Abstract
UDP-N-acetylglucosamine acyltransferase (LpxA) and UDP-3-O-(R-3-hydroxyacyl)-glucosamine acyltransferase (LpxD) catalyze the first and third steps of Lipid A biosynthesis, respectively. Both enzymes have been found to be essential for survival among Gram-negative bacteria which synthesize lipopolysaccharide, and are viable targets for antimicrobial development. Catalytically, both acyltransferases catalyze an acyl-acyl carrier protein (ACP) dependent transfer of a fatty acyl moiety to a UDP-glucosamine core ring. Herein, we exploit the single free-thiol unveiled on holo-ACP after transfer of the fatty acyl group to the glucosamine ring using the thiol specific labeling reagent, ThioGlo. The assay is continuously monitored as a change in fluorescence at λex = 379 nm and λem = 513 nm using a microtiter plate reader. This assay marks the first continuous and non-radioactive assay for either acyltransferase.
Keywords: Lipid A, acyltransferase, fluorescense assay, acyl carrier protein, Thioglo
Introduction
The outer cell wall of gram-negative bacteria is an asymmetrical bilayer comprised of phospholipids in the inner monolayer and lipopolysaccharide in the outer monolayer [1–3]. LPS contains the glycolipid, lipid A, which anchors it to the membrane through fatty acyl chains, a core polysaccharide region and an O-antigen repeat. Lipid A is an essential moiety necessary for survival of the bacterium [4], and further plays a crucial role in natural antibacterial resistance and bacterial sepsis [5]. Thus, lipid A biosynthesis should provide optimal targets for antimicrobial chemotherapeutic discovery [6–8].
Lipid A biosynthesis entails nine constitutive enzymatic processes utilizing UDP-N-acetylglucosamine (UDP-GlcNAc) as the precursor [9]. UDP-GlcNAc is acylated at the 3-hydroxyl through a thermodynamically unfavorable reaction catalyzed by the type II ACP-dependent UDP-N-acetylglucosamine acyltransferase (LpxA) [10]. UDP-3-O-(R-3-hydroxyacyl)-GlcNAc is subsequently deacetylated by UDP-3-O-(R-3-hydroxyacyl)-GlcNAc deacetylase (LpxC), providing the first committed step of lipid A biosynthesis [11, 12]. The third step in the pathway involves acylation of the free amine of the glucosamine ring by UDP-O-3-(R-3-hydroxyacyl)-glucosamine acyltransferase (LpxD) which is also an ACP-dependent acyltransferase [13]. Six downstream enzymes catalyze the formation of the mature hexaacylated Kdo2-lipid A moiety of LPS [2].
LpxA and LpxD are structurally homologous as demonstrated by their unique left-handed β-helix (LβH) fold, which stems from an extensive hexapeptide repeat motif in their respective primary amino acid sequences [14–16]. As well, both are functionally similar as displayed by a common mechanism whereby conserved histidine residues mediate the deprotonation of the 3-hydroxyl (LpxA) or the 2-amine (LpxD) of the core glucosamine ring, allowing a subsequent nucleophilic attack upon the thioester of acyl-ACP [17, 18]. Holo-ACP is generated upon acyl group transfer, which contains a single free-thiol on its phosphopantetheine prosthetic arm [19].
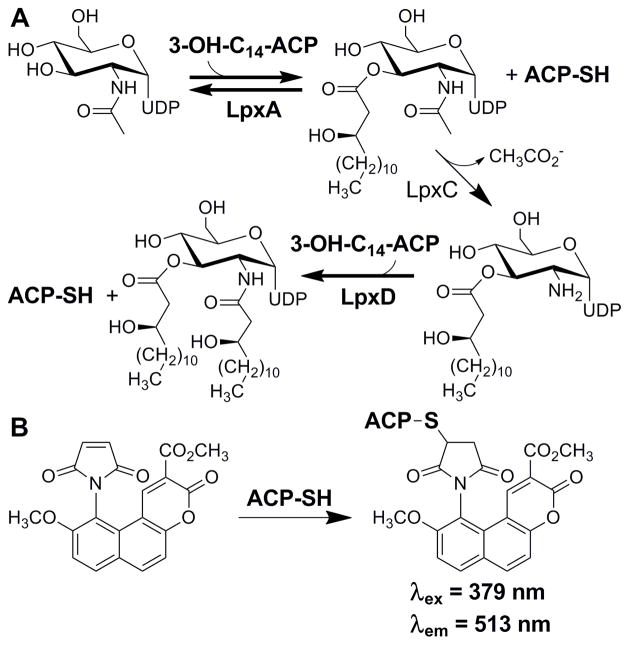
Although the LPS biosynthetic pathway has been established as a prime target for the development of novel antimicrobials, very few of the essential enzymes in the pathway have been subjected to high throughput screening efforts in an attempt to find novel inhibitors. Established assays for enzymes in this pathway are radioactivity based, and as such are not welcomed in many high throughput screening facilities. Herein we report the first non-radioactive assay for LpxA and LpxD. The assay uses a thiol-specific chemical reporter, ThioGlo, to continuously label and monitor holo-ACP generation over the course of LpxA and LpxD catalysis (Figure 1).
Figure 1.
A) Early enzymatic steps of LPS biosynthesis in E. coli and B) generation of the ThioGlo-ACP conjugate from holo-ACP produced in the acyltransferase catalyzed reactions.
