Abstract
HGF/c-Met signaling plays a pivotal role in hepatocyte survival and tissue remodeling during liver regeneration. HGF treatment accelerates resolution of fibrosis in experimental animal models. Here, we utilized Metfl/fl;Alb-Cre+/− conditional knockout mice and a carbon tetrachloride(CCl4)-induced liver fibrosis model to formally address the role of c-Met signaling in hepatocytes in the context of chronic tissue injury. Histological changes during injury (4 weeks) and healing phase (4 weeks) were monitored by immunohistochemistry; expression levels of selected key fibrotic molecules were evaluated by western blotting, and time-dependent global transcriptomic changes were examined using a microarray platform. Loss of hepatocyte c-Met signaling altered hepatic microenvironment and aggravated hepatic fibrogenesis. Greater liver damage was associated with decreased hepatocyte proliferation, excessive stellate cell activation and rapid dystrophic calcification of necrotic areas. Global transcriptome analysis revealed a broad impact of c-Met on critical signaling pathways associated with fibrosis. Loss of hepatocyte c-Met caused a strong deregulation of chemotactic and inflammatory signaling (MCP-1, RANTES, Cxcl10) in addition to modulation of genes involved in reorganization of the cytoskeletal network (Actb, Tuba1a, Tuba8), intercellular communications and adhesion (Adam8, Icam1, Itgb2), control of cell proliferation (Ccng2, Csnk2a, Cdc6, cdk10), DNA damage and stress response (Rad9, Rad52, Ercc4, Gsta1 and 2, Jun). Our study demonstrates that deletion of c-Met receptor in hepatocytes results in pronounced changes in hepatic metabolism and microenvironment, and establishes an essential role for c-Met in maintaining the structural integrity and adaptive plasticity of the liver under adverse conditions.
Keywords: c-met, fibrosis, microarray, CCl4
1. Introduction
Liver fibrosis occurs as a consequence of most chronic liver diseases [1]. Progressive accumulation of extracellular matrix (ECM) in response to excessive liver damage destroys tissue architecture and impairs liver function. Most common etiological factors in western countries are HCV, excessive alcohol consumption and development of non-alcoholic fatty liver (NAFLD)/non-alcoholic steato-hepatitis (NASH) [2-3]. These diseases, particularly NASH, affect more than 100 million patients in the USA alone and cause a rising incidence worldwide [4]. Liver fibrosis is generally a slow process [5] influenced by a variety of genetic factors leading to a broad spectrum of individual responses to the same etiological agent [6]. Despite increasing interest and substantial progress in unraveling the contributing genetic factors and associated key pathways (e.g. TGF-β) [7], the mechanisms of fibrogenesis remain poorly understood [8].
HGF/c-Met signaling is of major importance for liver development and regeneration with a mitogenic, motogenic and morphogenic effect on hepatocytes, hepatic progenitor cells and nonparenchymal liver cells [9-10]. HGF/c-Met signaling is also known to be involved in pathogenesis as well as resolution of fibrosis [11]. Administration of HGF has been shown to reverse fibrosis and decrease hepatocyte apoptosis in a rat model of cirrhosis [12]. Additionally, activation of HGF/c-Met signaling reduced levels of TGF-β, one of the key mediators of fibrogenesis, in several experimental models [13]. Besides the protective effects on hepatocytes, HGF administration caused apoptosis of activated hepatic stellate cells (HSC) and increased matrix metalloproteinase (MMP) expression [14-16]. However, despite the well documented protective role of HGF against fibrogenesis, contribution of HGF/c-Met signaling in hepatocytes in a setting of chronic liver damage is less known.
Given the essential role of HGF in liver homeostasis [10, 17], in this study we used a conditional mouse genetics and comprehensive gene expression profiling to define the molecular mechanisms underlying the anti-fibrotic effects of HGF/c-Met signaling in hepatocytes. Our results show that mice with a targeted inactivation of the c-met gene in hepatocytes were more prone to liver damage and to development of fibrosis when challenged with CCl4. The profibrotic effects of c-Met deficiency were mediated by increased expression of numerous inflammatory cytokines and oxidative stress response genes in addition to disruption of structural integrity and lack of compensatory regeneration. These studies support an essential role for HGF/c-Met in fibrogenesis and highlight a broad range of c-Met protective effects in chronic fibrotic liver diseases.
