Abstract
The immunostimulatory cytokine interleukin-2 (IL-2) is a growth factor for a wide range of leukocytes, including T cells and natural killer (NK) cells1–3. Considerable effort has been invested using IL-2 as a therapeutic agent for a variety of immune disorders ranging from AIDS to cancer. However, adverse effects have limited its use in the clinic. On activated T cells, IL-2 signals through a quaternary “high affinity” receptor complex consisting of IL-2, IL-2Rα (termed CD25), IL-2Rβ, and γc4–8. Naïve T cells express only a low density of IL-2Rβ and γc, and are therefore relatively insensitive to IL-2, but acquire sensitivity after CD25 expression, which captures the cytokine and presents it to IL-2Rβ andγc. Here, using in vitro evolution, we eliminated IL-2’s functional requirement for CD25 expression by engineering an IL-2 “superkine” (termed super-2) with increased binding affinity for IL-2Rβ. Crystal structures of super-2 in free and receptor-bound forms showed that the evolved mutations are principally in the core of the cytokine, and molecular dynamics simulations indicated that the evolved mutations stabilized IL-2, including a flexible helix in the IL-2Rβ binding site, into an optimized receptor-binding conformation resembling that when bound to CD25. The evolved mutations in super-2 recapitulated the functional role of CD25 by eliciting potent phosphorylation of STAT5 and vigorous proliferation T cells irrespective of CD25 expression. Compared to IL-2, super-2 induced superior expansion of cytotoxic T cells, leading to improved anti-tumor responses in vivo, and elicited proportionally less expansion of T regulatory cells and reduced pulmonary edema. Collectively, we show that in vitro evolution has mimicked the functional role of CD25 in enhancing IL-2 potency and regulating target cell specificity, which has implications for immunotherapy.
Results
Affinity maturation of IL-2 for IL-2Rβ
In order to engineer a CD25-independent version of IL-2, we displayed human IL-2 on the surface of yeast as a conjugate to Aga2p, and verified proper receptor binding properties with IL-2R β and γc ectodomain tetramers that were C-terminally biotinylated and coupled to phycoerythrin-conjugated streptavidin for use as a staining and sorting reagent9,10. Yeast-displayed IL-2 bound to γc in the presence of IL-2Rβ, recapitulating the cooperative assembly of the heterodimeric receptor complex as seen with soluble IL-2 (Fig. 1a, Supplementary Fig. 1). We proceeded to carry out two generations of in vitro evolution (Fig. 1b, Supplementary Fig. 2). Our first generation in vitro evolution strategy was to create an error-prone PCR library of the entire IL-2 coding sequence (Supplementary Fig. 2), which resulted in selection of a predominant IL-2 variant containing an L85V mutation (Fig 1c, Supplementary Fig. 3).
Figure 1. In vitro evolution of human IL-2 variants with high affinity for IL-2Rβ.
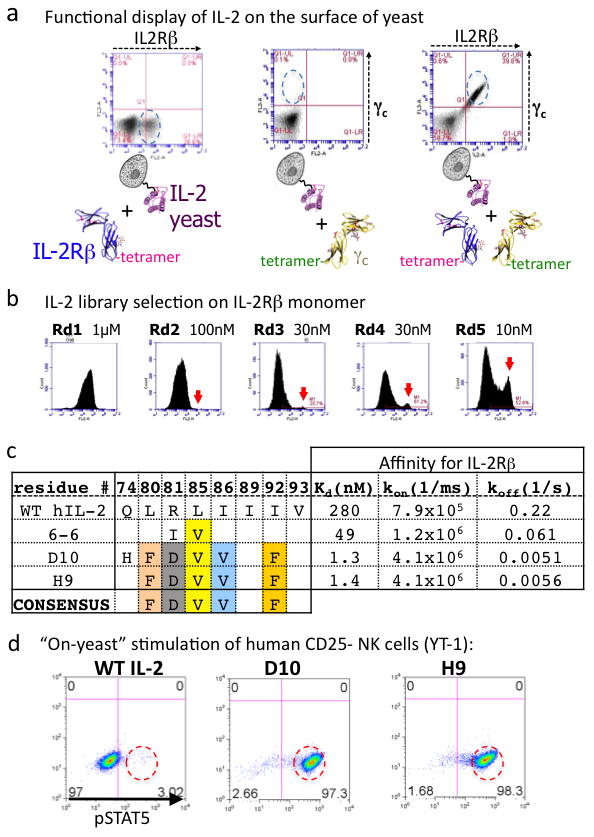