Materials and Methods
Materials
R-3-hydroxymyristic acid was purchased from Wako Chemicals. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), isopropanol, magnesium chloride and buffer reagents were purchased from Thermo Fisher Scientific. L-arabinose, ATP, and UDP-N-acetylglucosamine were purchased from Sigma-Aldrich. Benzonase, ThioGlo®1, and E. coli Rosetta(DE3)/pLysS cells were purchased from EMD Chemicals (Novagen). E. coli XL-1 Blue cells were purchased from Strategene. Isopropyl β-D-1-thiogalactopyranoside (IPTG), E. coli BL21-AI cells were purchased from Invitrogen. Bio-Gel P2 was purchased from Bio-Rad. All DNA modifying and restriction enzymes were purchased from New England Biolabs.
Cloning of E. coli lpxA, lpxC, lpxD, acpP, acpS and Vibrio harveyi aasS
PCR protocols were carried out under standard conditions utilizing Pfu DNA polymerase and DNA obtained from the E. coli K-12 strain MG1655 or the V. harveyi ATCC14126 strain. To perform the amplifications of the individual genes the following primers were used: lpxA (forward 5′-GCGCCATATGATTGATAAATCCGCCTTTGTGCATCCAACCGC, reverse 5′-CGCGCTCGAGTTAACGAATCAGACCGCGCGTTGAGCG); lpxC (forward 5′-GCGCCATATGATGATCAAACAAAGGACACT, reverse 5′-GCGCCTCGAGTGCCAGTACAGCTGAAGGCG); lpxD (forward 5′-CATCACCATCACCATCACGCTCAATTCGACTGGCTGATTTAGCG, reverse 5′-CGCGCTCGAGTTAGTCTTGTTGATTAACCTTGCGCTC); acpP (forward 5′-GCGCCATATGAGCACTATCGAAGAACGCGTTAAGAAAATTATC, reverse 5′-GCGCCTCGAGTTAACTTTCAATAATTACCGTGGCAC); acpS (forward 5′-CGCGTGGCATATGGCAATATTAGGTTTAG, reverse 5′-GCGCCTCGAGACTTTCAATAATTACCGTGGCACAAGC); V. harveyi aasS (forward 5′-GCGCCATATGAACCAGTATGTAAAT, reverse 5′-GCGCCTCGAGCAGATGAAGTTTACGCAG).
The PCR products for both lpxA, and acpP were cloned into pET24a using NdeI and XhoI restriction sites (underlined). The XhoI-restricted PCR product for lpxD was cloned into pET23d which had been NcoI restricted, T4 DNA polymerase filled-in, and then restricted with XhoI. Each of the PCR products for lpxC, acpS and aasS were cloned into pET23a using NdeI and XhoI restriction sites. All plasmids were transformed into E. coli XL-1 Blue cells for amplification and plasmids isolated from these cell lines were sequenced at the University of Michigan Sequencing Core Facility. From these confirmed plasmids the following E. coli expression strains were constructed: BL21-AI/pET24a::lpxA, Rosetta (DE3)/pLysS/pET23a::lpxC-his6, Rosetta (DE3)/pLysS/pET23d::his6-lpxD, BL21-AI/pET24a::acpP/pET23a::acpS-his6, BL21-AI/pET23a::aasS-his6. Genes containing a 6 histidine tag coding region are indicated by his6 in the above construct names. The his6 in front of the gene name denotes a 5′ histidine coding region, whereas the his6 after the gene name denotes a 3′ histidine coding region.
Cell cultures
Strains of interest were used to inoculate 500 mL LB (Lennox) or TB media containing the appropriate antibiotic(s), and incubated while shaking (250 rpm) at 37 °C until an OD600 of 0.6–1.0 was reached. The cultures were then induced with either 1 mM IPTG (Rosetta DE3/pLysS strains) or 0.2% L-arabinose/1 mM IPTG (BL21-AI strains). Unless otherwise noted, cells were induced at 37 °C and allowed to incubate at 37 °C for 4 hours post-induction. Cells were harvested by centrifugation at 5,000 × g for 10 min at 4 °C, suspended in 10 mL of buffer and stored at −80 °C. Cell suspensions were thawed and disrupted by French press at 20,000 psi. Cellular debris was removed by centrifugation at 20,000 × g for 30 min at 4 °C and the resultant crude cytosol used for protein purification.
Purification of LpxA
For LpxA purification, 10 mL of crude cytosol in 20 mM potassium phosphate (KPhos) buffer, 20% glycerol pH 7.0 was applied to a 10 mL Reactive Green 19 column, which had been pre-equilibrated in the same buffer. The column was washed successively with 50 mL of loading buffer containing 0 M, 0.5 M, and 1 M NaCl. LpxA eluted with the 1 M NaCl fractions and was dialyzed overnight at 4 °C against 4 L of 20 mM Tris-HCl, 10% glycerol pH 8.0. The enzyme was then loaded onto an 8 mL Source 15Q column, washed with 24 ml of loading buffer and eluted with a gradient from 0–500 mM NaCl. LpxA was desalted on a Bio-Gel P2 column equilibrated in 20 mM HEPES pH 8.0. The purified LpxA was analyzed by SDS-PAGE and its concentration determined by UV absorbance at 280 nm (ε = 9190 M−1 cm−1). The molecular weight of LpxA was confirmed by MALDI-TOF Mass spectrometry (MS) at the University of Michigan Protein Structure Facility.
