other specific porins that are abundant in this organism [25]. Mutants in which more OprF molecules fold into the open channel conformers are expected to grow faster in a minimal medium containing raffinose as the sole carbon source, and we tried to select for such strains in a chemostat. This effort, however, was not successful because the growth rate competition in chemostat did not work owing to the very strong tendency of P. aeruginosa to form biofilms. Even the use of mutants deficient in Psl polysaccharide [26], thought to play an essential role in biofilm formation, did not improve the situation significantly.
In the next stage of our effort, we abandoned the intact cell approach and tried to use the in vitro ribosomal display technology [27]. Here we used the trypsin resistance of the single-domain open conformer [17] as a method of selection. However, OprF proteins made in vitro apparently could not fold properly in the absence of the outer membrane bilayer and the factors needed for the assembly of outer membrane proteins, and we could not make the selection to work.
How are the divergent folding pathways regulated? Studies relying on site-directed or known mutations
Because of the failure of the random mutagenesis approach, we decided to carry out site-directed mutagenesis on selected residues of OprF as well as to use known mutants of outer membrane assembly pathway [28]. The site-directed approach obviously depends on making correct guesses on residues playing important roles in the folding pathway, and we must have missed some critical residues. Nevertheless, it generated interesting data [29].
As the experimental system, we chose to use an E. coli host, because so much is known in this organism concerning the folding of periplasmic and outer membrane proteins [28]. The OprF protein was expressed from a medium copy number vector, with a hexahistidine tag usually attached to the N-terminus of the mature sequence. It was purified by using the hexahistidine tag, and its pore-forming activity was assayed by the standard liposome swelling assay [29].
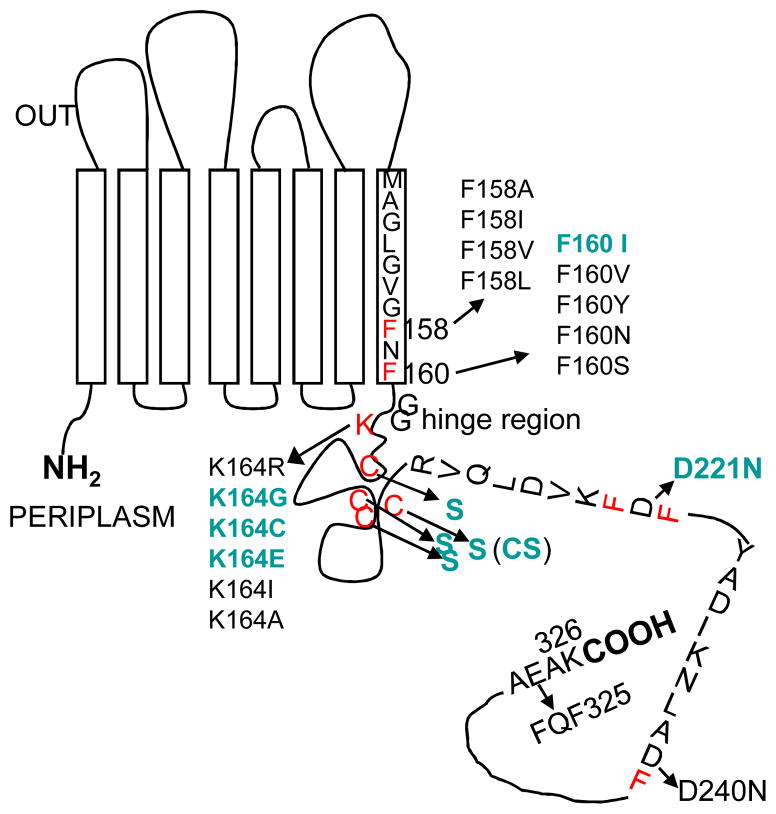
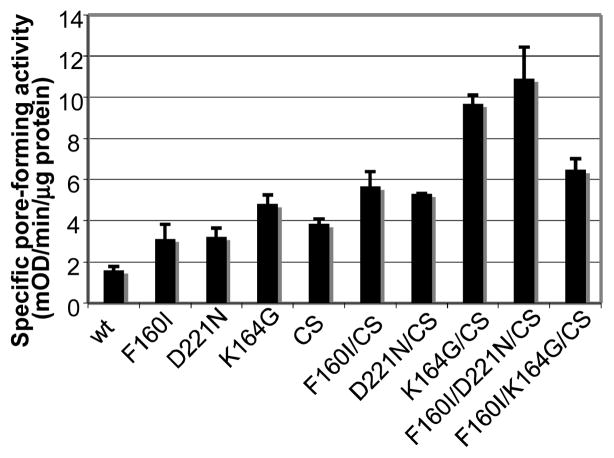
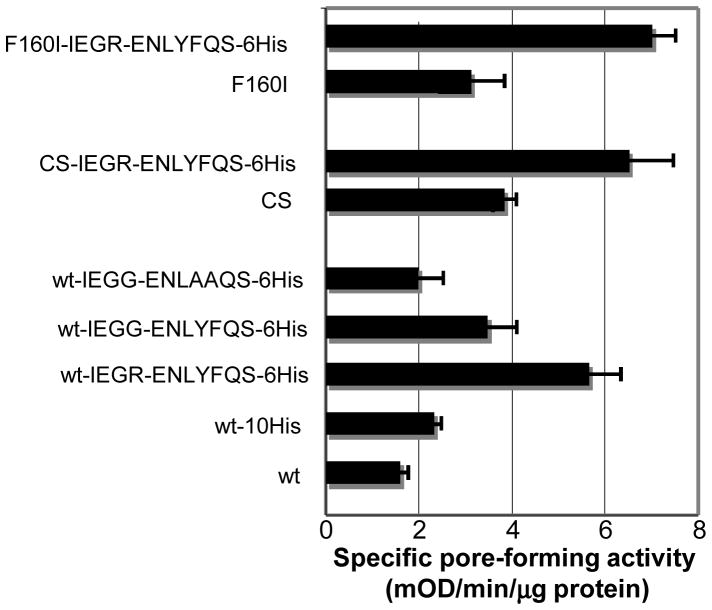
We first examined the effect of disulfide bond formation on folding (Fig. 4). This is because disulfide bond formation generally has a strong influence on the protein folding pathway [30] and because the four Cys residues in OprF are located right after the N-terminal β-barrel domain and are expected to exert a strong influence on the folding of the rest of the molecule, that is, whether it would fold as a continuation of β-barrel or as a separate globular domain. When OprF was expressed in an E. coli mutant deficient in the periplasmic disulfide oxidoreductase DsbA [31], the pore-forming