a, IL-2 displayed on yeast recapitulates cooperative receptor-binding activity. As measured by flow cytometry, IL-2 binds weakly to IL-2Rβ (left panel), undetectably to γc (middle panel), and cooperatively forms the IL-2Rβ/γc heterodimer (right panel). b, Enrichment of IL-2 variants on yeast by selection with progressively lower concentrations of IL-2Rβ. Arrows indicate an emerging population of high affinity IL-2Rβ binders (see also Supplementary Fig. 2). c, Sequences and affinities for IL-2Rβ of selected mutants from the first (mutant 6-6) and second (mutants D10 and H9) generation libraries (see Supplementary Fig. 3 for an extended table). d, On-yeast stimulation of YT-1 cells (human NK cell line) by wild-type (WT) IL-2-yeast and high affinity variants (super-2s) (see also Supplementary Fig. 4).
From inspection of the wild-type (WT) IL-2 structure, we were surprised to find that position 85 was not a direct IL-2Rβ contact residue, but rather resided on the internal face of the IL-2 C-helix, within the hydrophobic core of the cytokine (Fig. 2a). Thus, we surmised that L85V may affect the structure of helix C in a way that enhances binding to IL-2Rβ. Therefore, we carried out a second generation selection where we made a biased library that contained F/I/L/V at amino acids L80, L85, I86, I89, I92, and V93, which are contained within the hydrophobic core and linker region on helix C (Fig. 1b,c). To rapidly select the most active variants, we used the yeast-displayed cytokines themselves to stimulate STAT5 phosphorylation in the human NK cell line YT-1 by coincubation at varying yeast:YT-1 cell ratios (Fig 1d, Supplementary Fig. 4). Several clones stimulated substantially more STAT5 phosphorylation at lower yeast:cell ratios than yeast-displayed WT IL-2 (Fig 1d, Supplementary Fig. 4). Sequencing of a selected panel of high-affinity IL-2 clones revealed a consensus set of mutations L80F/R81D/L85V/I86V/I92F (Fig 1c, Supplementary Fig. 3).
Figure 2. Basis of affinity enhancement for IL-2Rβ from structural and molecular dynamics characterization of D10 super-2.
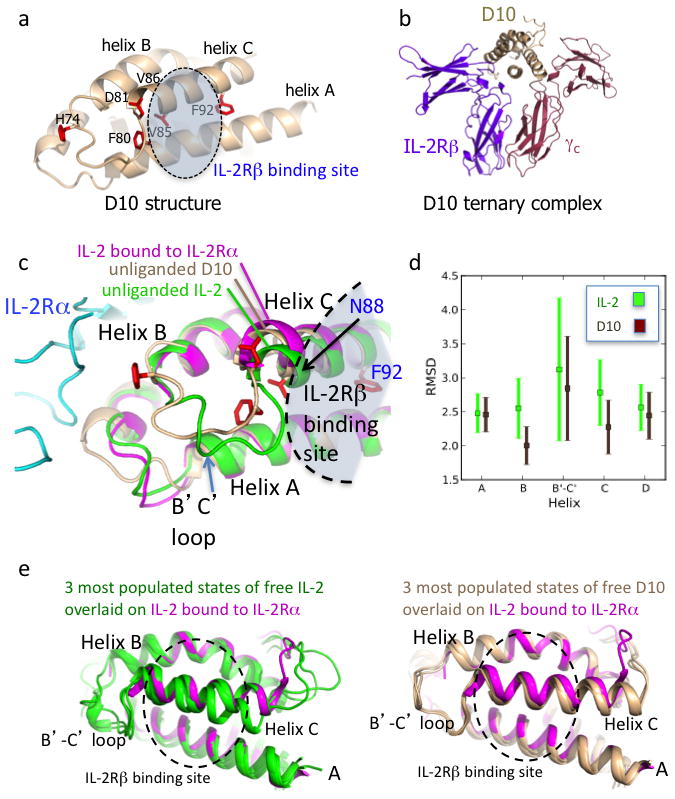
a, Crystal structure of the D10 super-2 at 3.1 Å with mutated residues in red (see also Supplementary Table 1 and Supplementary Fig. 7a). b, D10 in complex with human IL-2Rβ and γc preserves the WT receptor dimer geometry (see also Supplementary Fig. 7b). c, The unliganded D10 super-2 helix C (brown), moves towards its hydrophobic core compared to unliganded WT IL-2 (green, PDBID 3INK). This helix C position is more similar to that of helix C in IL-2 bound to IL-2Rα (purple, PDBID 1Z92) (see also Supplementary Fig. 8). d, A 40ns MD simulation shows a reduction of the average RMSD for the B and C helices, and the B-C loop in D10 versus IL-2 (see also Supplementary Fig. 8c). Error bars represent the standard error of the RMSD. e, Helix C in IL-2 (green, left panel) drifts during the MD simulation more than super-2 D10 (brown, right panel) when compared to IL-2 bound to IL-2Rα (purple).