2. Materials and methods
2.1 Animal Studies
Metfl/fl/Alb-Cre+/− and control Metwt/wt;Alb-Cre+/− mice were generated and genotyped as described [10]. Eight to nine week-old female mice received multiple i.p. injections of CCl4(1μl/g) in mineral oil (Sigma) twice a week for a period of 4 weeks (referred to as injury phase) and monitored for another 4 weeks (referred to as healing phase) (n=5 per genotype/time point) (Supplemental Figure S1). Mice were sacrificed each week, 2 days after the last injection. For acute studies, mice were injected once with CCl4 (1μl/g). Blood was collected from orbital venous plexus of 3 mice under isoflurane anesthesia before sacrifice, and the serum levels of albumin and aspartate aminotransferase (AST) were measured by Analytics, Inc. (Gaithersburg, MD). Animals were housed in an AAALAC facility and cared for in accordance with the guidelines from the Animal Care and Use Committee at the National Cancer Institute, NIH.
2.2 Histology, Immunhistochemistry and BrdU Labeling
Livers were fixed in 10% formalin overnight, embedded in paraffin and stained with H&E for routine histopathological examination, or processed for αSMA (Dako (M0851), 1:200), F4/80 (eBioscience (14-4801-81), 1:25) and CD47 (BD (555297), 1:100) immunohistochemistry. For the detection of BrdU incorporation, livers were fixed in alcoholic-formalin for 4–6 hr, and 5-μm paraffin sections were stained with ant-BrdU (1:100; Becton Dickinson). At least 1,000 cells were counted per animal to determine the percentage of BrdUrd-positive cells (labeling index).
2.3 Liver Collagen Content
Collagen content was assessed by morphometric analysis of Sirius red staining of liver sections and by hydroxyproline concentration [18]. The area of positive Sirius red staining was measured using a computerized analysis method. Hydroxyproline content was quantified colorimetrically from 0.1 g liver samples. The extent of liver fibrosis was determined on liver sections stained with Picrosirius red solution using ImageJ sofware. Histological grading of the fibrosis was determined using a scoring system adapted from Scheuer et al [19] (0, none; 1, enlarged, fibrotic central vein tracts; 2, pericentral septa, intact architecture; 3, fibrosis with architectural distortion, no cirrhosis; and 4, probable or definite cirrhosis).
2.4 Western Blotting
Whole cell lysates (100 μg) were prepared from frozen tissues using T-PER Tissue Extraction Buffer (Pierce) containing Complete Protease Inhibitor Cocktail (Roche), separated by SDS-PAGE and transferred onto Invitrolon PVDF (Invitrogen). Membranes were probed with specific antibodies against TGF-β1 (Santa Cruz (sc-146), 1:500), HGF-α (Santa Cruz (sc-1358), 1:200), CTGF (Santa Curz (sc-14939), 1:200), a-SMA (Dako (M0851), 1:200), Cd47 (BD (555297), 1:1000), Nrf2 (Santa Cruz (sc-722), 1:200), Keap1 (Santa Cruz (sc-33569), 1:200), γ-GCS (Santa Cruz (sc-22775), 1:500), p47 (Santa Cruz (sc-7660), 1:500), Nox2 (gp91 phox, Santa Cruz (sc-5826) 1:500), Nox4 (Santa Cruz (sc-21860), 1:200). Immune complexes were detected using the enhanced chemiluminescence system (Pierce).
2.5 Lipid Peroxidation Assay
Lipid peroxidation was assayed by the production of thiobarbituric acid-reactive components (TBARS) using spectrophotometry as described by Buege and Aust [20].
2.6 GSH Determination
GSH and GSSG content was determined by HPLC as described in [21].
2.7 Gene Expression Microarray and Data Analysis
Total RNA was isolated from frozen liver tissue with TRIzol reagent (Invitrogen) or RNeasy Kit (Qiagen) according to the manufacturer’s instructions. Whole liver transcriptomics were analyzed using Illumina Bead Chip Mouse Ref8-v2 arrays containing 25,600 well-annotated RefSeq transcripts, over 19,100 unique genes as described [22]. Four animals of each genotype were used for each time point (1 d, 1 w, 2 w and 3 w). Gene expression values were normalized by quantile normalization across all samples, referenced to the corresponding untreated livers and log2 transformed. The microarray datasets have been deposited to Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo, accession number GSE25583).