activity of OprF was not visibly changed. However, growth of this mutant in a medium containing 5 mM dithiothreitol increased about three times the activity of OprF expressed in this mutant [29], suggesting that the prevention of disulfide bond formation increases the formation of one-domain, open-channel conformer, as expected. Similarly, conversion of all four cysteines of OprF into serine by site-directed mutagenesis increased the pore-forming activity by a factor of 2.4 [29](CS[Cys-into-Ser] in Fig. 5). This is not due to changes in the pore size, which remained identical with that of the wild-type OprF.
Fig. 4.
Design of the site-directed mutagenesis experiments. The OprF protein is shown in its majority (two-domain) conformation. Residues thought to be important in affecting the folding pathway are shown in red, and the changes shown to increase permeability are indicated in green. From [29].
Fig. 5.
Specific pore-forming activity (determined by proteoliposome swelling using L-arabinose as the permeating solute) of some mutant OprF proteins. “CS” denotes the conversion of all four Cys in the native protein into Ser residues. From [29].
The N-terminal domain of OprF ends with the sequence GVGFNF160 (where the number in subscript shows the residue number) (Fig. 4), and this corresponds to the C-terminal β-barrel sorting sequence recognized by the outer membrane protein assembly factor YaeT (BamA), G(M/L/V)*(Y/F)(Q/N)F (where * indicates any amino acid), for E. coli [32]. The C-terminal F160 is thought to play an especially important role in this recognition [33]. When we changed F160 into various other amino acids, F160I mutation was especially effective in increasing the pore-forming activity (Fig. 5). It is unclear why substitution with other amino acids did not have similar effect.
We noted that the C-terminal half of OprF contains two segments that might act as the β-barrel termination signal: FDF222, and ADF241 (Fig. 4). However, these sequences are unlikely to be efficient, because the central position in this tripeptide sequence is never occupied by an acidic residue in the real β-barrel termination signals [32]. D240N mutation did not change the activity of OprF. However, the D221N mutation increased the pore-forming activity twofold, presumably by creating a β-barrel termination signal closer to the C-terminus of the protein, which could compete with the termination signal at position 160. The combination of this mutation with F160I mutation and the elimination of cysteine residues created the highest specific activity OprF mutant as shown in Fig. 5. Finally, we tried to create a β-barrel termination signal at the very end of the OprF sequence, by changing AEA325 sequence into FQF. However, there was no change in the pore-forming activity in this mutant.