Structure and comparison with WT IL-2
We expressed recombinant forms of several first and second-generation IL-2 clones in order to measure their binding affinities and kinetics for IL-2Rβ by surface plasmon resonance (SPR) (Fig. 1c, Supplementary Figs 3 and 5) and isothermal titration calorimetry (ITC) (Supplementary Fig. 6). By SPR, the affinity between IL-2 and IL-2Rβ was KD = 280 nM. The IL-2 superkines, termed “super-2s”, clustered into low, medium, and high affinity classes. The highest affinity mutants had KDs of 1.2–1.7 nM (D10, H9). The affinity increases were uniformly manifested in reductions in off-rates (Fig 1c, Supplementary Figs 3 and 5).
In order to understand the structural consequences of the evolved mutations, we crystallized the D10 super-2 (Fig. 2a, Supplementary Fig. 7a, and Supplementary Table 1). In the structure of D10 alone, five of the six mutations clustered on the B-C loop and within the C-helix core, in positions that did not contact IL-2Rβ. Notably, the B-C-helix linker region was ordered in the electron density map (Supplementary Figs 7a and 8a), compared to other IL-2 structures where this region is often disordered (Supplementary Fig. 8a). Collectively, the F80, V85, and V86 substitutions appeared to collapse into a hydrophobic cluster that stabilized the loop by pinning the C-helix into the core of the molecule. Only one of the five consensus mutations, I92F, was at a position that contacted IL-2Rβ in the receptor complex (Fig. 2a), but it was deeply inserted between the C and A helices, contributing only an additional 10Å2 of molecular surface buried by IL-2Rβ in the complex compared to Ile92. We also determined a low-resolution (3.8 Å) structure of the D10 ternary receptor complex to assess whether the mutations have perturbed the IL-2Rβ/γc receptor dimer geometry compared to the WT IL-2 complex (Fig. 2b, Supplementary Table 1). The overall IL-2Rβ/γc heterodimeric architecture and mode of cytokine/IL-2Rβ contact in the D10 ternary complex were essentially identical to the previously reported IL-2 quaternary assembly (RMSD = 0.43 Å) (Supplementary Fig. 7b).
Previously, we found that the C-helix of IL-2 appears to undergo subtle repositioning upon binding to IL-2Rα11 (Fig. 2c, Supplementary Fig. 8a). Inspection of three WT unliganded IL-2 structures revealed conformational variability in the C-helix position, consistent with higher crystallographic B-factors in this helix relative to the rest of the molecule (Supplementary Fig. 8b). We compared the structure of our D10 super-2 to that of an unliganded structure of IL-2, and IL-2 in the receptor complexes. We found that the C-helix in D10 was more similar to that seen in the two receptor-bound conformations of IL-2 than the free forms, having undergone a relatively small shift towards the helical core as a consequence of the stabilizing mutations (Fig. 2c).