Purification of LpxC-His6 and His6-LpxD
For His6-LpxD purification, Benzonase (Novagen) was added after cell lysis and the lysate was incubated for 30 min on ice prior to centrifugation at 20,000 × g. Ten milliliters of crude cytosol in 20 mM HEPES, 50 mM imidazole pH 8.0 was loaded onto 3 mL of Ni-NTA resin (Qiagen) equilibrated in the same buffer. The resin was washed with 10 column volumes of loading buffer containing 500 mM NaCl and then eluted with 20 mM HEPES, 250 mM imidazole pH 8.0. Purified His6-LpxD and LpxC-His6 were desalted on a Bio-Gel P2 column and analyzed by SDS-PAGE. Concentrations were determined by UV absorbance at 280 nm (ε = 22920 M−1 cm−1 for LpxC; ε = 27305 M−1 cm−1 for LpxD).
Purification of holo-ACP
A slightly modified protocol from Broadwater and Fox was utilized to prepare holo-ACP [20]. Holo-ACP was produced in cells expressing both apo-ACP (acpP gene product) and holo-ACP synthetase (acpS gene product). Protein was expressed as describe above (Cell culture) except cells were cooled to 18 °C prior to induction and then allowed to grow overnight at 18 °C. To the cellular lysate, in 20 mM HEPES, 1 mM TCEP pH 8.0, 10 mL of cold isopropanol was added slowly and while gently mixing for 1 hour at 4 °C. The resulting suspension was centrifuged at 20,000 × g for 30 min at 4 °C. The supernatant was removed and to this 20 mL of 20 mM HEPES, 1 mM TCEP pH 8.0 was added. This solution was loaded onto a Source 15Q column (8 mL). A gradient of 20 mM HEPES, 1 mM TCEP, pH 8.0 containing 0 – 500 mM NaCl was performed and holo-ACP eluted at approximately 300 mM NaCl as judged by SDS-PAGE analysis. The protein was subsequently desalted on a P2 Bio-Gel column in 20 mM HEPES, 1 mM TCEP, pH 8.0. The protein was freeze dried and stored at −20 °C. Protein concentrations were measured via BioRad protein assay. The molecular weight of holo-ACP was confirmed by MALDI-TOF Mass spectrometry (MS) at the University of Michigan Protein Structure Facility.
Purification of V. harveyi AasS-His6
Expression and purification of the soluble acyl-ACP synthetase (AasS) was carried out under the same conditions as His6-LpxD and LpxC-His6. Following elution from the Ni-NTA resin, AasS was desalted on a Bio-Gel P2 column in 20 mM Tris-HCl pH 7.5, 10% glycerol, 1 mM EDTA, 0.1 mM TCEP and 0.002% Triton-X100 as previously described for optimal storage [21]. The desalted protein was aliquoted into microcentrifuge tubes and stored at −80 °C. Protein concentration was determined by UV absorbance 280 nm (ε = 65780 M−1 cm−1).
Acylation of holo-ACP
Acylation was carried out as previously described with slight modifications [21]. Holo-ACP was reduced for 1 hour at room temperature in the presence of two equivalence of TCEP prior to the loading of the fatty acid. The acylation reaction contained, in a final volume of 10 ml, 40 μM reduced holo-ACP, 5 mM ATP, 5 mM MgCl2, 100 μM TCEP, 0.01% Triton X-100, 100 μg of AasS, and 150 μM R-3-hydroxymyristic acid in 100 mM Tris, pH 7.5. The reaction was incubated at 30 °C for 45 min and another 50 μg of AasS was added. After 20 min of additional incubation at 30 °C, the reaction was cooled and loaded directly onto a Source 15Q column (8 mL) equilibrated in 20 mM HEPES pH 8.0. The column was washed with 3 column volumes of equilibration buffer and eluted with an 80 mL linear gradient of 0 – 500 mM NaCl. Acyl-ACP eluted at approximately 300 mM NaCl and was subsequently desalted via a Bio-Gel P2 column and lyophilized.
Synthesis and purification of ThioGlo-ACP conjugate
A 1 mL suspension of 100 μM holo-ACP, 1 mM TCEP, and 132 μM ThioGlo, in a final buffer composition of 20 mM HEPES pH 8, 5% DMSO, was incubated at 25 °C for 25 min. An additional 132 μM of ThioGlo was added after the initial reaction period and the suspension mixed by pipetting up and down multiple times. The reaction was incubated for an additional 35 min. The mixture was desalted on a Bio-Rad P2 size-exclusion column equilibrated in 20 mM HEPES pH 8. Protein concentrations were determined using Bio-Rad Protein assay with R-3-hydroxymyristoyl-ACP as a standard.