Isolated hepatocytes microarrays were performed on Mm_MEEBO-v1p6 containing 37,099 probes printed at the Advanced Technology Center (NCI/NIH) following a dye-swapping design as described [23]. Hepatocytes were isolated 2 days after the second exposure to CCl4 (1 week time point) from three mice for each experiment performed in duplicates. Total RNA pooled from ten B6/129 strain mouse livers was used as universal hybridization reference. Arrays were scanned with a GenePix 4000A scanner (Axon Instruments, Ltd.) to achieve an optimal signal intensity at both channels with <1% saturated spots. The image spots with a diameter of <10 or >300 μm or a signal intensity below background intensity for any of the two fluorescent channels were excluded. After excluding the invalid features, all arrays were normalized to the 50th percentile of the median signal intensity using the mAdb data analysis suite.1. Unsupervised cluster analysis was done with the Cluster and TreeView programs Michael Eisen Laboratory, Lawrence Berkeley National Laboratory and University of California, Berkeley; http://rana.lbl.gov/eisen/)). The 2-factor time course analysis tool of the BRB ArrayTools V3.3.0 software package (Biometric Research Branch, National Cancer Institute) was used for the analysis of time course expression data. All tests are performed at a false discovery rate (FDR) threshold of 0.05. Only genes with a fold change ≥2 were included in the analyses. Selected genes were validated using RT-qPCR (Supplemental Figure 4). Functional classification and network analysis were performed using Ingenuity Pathway Analysis (Ingenuity Systems Inc.) and the GeneGo pathways analysis (Pathway Analysis MetaCore - GeneGo Inc., St. Joseph, MI) tools.
2.8 Statistical Analysis
Statistical analysis was performed using Student’s t-test. P-values ≤0.05 were considered statistically significant. Results are presented as mean ± SD or mean ± SEM as indicated.
3. Results
3.1 c-Met Deletion in Hepatocytes Results in Increased Activity of Stellate Cells and Rapid Progression of Fibrosis
To induce liver fibrosis, we used a model of toxic liver injury with repeated injections of CCl4 and compared the cellular, biochemical and molecular responses in c-Met deficient and control mice. Mice received biweekly injections of CCl4 during 4 weeks and were followed for 4 consecutive weeks (Supplemental Figure S1).
We confirmed that the basal expression of Cyp2e was comparable between genotypes (Supplemental Figure S2). After a challenge with a single dose of CCl4, both the serum AST levels and areas of necrosis increased more rapidly and reached higher levels in mutant livers, although the differences did not reach a statistical significance. c-Met deficient livers were capable of clearing necrotic tissue and to repopulate the affected area albeit after some delay suggesting that compensatory liver regeneration was not significantly affected (Supplemental Figure S2, A-C) [10]. The latter appears to contradict the mitogenic role of HGF during acute liver injury with CCl4 [24-25]. This seeming discrepancy may be attributed to the differences between the employed experimental models. In mice with a targeted disruption of c-Met expressing the Cre-recombinase under the control of Alb-Cre promoter, the gradual c-met gene deletion is taking place after birth and is completed by 6 weeks of age [10, 26-27] causing an adaptive up-regulation of alternative survival and proliferating signaling [21, 23, 28]. However, upon a greater CCl4-induced liver injury, e.g. caused by phenobarbital pre-treatment [10] or chronic toxic liver insult [29], Metfl/fl;Alb-Cre+/- hepatocytes were unable to maintain an adequate level of proliferation. The decrease in proliferation was paralleled by a considerable decrease in serum albumin levels (Figure 1A-D) thus highlighting the non-redundant role of HGF/c-Met signaling for efficient regeneration of the diseased liver.