We used molecular dynamics (MD) simulations of IL-2 and D10 to further interrogate the mechanism responsible for higher binding affinity to IL-2Rβ by super-2 (Fig. 2d,e). We constructed an atomically detailed Markov state model (MSM) in order to directly probe the relative conformational flexibility of IL-2 versus D10. Analysis of the MSM clearly demonstrated that D10 was more stable than IL-2, and that IL-2 visited nearly twice as many clusters as D10. For example, D10’s most populated state had an equilibrium probability of ~0.20, compared to ~0.05 for IL-2, demonstrating that D10’s equilibrium population was far more localized than IL-2. Helix B, the B-C loop, and helix C appeared rigidified in D10 compared to IL-2 as evidenced by reduced RMSD from the starting conformations (Fig. 2d, Supplementary Movies 1, 2). F92 appeared to act as a molecular wedge between helix C and helix A, stabilizing the more C-terminal end of the helix (Fig. 2a). We also simulated both D10 and IL-2 starting in a receptor-bound-like structure and monitored the divergence in RMSD of the B-C loop and helix C from the actual receptor-bound structure. IL-2 (Fig. 2e-left, Supplementary Fig. 8c) quickly “wandered” from the receptor conformation and experienced drastic fluctuations compared to D10 (Fig. 2e-right, Supplementary Fig. 8c, Supplementary Movies 1, 2). Based on these observations, we propose a mechanism whereby the reduced flexibility of helix C in super-2, as a result of its improved core packing with helix B, results in a superior receptor-binding poise that increases its affinity for IL-2Rβ, and consequently mimics a functional role of CD25. The structural and MD results suggest that the evolved mutations in super-2 cause a conformational stabilization of the cytokine, that reduces the energetic penalties for binding to IL-2Rβ.
Stimulation of NK cells
We asked if the super-2s demonstrated signaling potencies on cells in accordance with their IL-2Rβ binding affinities, and whether their activities depended on cell surface expression of CD25. We determined the dose-response relationships of WT IL-2 versus the super-2s 6-6, D10, and H9 on both CD25− and CD25+ human YT-1 NK cells by assaying STAT5 phosphorylation with flow cytometry (Fig. 3a–d, Supplementary Fig. 9). On CD25− YT-1 cells, the EC50 of H9 and D10 were decreased over 10-fold (EC50 = 2.5 and 1.8 ng/mL, respectively) compared to IL-2 (EC50 = 39 ng/mL), with the 6-6 mutein yielding an EC50 intermediate between IL-2 and H9/D10 (EC50 = 15 ng/mL), consistent with the improved affinity of the super-2s for IL-2Rβ (Fig. 3a). On CD25+ YT-1 cells, the EC50 of IL-2 decreased over 50-fold relative to CD25− YT-1 cells, from 39 to 0.66 ng/mL (Fig. 3b). In contrast, the EC50 of H9 and D10 improved only modestly in the presence of CD25 (EC50 of 0.47 and 0.52 compared to 2.5 and 1.8 ng/mL, respectively) (Fig. 3b).
Figure 3. Functional properties of super-2 on human NK cells in vitro.