Synthesis of UDP-3-O-(R-3-hydroxmyristoyl)-GlcN
50 μM holo-ACP was reduced for 1 hour at room temperature with 3 mM TCEP in 100 mM Tris-HCl pH 7.6 containing 0.01% Triton-X100. Following reduction of ACP, 3 mM ATP, 3 mM MgCl2, 150 μM R-3-hydroxymyristic acid, and 150 μg of AasS were added to the centrifuge tube at a final volume of 5 mL and the tube was incubated at 37 °C for 15 min. Substrate synthesis was initiated by the addition of LpxA, LpxC and UDP-GlcNAc at concentrations of 30 μg/mL, 50 μg/mL and 500 μM and the solution was incubated for 45 min at 37 °C. Following the 45 min incubation, 100 μM of R-3-hydroxmyristic acid, 30 μg/mL LpxA, 30 μg/mL AasS, and 50 μg/mL LpxC were added and incubated for an additional 30 min. The solution was added to a round bottom flask containing silica gel (20 mg) and the water removed by rotary evaporation. The dried silica gel was added to a silica column (2 g) pre-equilibrated in 75:25 hexanes/ethyl acetate (EtOAc). The silica was washed with 100 mL 75:25 hexanes/EtOAc to remove the R-3-hydroxmyristic acid. The plug was subsequently washed with 50 mL of 25:15:4:2 v/v/v/v dichloromethane (DCM)/methanol (MeOH)/water/acetic acid. The solvent was evaporated off and the resulting solid was resuspended in 1 mL of DMSO and filtered through a .2 micron filter to remove any silica gel. UDP-3-O-(R-3-hydroxmyristoyl)-GlcN was purified by HPLC using 0.05% ammonium acetate and acetonitrile on a reverse phase C18 semi-prep column as reported by Anderson and Raetz [22]. Purified UDP-3-O-(R-3-hydroxymyristoyl)-GlcN was lyophilized, dissolved in water and frozen at −80 °C until further use. The concentration was measured by UV absorbance at 262 nm (ε = 9900 M−1 cm−1) [17]. The purified product was subjected to analytical HPLC and negative ion electrospray ionization mass spectrometry (ESI-MS) for characterization.
Fluorescent enzyme assay for LpxA and LpxD
Assays were performed at 25 °C in Corning black, 96-well half-area plates and all solutions were made up in 20 mM HEPES pH 8.0 with a final assay volume of 100 μL. A SpectraMax M5 (Molecular Devices) plate reader was used to monitor fluorescence, with PMT sensitivity set to low to prevent saturation and number of readings set to 100. First, 50 μL of 20 μM ThioGlo solution was added to 20 μL of various concentrations of 3-hydroxymyristoly-ACP and 20 μL of various concentrations of UDP-GlcNAc or UDP-3-O-(R-3-hydroxymyristolyl)-GlcN. This mixture was incubated in the dark at 25 °C for 5 min to allow any unacylated ACP to react with the ThioGlo solution. To initiate the reaction, 10 μL of 100 nM acyltransferase solution was added directly to the well, mixed gently and the plate was read continuously at λex = 379 nm and λem = 513 nm for 10 min at 15 s intervals. It should be noted that 100 nM LpxA or LpxD solutions were made fresh between each run from 1 mg/mL stock solution of purified enzymes. All reactions were performed in triplicate.
Data Analysis
Initial velocities were calculated through linear regression analysis during the first 2 min of the assay. Km’s were determined by non-linear regression analysis using KaleidaGraph software (Synergy Software), from plots of initial velocities versus substrate concentrations, while holding the other substrate at saturating conditions (eq. 1). Control reactions lacking individual substrate or enzyme components were performed to demonstrate that increase in fluorescence was both enzyme and substrate dependent, and that no individual substrate or enzyme was causing an increase in relative fluorescence over time. Linear dependence of enzyme concentration with respect to initial velocity was established through altering enzyme concentration while holding both substrates constant.
| (1) |
Results & Discussion
The enzymes in early lipid A biosynthesis are highly conserved in Gram-negative pathogens and represent potential targets for antibacterial development. Both biochemical and genetic studies have demonstrated the essential nature of LpxA and LpxD acyltransferases in Gram-negative organisms [13, 23, 24]. For the most part, current assays utilized for assaying enzymes within the lipid A biosynthetic pathway use radiolabeled substrates coupled with thin layer chromatography and phosphorimaging development. These assays have endured over the years due to their reliability and the inherent need for sensitivity as a result of scarcity and solubility of substrates. Fluorescence-based enzymatic assays have proven to be highly sensitive and robust, and unlike radioactivity-based assays, have been used widely for high throughput screening (HTS) [25]. The continuous fluorescent assays established herein are initial efforts to facilitate the identification and evaluation of LpxA/D inhibitors to be used as research probes and/or antimicrobial leads.
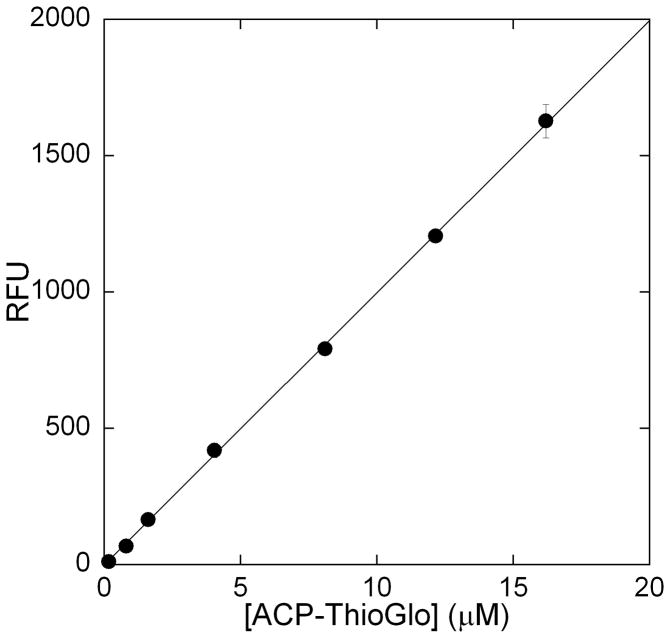
ThioGlo®1 (methyl 10-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-9-methoxy-3-oxo-3H-benzo[f]chromene-2-carboxylate) has been used in the fluorescent detection of free thiol containing compounds in analytical HPLC, biological tissue samples, cell free extracts, and HTS [25–28]. This reagent has been used to determine protein thiol concentrations as low as 10 nM [29]. Therefore, we sought to utilize this reagent to monitor holo-ACP generation during LpxA and LpxD catalysis. With this assay the rate of holo-ACP production can be monitored continuously by detection of the fluorescent ThioGlo-ACP conjugate (λex = 379 nm and λem = 513 nm). A standard fluorescent curve was generated by reading the fluorescence of various concentrations (0.1–13 μM; 100 μL) of ThioGlo-ACP conjugate in a Corning black, 96-well half-area plate (Figure 2). No deviation from linearity was noted upon adding ThioGlo at concentrations up to 10 μM. Under these conditions 1 μM ThioGlo-ACP gives 99.8 RFU.