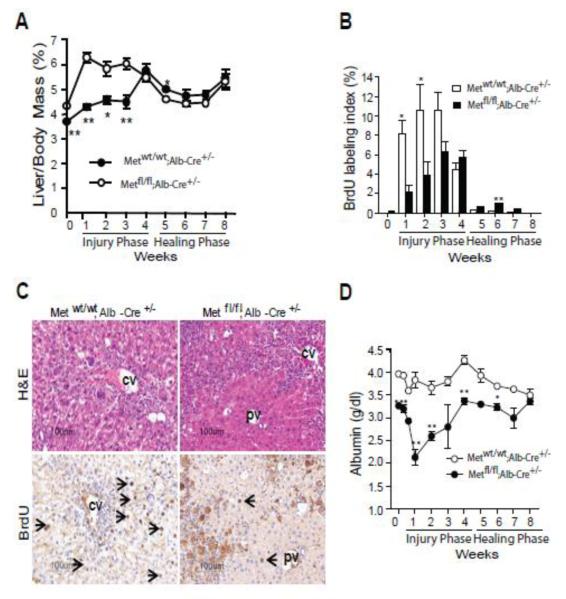
Fig. 1.
Reduced DNA replication and impaired functional performance during CCl4-induced fibrosis in Metfl/fl;Alb-Cre+/− mice. (A) Time course changes in liver-to-body mass ratio. The data are presented as means ± SE (n=5). (B) BrdU labeling index. Mice received 150 μg/g of BrdU 2 h before tissue collection. The labeling index was determined after scoring of at least 1000 hepatocytes per animal and expressed as percent of BrdU-positive cells. The data are presented as means ± SE (n=5). (C) Representative images of BrdU immunostaining after two injections of CCl4. (D) Changes in albumin serum levels. The data are presented as means ± SD (n=4). *P<0.05; **P<0.01; ***P<0.001.
When challenged with repeated CCl4 injections, the mutant livers rapidly became stiff and acquired irregular surface characteristic of fibrotic liver disease (Figure 2A). Already after 2 injections of CCl4, the concentration of hydroxyproline, a major component of collagen, was increased in c-Met deficient livers about 2-fold (P < 0.05) as compared to similarly treated Metwt/wt;Alb-Cre+/− controls (Figure 2B). Quantification of Sirius red staining of collagen fibers and grading of fibrosis using a scoring system adapted from Scheuer et a l [19] confirmed a rapid accumulation of extracellular matrix (ECM) (Figure 2C-E). Histologically, mutant livers showed a quick activation of hepatic stellate cells, a key contributor to fibrosis [30], and expressed considerably higher levels of α-SMA, a marker of activated stellate cells, as shown by immunohistochemistry and Western blotting (Figure 2, F and G). Furthermore, resolution of fibrosis induced by chronic CCl4 exposure occurred slower in knockout mice. The fibrotic septa and uneven liver surface were still visible after four weeks of healing in Metfl/fl;Alb-Cre+/− mice (Figure 2A). In contrast, control mice showed only a rudimentary collagen accumulation which did not result in distortion of lobular architecture and regressed rapidly during the healing phase.
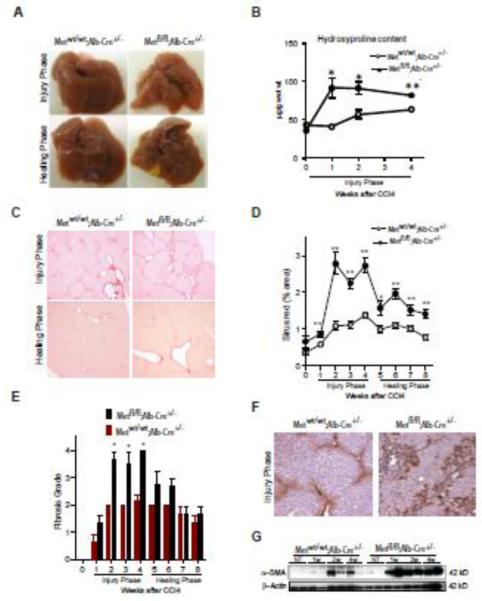
Fig. 2.