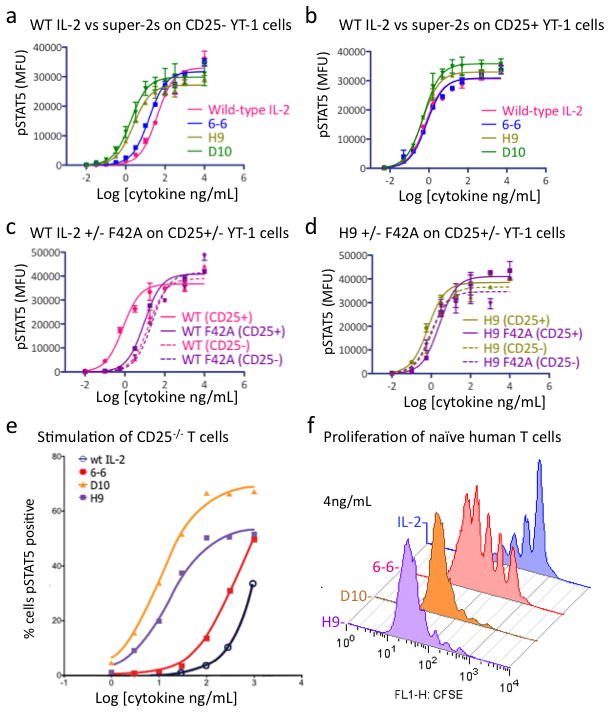
Dose-response curves of STAT5 phosphorylation on (a) CD25− and (b) CD25+ YT-1 cells with WT IL-2 and three super-2s. c, Dose-response curves of STAT5 phosphorylation on CD25+ (solid curve) and CD25−(dashed curve) YT-1 cells with WT IL-2 (pink curves) and IL-2-F42A mutation (purple curves). d, Dose-response curves of STAT5 phosphorylation on CD25+ (solid curve) and CD25−(dashed curve) YT-1 cells with H9 (green curves) and H9-F42A mutation (purple curves). e, Super-2s have superior potency over IL-2 on T cells derived from CD25−/− mice as demonstrated by dose-response curves for STAT5 phosphorylation on T cells demonstrating that potency correlates with IL-2Rβ affinity (see also Supplementary Fig. 10). f, Proliferation of human naïve CD4+ T cell (CD25low) reveals similar potency profiles as seen with CD25−/− T cells. Proliferation was measured by CFSE dilution on day 5 (see also Supplementary Fig. 10). Error bars in a–d represent SEM of mean fluorescence units for each sample at the indicated cytokine concentration.
We sought to further probe the CD25-independence of the super-2s by taking advantage of a previously characterized mutation in IL-2, Phe42 to Ala (F42A), which showed reduced binding to CD25 by ~220-fold for H9 (KD 6.6 nM versus 1.4 μM) and ~120-fold for IL-2 (KD 6.6 nM versus 0.8 μM) (Supplementary Fig. 10)12,13. The F42A mutation is an alternative diagnostic probe of the relative CD25 dependence of IL-2 and super-2. The F42A mutation right-shifted the dose-response curve of WT IL-2 on CD25+ cells by ~1 log, but had no effect on CD25− cells (Fig. 3c). In contrast, H9 was far less sensitive to the F42A mutation, with the dose-response curves of H9 versus H9 F42A being very similar on both CD25+ and CD25− cells (Fig. 3d).
Activity of super-2 on T cells
We assessed the activity of several super-2s on T cells that were either deficient in, or expressed CD25. For the former experiment, CD4+ T cells were isolated from CD25-knockout mice, followed by stimulation by either WT IL-2 or six super-2s and assaying for STAT5 phosphorylation at a range of cytokine concentrations (Fig 3e, Supplementary Fig. 11). CD25−/− CD4+ T cells responded poorly to exogenous WT IL-2 stimulation, but super-2s induced STAT5 phosphorylation in these cells proportional to their affinity for IL-2Rβ.
The principle functional effect of IL-2 is to promote T cell proliferation, particularly for naïve T cells. Human naïve CD4+ T cells were isolated and left either unstimulated or stimulated with plate-bound anti-CD3 antibody with or without the different IL-2 variants (Fig 3f, Supplementary Fig. 12). Increased proliferation effects on naïve human T cells correlated with increased affinity for IL-2Rβ and STAT5 phosphorylation shown earlier, as the rank order of potency was D10 = H9 > 6-6 > WT IL-2 (see Supplementary Fig. 12 for the complete titration).
We next tested the IL-2 variants for their ability to induce STAT5 phosphorylation on experienced human CD4+ T cells (Supplementary Fig. 13), which highly express the trimeric IL-2Rαβγ complex. Human CD4+ T cells were in vitro activated by T cell receptor (TCR) stimulation and rested to generate “experienced” human CD4+ CD25+ T cells. As for the CD25+ YT-1 cells, we observed a much smaller difference between IL-2 and the super-2s.