Other effective modifications of OprF sequence were found by accident. K164, which is only a few residues away from the barrel termination signal at position 160 in the middle of the protein, turned out to be important because changing it to Gly, Cys, or Glu increased the pore-forming activity about twofold. Another fortuitous finding was that the insertion of the IEGRENLYFQS sequence, containing cleavage sites for factor X and Tobacco Etch Virus protease, at the C-terminus of OprF increased the pore-forming activity more than twofold. Changing the Tyr-Phe in this sequence into Ala-Ala abolished this effect, and thus the original sequence seems likely to act as a β-barrel termination sequence (Fig. 6). In any case, these accidental discoveries emphasize the shortcoming of the site-directed mutagenesis approach: because we know so little about the folding process of these proteins we are bound to overlook some residues that are important in this process.
Fig. 6.
Specific pore-forming activity of OprF proteins containing TEV-Factor × cleavage sequence at its C-terminus. “IEGR-ENLYFQS” and its variants denote the TEV cleavage site added to the C-terminus. From [29].
Finally we examined the folding of OprF in mutants deficient in the known components of the outer membrane protein assembly pathway. The results were not always easy to interpret, because defects in such factors, even when they are not lethal, tend to produce shock responses. However, deletion of the periplasmic chaperone Skp appeared to increase the pore-forming activity of no-cysteine mutant of OprF. This is interesting because Skp, which has a cup-like shape [34], is thought to bind the N-terminal domain of OmpA [35] while the C-terminal half of OmpA is folded into its native globular shape outside the cavity of Skp [36, 37]. Perhaps the absence of Skp prevents the early folding of the C-terminal half of OprF into an independent domain, and thus favors the formation of one-domain, open-channel conformer. If this interpretation is correct, our past effort, concentrating on preventing the formation of N-terminal eight-stranded β-barrel, might not have been the best strategy. For encouraging the formation of one-domain conformers, it might possibly be more important to prevent the early assembly of the globular C-terminal domain, and this could be one of our future strategies.
Although we have been successful in altering the folding paths of OprF in E. coli, there is no guarantee that its folding in P. aeruginosa, probably with a different set of periplasmic proteins, will follow the same pattern. Thus we expressed OprF from a plasmid in a oprF-deficient mutant of P. aeruginosa, and measured the pore-forming activity of purified OprF by liposome swelling. We found that the product of wild-type oprF gene had about fivefold higher pore-forming activity when expressed in the presence of 5 mM dithiothreitol. Furthermore in the absence of dithiothreitol, the “cysteineless” mutant OprF, expressed in P. aeruginosa, was more than sixfold active in pore formation than the wild-type OprF (E. Sugawara and H. Nikaido, unpublished results). These results give us confidence that our findings are valid, at least in outlines, in P. aeruginosa as well as in E. coli.
Comparison with OmpA
Because E. coli OmpA and P. aeruginosa OprF are the best studied members of this protein family, it would be useful to compare their behaviors. Both serve, in their majority conformation, as a physical linker between the peptidoglycan and the outer membrane, as mentioned above. Both contain minority conformers that produce open channels [16, 17]. The closed conformer of OmpA was reported to become converted to the open conformer in planar bilayers by temperatures as low as 42 °C [38]. We have tried to see a similar temperature-induced transition in OprF, reconstituted into liposomes, but have not so far observed any indication of such changes, and in our hands both the open and closed conformers appear to be quite stable. We note, in the OmpA study [38], that the protein was extracted with the denaturing detergent lithium dodecylsulfate. Under such conditions, the C-terminal half becomes completely denatured, as we have shown by using the CD spectrum [14]. The addition of this preparation to a planar bilayer leads to the insertion of the N-terminal β-barrel into the bilayer, but the C-terminal half most likely remains in the denatured state. Perhaps this structure is more easily converted into an open-channel, single-domain conformer, because the C-terminal half has not been stabilized by folding into a globular domain. Even if the C-terminal domain were correctly folded, the transition would have little biological significance, because in intact cells, the C-terminal domain would be bound strongly to the underlying peptidoglycan [19, 20], and would not easily become detached to participate in the formation of the single-domain conformer.
In any case, comparison of OmpA and OprF sequences reveal that the several residues after the end of the β-barrel domain are quite similar but then OprF contains an approximately 30-residue sequence, with all of the four Cys residues, which does not exist in OmpA. Thus it may not be surprising to find that the folding pathways of these two proteins are significantly different.