Genetic loss of c-Met receptor in hepatocytes results in excessive HSC activation and progression of CCl4-induced fibrosis. (A) Macroscopic appearance of liver. (B) Hydroxyproline content. Values are mean ± SE (n=5). (C) Representative images of Sirius red staining during injury (3 w) and healing phase (8 w) in Metfl/fl;Alb-Cre+/− and Metwt/wt;Alb-Cre+/− mice. (D) Quantification of Sirus red-positive areas using NIH ImageJ software. The data are presented as means ± SE (n=5). (E) Histological grading of fibrosis (adapted from Scheuer et al[19]). The data are presented as means ± SE (n=5). (F) Representative images of α-SMA immunohistochemistry during injury phase (3 w). (G) Western blotting of whole cell lysates prepared from Metfl/fl;Alb-Cre+/− and Metwt/wt;Alb-Cre+/− livers using anti-α-SMA. β-Actin served as a loading control. * P<0.05; **P<0.01.
3.2 Disruption of Hepatic Wound Healing Response in c-Met Deficient Mice
The hallmark feature of CCl4-induced liver fibrosis in c-Met deficient mice was disruption of coordinated hepatic wound-healing response leading to chronic injury. While control livers showed repetitive cycles of hepatic damage and repair, c-Met-deleted livers displayed persistence of centrolobular injury a long time after the first CCl4 administration (Supplemental Figure S3A-G). The repair impediment cannot be explained only by a diminished proliferative response (Figure 2) but rather due to changes in the necrotic milieu affecting the dynamics of healing. Histological markers of unresolved healing in Metfl/fl;Alb-Cre+/− mice were rapid dystrophic calcification of necrotic areas and massive ‘foreign body reaction’ as manifested by von Kossa staining and accumulation of multinucleated giant cells. Multinucleated cells were positive for F4/80, a marker of infiltrating macrophages, and expressed CD47, known to contribute to macrophage fusion thought to increase the capacity to remove or sequester necrotic material (Figure 3, B and C) [31]. Together these results imply that the function of macrophages/Kupffer cells was compromised in c-Met deficient livers which could contribute to the delayed resolution of inflammation and fibrosis.
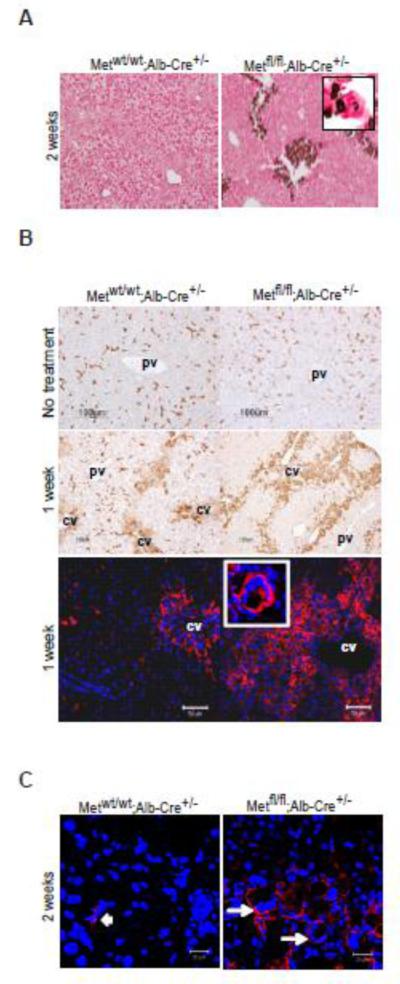
Fig. 3.
Abnormalities of liver remodeling after chronic injury with CCl4 caused by hepatocyte c-met deletion. (A) Dystrophic calcification of pericentral necrotic areas in Metfl/fl;Alb-Cre+/− livers. Van Kossa staining, 2 w of injury. Original magnification, X200. (B) Representative images of F4/80 immunhistochemistry (upper and middle panels) and F4/80 imunnofluorescence (bottom panels). Inset shows a multinucleated F4/80-positive cell at higher magnification. (C) Expression of Cd47 in multinucleated giant cells present in Metfl/fl;Alb-Cre+/− livers at 2 weeks of CCl4-induced injury) (white arrows). Metwt/wt;Alb-Cre+/− livers displayed only weak Cd47 positivity detected in a few mononucleated cells within the inflammatory infiltrate (arrowhead).Nuclei were counterstained with DAPI. Scale bars, 50 μM (B) and 20 μM (C). pv, portal vein; cv, central vein.