In vivo properties of super-2
We assessed the potency of the super-2 H9 on expansion of CD25low versus CD25high T cells, in comparison to WT IL-2 and IL-2/anti-IL-2 monoclonal antibody (mAb) complexes, which have been shown to exert reduced pulmonary edema yet very potent anti-tumor responses in vivo14–16. On antigen-experienced (memory-phenotype, MP) CD8+ T cells, expressing only low levels of CD25 but high levels of IL-2Rβγ, H9 induced more than three times the rate of proliferation and expansion as WT IL-2 (Fig 4a, Supplementary Fig. 14a). However, on CD4+ CD25high T regulatory (Treg) cells, we found that the CD25-competent WT IL-2 and H9 achieved comparable maximal expansion, demonstrating again that expression of CD25 mitigates the difference between super-2 and WT IL-2 (Fig. 4a, S14b). Thus, the H9 exhibits the desired property that it shows enhanced stimulation towards CD8+ T cells, but not towards Treg cells compared to WT IL-2.
Figure 4. Functional and anti-tumor activities of super-2 in vivo.
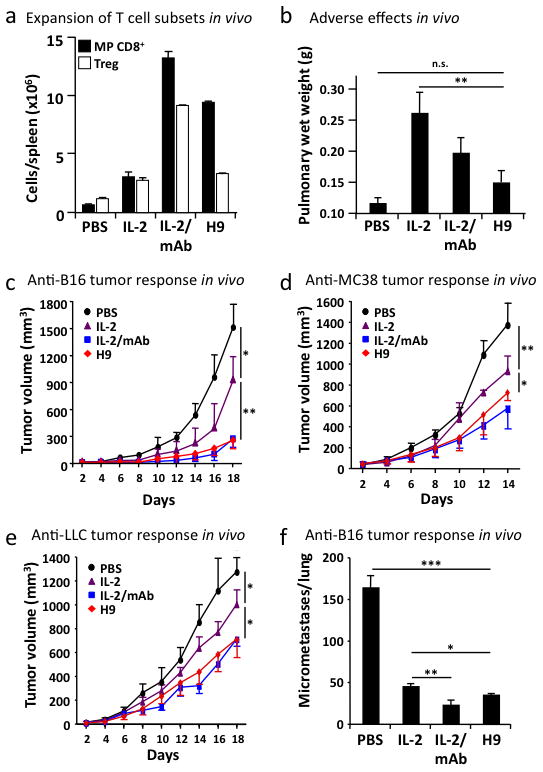
a, Total cell counts of host CD3+ CD8+ CD44high memory-phenotype T cells (MP CD8+, closed bars), and host CD3+ CD4+ CD25high T cells (Treg, open bars) was determined in the spleens of mice receiving either PBS, 20 μg IL-2, 1.5 μg IL-2/anti-IL-2 mAb complexes (IL-2/mAb), or 20 μg H9 (see also Supplementary Fig. 14). b, Pulmonary edema (pulmonary wet weight) served to assess adverse toxic effects following IL-2 treatment, and was determined by weighing lungs before and after drying. c–f, C57BL/6 mice (n=3–4 mice/group) were injected either subcutaneously with 106 B16F10 melanoma cells (B16, c), 2.5× 106 murine colon carcinoma 38 (MC38, d), 106 Lewis lung carcinoma (LLC, e), or mice received 3× 105 B16F10 melanoma cells intravenously (B16, f), followed by daily injections of either PBS, 20 μg IL-2, 1.5 μg IL-2/mAb complexes, or 20 μg H9 for five days once subcutaneous tumor nodules became visible and palpable or from day three on for intravenously-injected tumors (see also Supplementary Fig. 15). Shown is mean tumor volume in mm3 (+/− SD) vs. time upon tumor inoculation. Error bars represent SEM. P values refer to comparisons of WT with the other treatment modalities. *, p<0.05; **, p<0.01; ***, p<0.001.
As previously reported, administration of high-dose WT IL-2 for five days induced substantial pulmonary edema, which is known to be CD25-dependent15 (Fig. 4b). Although significantly more stimulatory for cytotoxic CD8+ T cells (Fig. 4a), H9 super-2 caused substantially less pulmonary edema (Fig. 4b).