Figure 2.
Fluorescence vs. [ThioGlo-ACP] plot. The fluorescence of various concentrations of purified ThioGlo-ACP conjugate, in 20 mM HEPES pH 8 (total volume 100 μL), was determined at 25 °C using a SpectraMax M5 plate reader.
Optimization of the reagents utilized for the kinetic analysis needed to be performed prior to kinetic characterization of LpxA. Final concentrations of LpxA were found to be optimal at 10 nM, where linear velocities could be measured within the first 2 min of a 10 min assay. It should be noted that when LpxA was diluted in assay buffer alone to 100 nM stock solution, the enzyme lost activity over time while on ice. Therefore, enzyme from a 1 mg/mL stock was diluted fresh to 100 nM for each assay and used immediately.
Since the free thiol generated upon holo-ACP generation is prone to oxidation and disulfide formation, which would interfere with ThioGlo conjugation, initially TCEP (a reducing agent) was employed to mitigate this possibility. However, TCEP proved to interfere with the reproducibility of labeling holo-ACP with ThioGlo in agreement with previous protein labeling studies with maleimide reagents in the presence of TCEP [30]. Exclusion of reducing agent resulted in greatly enhanced reproducibility and robustness of the assay, and thiol oxidation over the course of the assay time was not an issue.
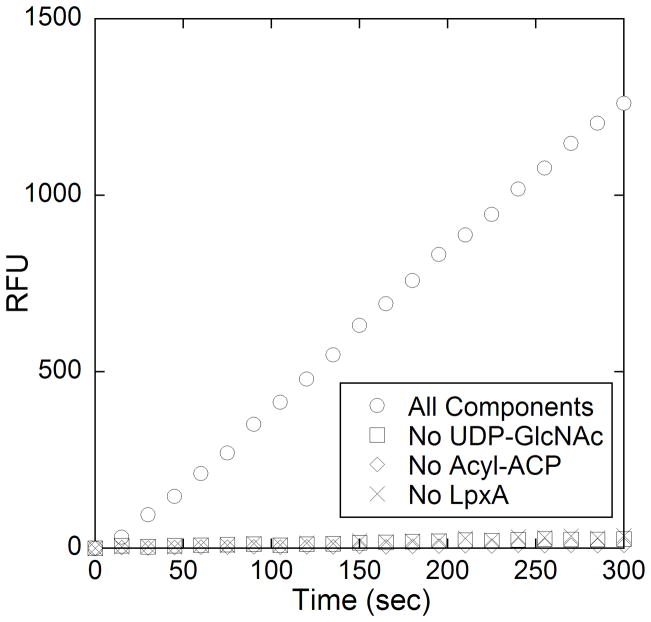
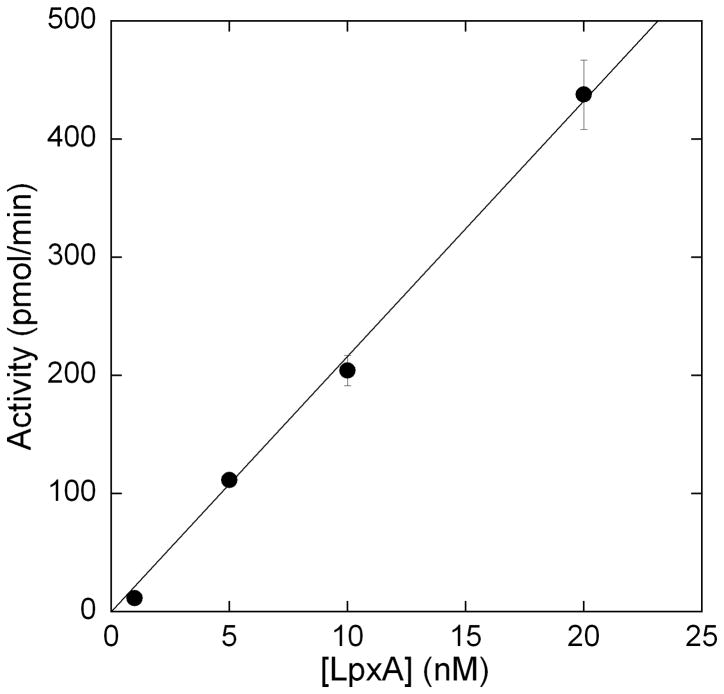
Once conditions were optimized time dependent-turnover of LpxA catalyzed acyl group transfer was demonstrated (Figure 3). Control reactions where individual components were omitted showed no increase in fluorescence, demonstrating that the generation of holo-ACP was being measured. The assay shows a linear dependence on enzyme concentration up to 20 nM which further establishes that the kinetics of LpxA are being measured, rather than the kinetics of ThioGlo conjugation to ACP (Figure 4). The specific activity for LpxA under our reaction conditions (25 °C) is 7.7 μmol/min/mg (kcat = 3.6 s−1). This value is reasonably lower (36 %) than literature values where assays were performed at a higher temperature (30 °C) [10].