Other organisms that use OprF homologs as the major porin
The results we have obtained with P. aeruginosa have a wider relevance, because there are very large numbers of organism that appear to use OprF homologs as the major porin. Because most early studies on porins were done with E. coli and salmonella, we were led to believe that a typical Gram-negative bacterium would contain highly permeable, always open porin channels exemplified by OmpF and OmpC. However, these are organisms that live in a highly specialized ecological niche, the upper intestinal tract of higher animals. Much more numerous species of Gram-negative organisms live in the “environment,” typically in soil, where they are constantly exposed to antimicrobial compounds produced by competing microorganisms. For them, the highly permeable, always open channels of OmpF/C-like porins would be suicidal. Thus such organisms live with porins with extremely low permeability, and the ubiquitous OmpA family proteins most often seems to play the role of the major porin.
This is certainly true of organisms belonging to the genus Pseudomonas. Interestingly, growth temperature affected the ion conductivity of OprF from psychrotrophic P. fluorescens strains, the protein from organism cultured at 28 °C showing much higher single-channel conductivity than that from that grown at 8 °C [39]. If the former is enriched for a single domain conformer, one would expect increased resistance to trypsin digestion, which tends to cut first in the “linker” region between the domains in the two-domain conformer. Indeed this was what was observed.
One species that are becoming an important nosocomial pathogen is Acinetobacter baumanii [40], where high-level resistance to a number of antibiotics is often encountered. Although not much is known about the porins in this species [41], the outer membrane of the related species A. calcoaceticus was shown, upon reconstitution into proteoliposomes, to produce permeability that is about two orders of magnitude lower than that of E. coli outer membrane [42]. We have shown by gene knockouts that the OmpA homolog in A. baumanii is the major porin in this species, and that this protein produces a very low permeability similar to P. aeruginosa OprF (E. Sugawara and H. Nikaido, unpublished results). Since the low outer membrane permeability amplifies the resistance-elevating capacity of multidrug efflux pumps [43], this is another protein that warrants close studies in view of the clinical importance of the organism.
Fig. 2.
Separation of Open and Closed Conformers of E. coli OmpA. A. Principles of separation. The isoosmotic gradient contains higher concentrations of urea (+) at the top, and higher concentration of sucrose (•) at the bottom. When unilamellar vesicles containing only a few molecules of OmpA, made in 0.3 M urea, are centrifuged through this gradient, those containing only closed conformers will remain on top, but those containing open conformer(s) will sediment because the influx of sucrose will make the vesicles heavier. B. The experimental result obtained by Sugawara and Nikaido in 1994 [16]. This figure is after that published in [44].
Acknowledgments
Studies in our laboratory were supported in part by a grant (AI-09644) from the National Institutes of Health.
References
- 1.Diekema DJ, Pfaller MA, Jones RN, Doern GV, Kugler KC, Beach ML, Sader HS. Trends in antimicrobial susceptibility of bacterial pathogens isolated from patients with bloodstream infections in the USA, Canada and Latin America. SENTRY Participants Group. Int J Antimicrob Agents. 2000;13:257–71. doi: 10.1016/s0924-8579(99)00131-4. [DOI] [PubMed] [Google Scholar]
- 2.Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982;152:636–42. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 4.Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–53. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsueh PR, Tseng SP, Teng LJ, Ho SW. Pan-drug-resistant Pseudomonas aeruginosa causing nosocomial infection at a university hospital in Taiwan. Clin Microbiol Infect. 2005;11:670–3. doi: 10.1111/j.1469-0691.2005.01196.x. [DOI] [PubMed] [Google Scholar]
- 6.Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect. 2007;13:560–78. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 7.Hancock RE, Decad GM, Nikaido H. Identification of the protein producing transmembrane diffusion pores in the outer membrane of Pseudomonas aeruginosa PAO1. Biochim Biophys Acta. 1979;554:323–31. doi: 10.1016/0005-2736(79)90373-0. [DOI] [PubMed] [Google Scholar]
- 8.Decad GM, Nikaido H. Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J Bacteriol. 1976;128:325–36. doi: 10.1128/jb.128.1.325-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellido F, Martin NL, Siehnel RJ, Hancock RE. Reevaluation, using intact cells, of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J Bacteriol. 1992;174:5196–203. doi: 10.1128/jb.174.16.5196-5203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–9. [PubMed] [Google Scholar]
- 11.Ried G, Koebnik R, Hindennach I, Mutschler B, Henning U. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol Gen Genet. 1994;243:127–35. doi: 10.1007/BF00280309. [DOI] [PubMed] [Google Scholar]