3.3 Loss of c-Met Aggravates Fibrosis through Mechanisms Dependent on Oxidative Stress
Oxidative damage is a well established component of CCl4-induced hepatic injury. In particular, mediators released from damaged hepatocytes, such as reactive oxygen species (ROS) and lipid peroxidation products have been identified as strong inducers of HSC activation [1, 32-34]. Previously, we have shown that loss of c-Met receptor in hepatocytes creates a prooxidant state which is compensated by constitutive activation of key transcription factors protecting against oxidative and xenobiotic stress, including nuclear factor (erythroid-derived 2)-like 2 (Nrf2) and NF-kB [21, 23]. Furthermore, although hepatocyte c-Met conditional knockout mice did not exhibit an obvious pathology, these mice were more susceptible to increased oxidative load, e.g. caused by Fas or DEN exposure [10, 21, 28].
To address the possibility whether the loss of c-Met receptor in hepatocytes may contribute to fibrosis through modulation of ROS generation, we first measured the rate of production of thiobarbituric acid-reactive substances (TBARS). The lipid oxidation was considerably increased in Metfl/fl;Alb-Cre+/− mice, both untreated and challenged with CCl4 (Figure 4A). In addition, we found a statistically significant decrease in the GSH/GSSG ratios reflecting a diminished capacity of the major antioxidant defense system in the absence of hepatocyte c-Met (Fig. 4B). Furthermore, while control mice showed a strong upregulation of Nrf2 which was paralleled by a decreased expression of Keap1, a repressor of Nrf2 activity, hepatocyte c-Met knockout mice displayed reduced protein levels of Nrf2 as well as SOD1 and γ-GCS, two well known targets of Nrf2 signaling (Figure 4C). Conversely, the expression of several components of ROS generating NADPH oxidase complex, in particular p47 and Nox4, were increased in c-Met deficient livers as compared to CCl4-treated controls (Figure 4C). Western blotting confirmed that the protein levels of TGF-β, one of the major profibrotic factors as well as the duration of activation were increased in Met-deficient livers, while the expression levels of HGF and CTGF were comparable in Metfl/fl;Alb-Cre+/− and control mice subjected to multiple CCl4 injections (Figure 4D). Consistent with previous reports, an initial increase in Hgf mRNA levels in response to the CCl4-mediated liver injury was followed by a rapid decline (not shown) possibly due to TGF-β upregulation [35-37]. Together, these results suggest that a greater CCl4-mediated liver toxicity in hepatocyte specific c-Met knockout livers may be related to increased oxidative stress.
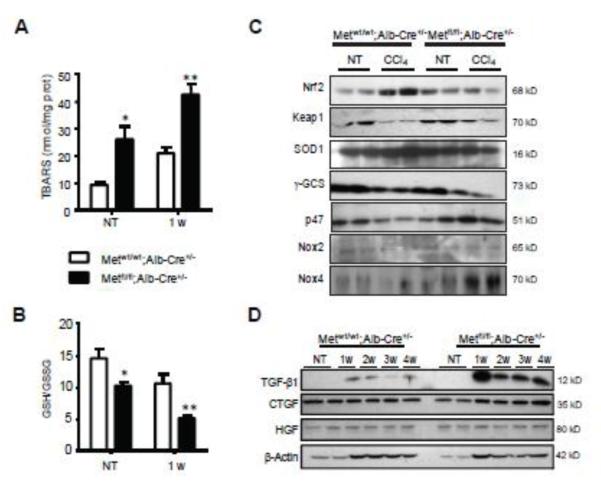
Fig. 4.
Increased oxidative stress in livers of CCl4-exposed c-Met deficient livers. (A) The levels of TBARS, a byproduct of lipid peroxidation, were detected by the TBARS assay using thiobarbituric acid as a reagent and expressed as nmol/mg protein. The data are presented as means ± SD (n=3). (B) The ratios of reduced glutathione (GSH) to oxidized glutathione (GSSG) determined by HPLC. The data are presented as means ± SD (n=3). (C and D) Western blotting of whole cell lysates prepared from Metfl/fl;Alb-Cre+/− and Metwt/wt;Alb-Cre+/− livers using indicated antibodies. β-Actin served as a loading control. * P<0.05; **P<0.01.