Given the more favorable properties of H9 in comparison to IL-2, we assessed its ability to stimulate effector functions of cytotoxic T cells in four different tumor models in vivo, where high-dose IL-2 administration has been previously shown to result in tumor regression15,17. To this end, C57BL/6 mice were injected subcutaneously with B16F10 melanoma cells, followed by administration of either high-dose IL-2, IL-2/anti-IL-2 mAb complexes, or H9 super-2, once tumor nodules became visible and palpable. PBS-treated control mice rapidly developed large subcutaneous tumors reaching a volume of about 1500 mm3 on day 18 (Fig. 4c). As previously shown, high-dose IL-2 treatment was able to delay tumor growth by as much as 39% on day 18 (p<0.05), while IL-2/anti-IL-2 mAb complexes exerted very effective tumor control, reducing tumor growth by >80% on day 18 (p<0.005) (Fig. 4c). Significantly, similar to IL-2/anti-IL-2 mAb complexes, mice receiving high-dose H9 showed a dramatic decrease of tumor load on day 18, which was reduced by >80% compared to PBS (p<0.005) and by >70% compared to WT IL-2 (p<0.005) (Fig. 4c). Similar results were obtained using three other tumor models, including murine colon carcinoma and Lewis lung carcinoma injected subcutaneously (Fig. 4d,e) and B16F10 cells administered intravenously (Fig. 4f, Supplementary Fig. 15). Collectively, these data show that the H9 super-2 is very effective against different tumors, albeit inducing reduced pulmonary edema.
The practical implications are that this conformational nuance in IL-2 can be exploited for therapy. Super-2 robustly activates cytotoxic CD8+ T cells and NK cells for potent anti-tumor immune responses, yet it elicits minimal toxicity, suggesting that super-2 could warrant reconsideration for clinical applications of IL-2.
Methods Summary
Yeast display and selection of IL-2
Error-prone and site-directed libraries of IL-2 were displayed on yeast as previously described18 and stained with biotinylated IL-2Rβ at successively decreasing concentrations. Staining was detected with streptavidin-PE and yeast were separated using paramagnetic anti-PE microbeads (Miltenyi; MACS). Enrichment of positively-staining yeast was monitored by flow-cytometry.
Protein expression, purification, and structural determination
Human IL-2 variants and the ectodomains of IL-2Rβ, γc, and CD25 were expressed in Hi5 cells and purified as previously described11. Proteins were concentrated to 8–20 mg/mL and crystallized by vapor diffusion in sitting drops. Diffraction studies were performed at the Stanford Synchrotron Radiation Laboratory and the Advanced Light Source. Crystal structures were solved by molecular replacement with PHASER19 and refined using PHENIX20 and COOT21.
Mice
C57BL/6 and Thy1.1-congenic mice on a C57BL/6 background were maintained under specific pathogen-free conditions and used at 3–6 months of age. Experiments were performed in accordance with the Swiss Federal Veterinary Office guidelines and approved by the Cantonal Veterinary Office.
In vivo T cell proliferation
2–3×106 CFSE-labeled CD44high CD8+ T cells from Thy1.1-congenic mice were injected intravenously to Thy1.2-congenic animals. Mice received daily intraperitoneal (i.p.) injections of PBS, 20 μg IL-2, 1.5 μg IL-2/anti-IL-2 mAb complexes, or 20 μg H9 for 5 days. On the sixth day, spleens were removed and analyzed by flow cytometry.
Toxicity
Pulmonary edema was determined by measurement of pulmonary wet weight on the sixth day after five daily i.p. injections of PBS, 20 μg IL-2, 1.5 μg IL-2/anti-IL-2 mAb complexes, or 20 μg H9 as previously described15.