Figure 3.
Reaction progress curves of the complete LpxA reaction (○), and control reactions without either nucleotide (□), acyl-ACP (◇), or acyltransferase (x). The complete LpxA assay mixture contained 20 mM HEPES (pH 8), 40 μM R-3-hydroxymyristoly-ACP, 4 mM UDP-GlcNAc, 10 μM ThioGlo, and 10 nM LpxA (added 5 minutes after ThioGlo) in a final volume of 100 μL. The reaction was incubated at 25 °C and its progress was monitored continuously at λex = 379 nm and λem = 513 nm for 10 min at 15 s intervals. Control reactions were run in similar fashion with the omission of substrate or enzyme, as indicated.
Figure 4.
Linear dependence of initial reaction velocity with varying LpxA concentrations using the ThioGlo coupled fluorescence assay. The assays were run as described under Figure 3 caption, with varying amounts of LpxA being added. The assay showed a linear relationship between activity and enzyme concentration up to 20 nM LpxA.
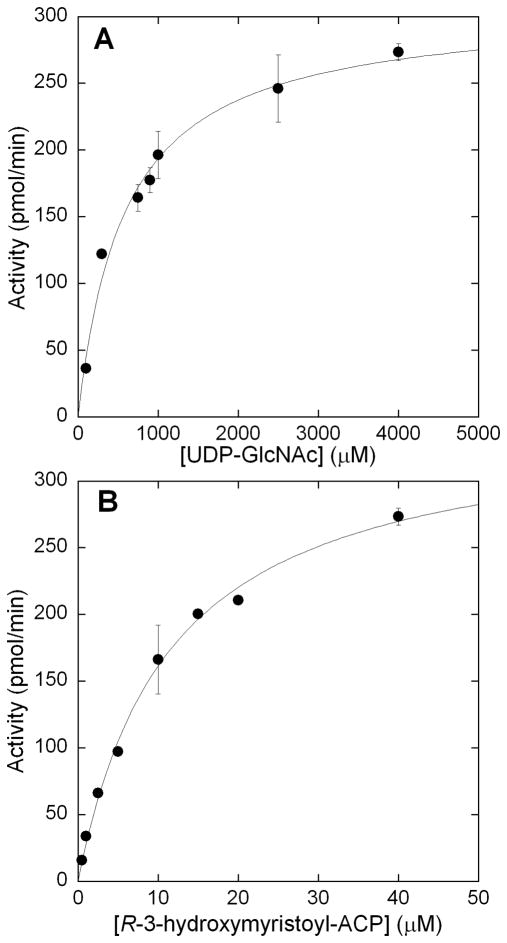
The steady state kinetic parameters of LpxA were analyzed to determine the utility of the assay. The Michaelis-Mentin constant for UDP-GlcNAc was established by holding acyl-ACP constant at 40 μM and varying UDP-GlcNAc between concentrations of 150 μM – 4.0 mM (Figure 5A). The Km was found to be 600 ± 80 μM, which was in good accordance with the previously published values of 100–800 μM [10, 18]. Next the Km for acyl-ACP was established by holding UDP-GlcNAc constant at 4.0 mM concentration, while varying acyl-ACP between 1–40 μM and plotting the initial velocity versus acyl-ACP concentration (Figure 5B). The Km of acyl-ACP was determined to be 10 ± 1 μM. Estimates of acyl-ACP Km using the radioactivity assay ranged from 1.5–5 μM [10, 31]. However, LpxA has an unfavorable equilibrium constant in the forward direction (Keq = 0.01), and this has made it difficult to precisely measure the conversion of radiolabeled UDP-GlcNAc to acylated product under high nucleotide and low acyl-ACP concentrations [18]. With our current assay ThioGlo is able to sequester the holo-ACP product, thus mitigating the unfavorable Keq and allowing accurate determination of the Km of acyl-ACP.
Figure 5.
Saturation kinetics of LpxA for A) UDP-GlcNAc and B) R-3-hydroxymyristoyl-ACP. A) UDP-GlcNAc was varied between 150–4000 μM, while holding acyl-ACP constant at 40 μM. B) R-3-hydroxymyristoyl-ACP was varied over a concentration range of 1–40 μM, while holding UDP-GlcNAc constant at 4000 μM. Assays were run in triplicate.