- 12.Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–7. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 13.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol. 2004;51:1027–37. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 14.Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–9. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong RS, Wirtz RA, Hancock RE. Pseudomonas aeruginosa outer membrane protein OprF as an expression vector for foreign epitopes: the effects of positioning and length on the antigenicity of the epitope. Gene. 1995;158:55–60. doi: 10.1016/0378-1119(95)00155-y. [DOI] [PubMed] [Google Scholar]
- 16.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–7. [PubMed] [Google Scholar]
- 17.Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem. 2006;281:16220–9. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestorovich EM, Sugawara E, Nikaido H, Bezrukov SM. Pseudomonas aeruginosa porin OprF: properties of the channel. J Biol Chem. 2006;281:16230–7. doi: 10.1074/jbc.M600650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–4. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 20.Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–70. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 21.Manchur MA, Kikumoto M, Kanao T, Takada J, Kamimura K. Characterization of an OmpA-like outer membrane protein of the acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans. Extremophiles. 2011;15:403–410. doi: 10.1007/s00792-011-0371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff WA, Hancock RE. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J Bacteriol. 1989;171:3304–9. doi: 10.1128/jb.171.6.3304-3309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–5. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawling EG, Martin NL, Hancock RE. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect Immun. 1995;63:38–42. doi: 10.1128/iai.63.1.38-42.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock RE, Brinkman FS. Function of Pseudomonas porins in uptake and efflux. Annu Rev Microbiol. 2002;56:17–38. doi: 10.1146/annurev.micro.56.012302.160310. [DOI] [PubMed] [Google Scholar]
- 26.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–21. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanes J, Jermutus L, Pluckthun A. Selecting and evolving functional proteins in vitro by ribosome display. Methods Enzymol. 2000;328:404–30. doi: 10.1016/s0076-6879(00)28409-7. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 29.Sugawara E, Nagano K, Nikaido H. Factors affecting the folding of Pseudomonas aeruginosa. OprF porin into the one-domain open conformer. mBio. 2010;1:e00228–10. doi: 10.1128/mBio.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creighton TE. Protein folding coupled to disulphide bond formation. BiolChem. 1997;378:731–44. doi: 10.1515/bchm.1997.378.8.731. [DOI] [PubMed] [Google Scholar]
- 31.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–35. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 32.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–8. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 34.Korndorfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11:1015–20. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- 35.Qu J, Behrens-Kneip S, Holst O, Kleinschmidt JH. Binding regions of outer membrane protein A in complexes with the periplasmic chaperone Skp. A site-directed fluorescence study. Biochemistry (Mosc) 2009;48:4926–36. doi: 10.1021/bi9004039. [DOI] [PubMed] [Google Scholar]
- 36.Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci U S A. 2009;106:1772–7. doi: 10.1073/pnas.0809275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–74. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Zakharian E, Reusch RN. Kinetics of folding of Escherichia coli OmpA from narrow to large pore conformation in a planar bilayer. Biochemistry (Mosc) 2005;44:6701–7. doi: 10.1021/bi047278e. [DOI] [PubMed] [Google Scholar]
- 39.De E, Orange N, Saint N, Guerillon J, De Mot R, Molle G. Growth temperature dependence of channel size of the major outer-membrane protein (OprF) in psychrotrophic Pseudomonas fluorescens strains. Microbiology. 1997;143(Pt 3):1029–35. doi: 10.1099/00221287-143-3-1029. [DOI] [PubMed] [Google Scholar]
- 40.Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med. 2008;358:1271–81. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 41.Vila J, Marti S, Sanchez-Cespedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2007;59:1210–5. doi: 10.1093/jac/dkl509. [DOI] [PubMed] [Google Scholar]
- 42.Sato K, Nakae T. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J Antimicrob Chemother. 1991;28:35–45. doi: 10.1093/jac/28.1.35. [DOI] [PubMed] [Google Scholar]
- 43.Coyne S, Courvalin P, Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55:947–53. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara E, Nikaido H. OmpA/OprF: Slow porins or channels produced by alternative folding of outer membrane proteins. In: Benz R, editor. Bacterial and Eukaryotic Porins. Weinheim: 2004. pp. 119–138. [Google Scholar]