Tumor models
B16F10 melanoma cells, Lewis lung carcinoma, or murine colon carcinoma 38 cells were injected into mice (3–4 mice/group), as previously reported15,17. Treatment consisted of five daily i.p. injections of PBS, 20 μg IL-2, 1.5 μg IL-2/anti-IL-2 mAb complexes, or 20 μg H9.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge W. Leonard, R. Levy, and R. Schwendener for reagents and discussion. This work was supported by NIH-RO1AI51321 (to K.C.G.), PP00P3-128421 from the Swiss National Science Foundation and KFS-02672-08-2010 from the Swiss Cancer League (both to O.B.), NIH R01-GM062868 (to V.P.), MRI-R2 [This award is funded under the American Recovery and Reinvestment Act of 2009 (Public Law 111-5)] (to V.P.), NIH-AR050942 (to J.T.L.), NIH U01 DK078123 (to C.G.F.), and NIH U19 AI 082719 (to C.G.F.). A.M.R. was supported by the Stanford Medical Scientist Training Program (NIH- GM07365). K.C.G. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions A.L. performed in vitro evolution and contributed to preparation of manuscript. D.L.B. produced recombinant proteins, determined crystal structures, and carried out surface plasmon resonance analysis. A.M.R. carried out cellular and signaling assays, biophysical measurements, and contributed to preparation of the manuscript. C.K. carried out in vivo experiments, analyzed data, and contributed to preparation of the manuscript, M.E.R. carried out in vivo experiments in mice. I. M. analyzed cell signaling data. G.R.B., P.N., and V.S.P. carried out and analyzed molecular dynamics simulations. J.T.L., L.S., and C.F.G. performed and analyzed T cell signaling experiments. O.B. designed and supervised in vivo experiments, analyzed data and contributed to preparation of manuscript. K.C.G conceived of project, analyzed data, supervised execution of project, and prepared the manuscript.
Author Information Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3QAZ and 3QB1. K.C.G., A.L., and A.R. declare competing financial interests due to submission of a pending patent application describing super-2. O.B. declares competing financial interests due to being a shareholder of Nascent Biologics Inc..
References
- 1.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9 (7):480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith KA. Interleukin-2: inception impact, and implications. Science. 1988;240 (4856):1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 3.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6 (8):595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 4.Cosman D, et al. Cloning, sequence and expression of human interleukin-2 receptor. Nature. 1984;312 (5996):768–771. doi: 10.1038/312768a0. [DOI] [PubMed] [Google Scholar]
- 5.Leonard WJ, et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature. 1984;311 (5987):626–631. doi: 10.1038/311626a0. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido T, et al. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature. 1984;311 (5987):631–635. doi: 10.1038/311631a0. [DOI] [PubMed] [Google Scholar]
- 7.Hatakeyama M, et al. Interleukin-2 receptor beta chain gene: generation of three receptor forms by cloned human alpha and beta chain cDNA’s. Science. 1989;244 (4904):551–556. doi: 10.1126/science.2785715. [DOI] [PubMed] [Google Scholar]
- 8.Takeshita T, et al. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257 (5068):379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 9.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15 (6):553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 10.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1 (2):755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310 (5751):1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 12.Mott HR, et al. The solution structure of the F42A mutant of human interleukin 2. J Mol Biol. 1995;247 (5):979–994. doi: 10.1006/jmbi.1994.0194. [DOI] [PubMed] [Google Scholar]
- 13.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci U S A. 2006;103 (42):15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311 (5769):1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 15.Krieg C, Letourneau S, Pantaleo G, Boyman O. Improved IL-2 immunotherapy by selective stimulation of IL-2 receptors on lymphocytes and endothelial cells. Proc Natl Acad Sci U S A. 2010;107 (26):11906–11911. doi: 10.1073/pnas.1002569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letourneau S, et al. IL-2/anti-IL-2 antibody complexes show strong biological activity by avoiding interaction with IL-2 receptor alpha subunit CD25. Proc Natl Acad Sci U S A. 107(5):2171–2176. doi: 10.1073/pnas.0909384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg SA, Mule JJ, Spiess PJ, Reichert CM, Schwarz SL. Regression of established pulmonary metastases and subcutaneous tumor mediated by the systemic administration of high-dose recombinant interleukin 2. J Exp Med. 1985;161 (5):1169–1188. doi: 10.1084/jem.161.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Interleukin 2 (IL-2) variants engineered for increased IL-2 receptor alpha-subunit affinity exhibit increased potency arising from a cell surface ligand reservoir effect. Mol Pharmacol. 2004;66 (4):864–869. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40 (Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58 (Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60 (Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