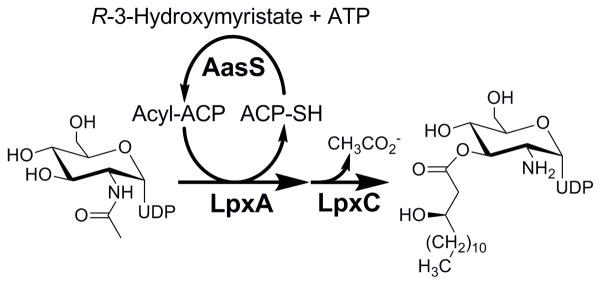
In order to assay LpxD the nucleotide substrate, UDP-3-O-(R-3-hydroxymyristoyl)-GlcN needed to be synthesized. The current method for obtaining LpxD substrate is through an LpxA and LpxC coupled enzymatic process, utilizing acyl-ACP and UDP-GlcNAc as substrates [13, 17]. To decrease the amount of acyl-ACP necessary for this coupled assay, the soluble V. harveyi acyl-ACP synthetase was utilized to produce and continuously regenerate R-3-hydroxymyristoyl-ACP in situ from holo-ACP within the LpxA/LpxC coupled reaction (Figure 6).
Figure 6.
Schematic representation of UDP-3-O-(R-3-hydroxymyristoyl)-GlcN enzymatic preparation. The LpxA catalyzed formation of UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc is coupled directly to LpxC to form UDP-3-O-(R-3-hydroxymyristoyl)-GlcN, while AasS catalyzes the acylation of holo-ACP to regenerate acyl-ACP.
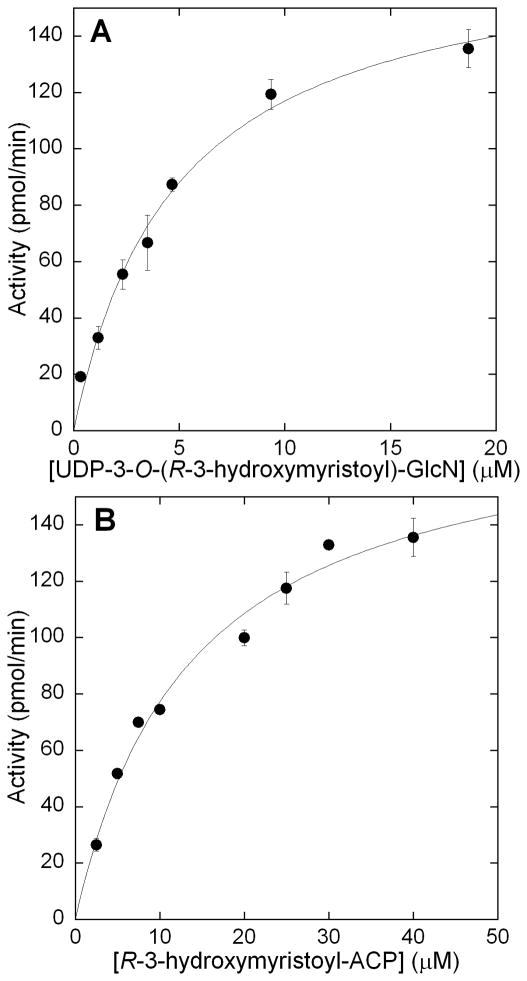
Upon obtaining UDP-3-O-(R-3-hydroxymristoyl)-GlcN, the kinetic parameters of His6-LpxD were next established. It should be noted that previous literature precedent has established there is no difference in activity between His6-LpxD and native LpxD [17]. Acyl-ACP was varied between 2.5–40 μM while holding UDP-3-O-(R-3-hydroxymyristoyl)-GlcN at 19 μM to determine the Km of acyl-ACP (Figure 7B). The Km was 12 ± 2 μM which is in good accordance with the previously published literature value of 3 μM [17]. To determine the Km of UDP-3-O(R-3-hydroxymyristoyl)-GlcN, the substrate was varied between 0.35–19 μM, while holding acyl-ACP constant at 40 μM (Figure 7A). Again the Km established, 4 ± 0.5 μM, correlated well to the reported value of 2.5 μM. The LpxD specific activity calculated from the Vmax and protein concentration used in the steady state analysis was determined to be 5.0 μmol/min/mg (kcat = 3.0 s−1). Previously determined specific activity for LpxD was 8.9 μmol/min/mg (30 °C) [17]. These results demonstrated the reproducibility and robustness for the ThioGlo assay for LpxD.
Figure 7.
Saturation kinetics of LpxD for A) UDP-3-O-(R-3-hydroxymyristoyl)-GlcN and B) R-3-hydroxymyristoyl-ACP. A) UDP-3-O-(R-3-hydroxymyristoyl)-GlcN was varied over a range of 0.35–19 μM while holding acyl-ACP constant at 40 μM. B) acyl-ACP was varied over a concentration range of 1–40 μM while holding UDP-3-O-(R-3-hydroxymyristoyl)-GlcN constant at 19 μM. Assays were run in triplicate.
We have developed the first non-radioactive kinetic assay for both LpxA and LpxD. The above results demonstrate the utility of the thiol-specific reagent ThioGlo for use in kinetic assays involving turnover of acyl-ACP to holo-ACP. We have established and optimized the conditions essential to monitor turnover, and with these optimized conditions have determined the Michaelis-Mentin constants for both E. coli LpxA and LpxD substrates. These values are in good agreement with previously reported literature values. The assay is fast, continuous, and can be adapted to 384-well plates for high-throughput screening, depending on the readiness of noncommercially available substrates (acyl-ACP and UDP-3-O-(R-3-hydroxymyristoyl)-GlcN). This assay will be instrumental in the discovery and characterization of small-molecule inhibitors of LpxA and LpxD. Beyond LPS biosynthesis, holo-ACP is involved in a multitude of biochemical pathways such as membrane phospholipid biosynthesis [32], fatty acid biosynthesis [33], polyketide biosynthesis [34], and quorum sensing [35], to name but a few. Therefore, this assay may find broad utility across various research communities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muhlradt PF, Golecki JR. Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur J Biochem. 1975;51:343–52. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 4.Meredith TC, Aggarwal P, Mamat U, Lindner B, Woodard RW. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem Biol. 2006;1:33–42. doi: 10.1021/cb0500015. [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 6.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CR. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry. 2005;44:16574–83. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaara M. Lipid A: target for antibacterial drugs. Science. 1996;274:939–40. doi: 10.1126/science.274.5289.939. [DOI] [PubMed] [Google Scholar]
- 8.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CR. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–2. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 9.Anderson MS, Bulawa CE, Raetz CR. The biosynthesis of gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J Biol Chem. 1985;260:15536–41. [PubMed] [Google Scholar]
- 10.Anderson MS, Bull HG, Galloway SM, Kelly TM, Mohan S, Radika K, Raetz CR. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J Biol Chem. 1993;268:19858–65. [PubMed] [Google Scholar]
- 11.Jackman JE, Raetz CR, Fierke CA. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry. 1999;38:1902–11. doi: 10.1021/bi982339s. [DOI] [PubMed] [Google Scholar]
- 12.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CR, Hyland SA, Anderson MS. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem. 1995;270:30384–91. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 13.Kelly TM, Stachula SA, Raetz CR, Anderson MS. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–74. [PubMed] [Google Scholar]
- 14.Buetow L, Smith TK, Dawson A, Fyffe S, Hunter WN. Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis. Proc Natl Acad Sci U S A. 2007;104:4321–6. doi: 10.1073/pnas.0606356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 16.Vuorio R, Harkonen T, Tolvanen M, Vaara M. The novel hexapeptide motif found in the acyltransferases LpxA and LpxD of lipid A biosynthesis is conserved in various bacteria. FEBS Lett. 1994;337:289–92. doi: 10.1016/0014-5793(94)80211-4. [DOI] [PubMed] [Google Scholar]
- 17.Bartling CM, Raetz CR. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyckoff TJ, Raetz CR. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J Biol Chem. 1999;274:27047–55. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 19.Majerus PW, Alberts AW, Vagelos PR. Acyl Carrier Protein. Iv. the Identification of 4′-Phosphopantetheine as the Prosthetic Group of the Acyl Carrier Protein. Proc Natl Acad Sci U S A. 1965;53:410–7. doi: 10.1073/pnas.53.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broadwater JA, Fox BG. Spinach holo-acyl carrier protein: overproduction and phosphopantetheinylation in Escherichia coli BL21(DE3), in vitro acylation, and enzymatic desaturation of histidine-tagged isoform I. Protein Expr Purif. 1999;15:314–26. doi: 10.1006/prep.1998.1016. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y, Chan CH, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry. 2006;45:10008–19. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 22.Anderson MS, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem. 1987;262:5159–69. [PubMed] [Google Scholar]
- 23.Vuorio R, Vaara M. Comparison of the phenotypes of the lpxA and lpxD mutants of Escherichia coli. FEMS Microbiol Lett. 1995;134:227–32. doi: 10.1111/j.1574-6968.1995.tb07942.x. [DOI] [PubMed] [Google Scholar]
- 24.Galloway SM, Raetz CR. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem. 1990;265:6394–402. [PubMed] [Google Scholar]
- 25.Bulfer SL, McQuade TJ, Larsen MJ, Trievel RC. Application of a high-throughput fluorescent acetyltransferase assay to identify inhibitors of homocitrate synthase. Anal Biochem. 2011;410:133–40. doi: 10.1016/j.ab.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Ferrance JP, Landers JP, Weber SG. Integration of a precolumn fluorogenic reaction, separation, and detection of reduced glutathione. Anal Chem. 2010;82:7267–73. doi: 10.1021/ac101182r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi Z, Hong B, Mirnics ZK, Tyurina YY, Kagan VE, Liang Y, Schor NF. Bcl-2-mediated potentiation of neocarzinostatin-induced apoptosis: requirement for caspase-3, sulfhydryl groups, and cleavable Bcl-2. Cancer Chemother Pharmacol. 2006;57:357–67. doi: 10.1007/s00280-005-0054-z. [DOI] [PubMed] [Google Scholar]
- 28.Mare S, Penugonda S, Ercal N. High performance liquid chromatography analysis of MESNA (2-mercaptoethane sulfonate) in biological samples using fluorescence detection. Biomed Chromatogr. 2005;19:80–6. doi: 10.1002/bmc.420. [DOI] [PubMed] [Google Scholar]
- 29.Wright SK, Viola RE. Evaluation of methods for the quantitation of cysteines in proteins. Anal Biochem. 1998;265:8–14. doi: 10.1006/abio.1998.2858. [DOI] [PubMed] [Google Scholar]
- 30.Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- 31.Williams AH, Immormino RM, Gewirth DT, Raetz CR. Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. Proc Natl Acad Sci U S A. 2006;103:10877–82. doi: 10.1073/pnas.0604465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YM, Rock CO. Thematic review series: Glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–74. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell JW, Cronan JE., Jr Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol. 2001;55:305–32. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 34.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 35.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci U S A. 1999;96:4360–5. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]