3.4 Transcriptomic Changes during Fibrogenesis
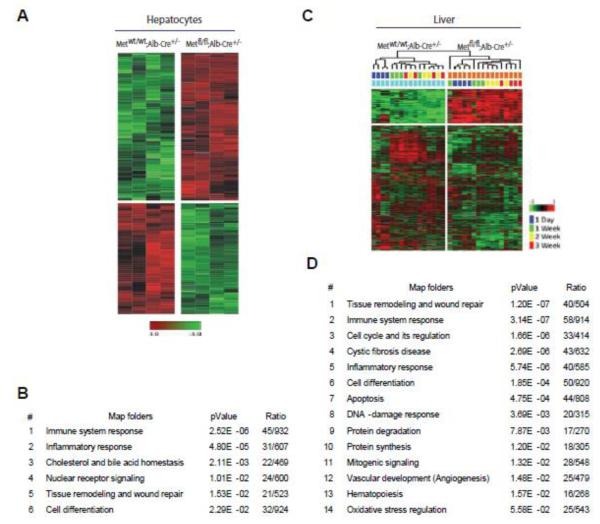
To further attribute the involved functional processes directly to c-Met deficiency in hepatocytes, we performed microarray analysis of hepatocytes isolated from Metfl/fl;Alb-Cre+/− and Metwt/wt;Alb-Cre+/− livers two days after the second exposure to CCl4, a time point of prominent impairment of hepatocyte function and rapid activation of stellate cells in c-Met deficient livers (Figure 1D and Figure 2, F and G). An analysis using complementary DNA microarrays identified 406 transcripts which were differentially (175 down; 231 up) expressed in c-Met knockout hepatocytes with more than 2-fold expression changes (Bootstrap t-test with a P-value cut-off of 0.01). These genes efficiently separated two genotypes in the cluster analyses (Figure 5A). The major associated functional networks were centered on plethora of genes important for immune response and inflammation (Figure 5B). Significantly, exposure to CCl4 increased mRNA levels of inflammatory mediators strongly linked to activation of macrophages and stellate cells including Ccl2, Ccl4, Ccl5, Cd14, , Cxcl10, Icam1, Itgb2, Timp1. Also, acute phase protein App, complement component C1qa and the pattern recognition receptor CD14, involved in the formation and transport of MHC class II protein, as well as regulators of cell-to cell communication and adhesion (Adam8, Icam1, Itgax, Itgb2, CD48), intracellular trafficking and calcium ion channel formation (Annexin A2, CD93, CD97) were among the genes with increased expression. Additionally, signaling associated with survival, cell death and stress response (App, Aqp2, Bax, Bcl3, C1qa, Capn1, Cd14, Cdkn1a, Cyba, Cyfip2, Dnmt3a, Egr1, Fasn, Fgr, Hif1a, Hspa5, Icam1, Lmo2, Ly86, Max, Ncf1, Ncf2, Ncf4, Pik3r1, Plg, Rcan1, S100a11, Serpine1, Soat1, Tasp1, Timp1, Tnfaip8, Tnfrsf1b, Tyrobp, Usp18, Xrcc4, Zfp36) as well as tissue remodeling and wound repair (B4galt1, Col3a1, Col5a1, Itgb2, Mfap2, Pdfga, Plg, Serpine1) were affected by the loss of c-Met in hepatocytes. Consistently, genes essential for hepatocyte function such as cholesterol and bile acid homeostasis (Dhcr7, Fdps, Idi1, Sqle, Cyp8b1, Lipc, Pklr, Slc22a5, Slc22a7) and metabolism (Aldh6a1, Cyp17a1, Cyp2b10, Cyp2c55, Cyp2c68, Cyp2f2, Cyp2j5, Cyp4a10, Cyp4f14) were downregulated in Metfl/fl;Alb-Cre+/−hepatocytes. However, CCl4 treatment increased mRNA levels of several components of the superoxide-producing NADPH-oxidase, including Ncf1, Ncf2, and Ncf4, supporting our earlier findings in c-Met-deficient hepatocytes [21].
Fig. 5.
Microarray analysis reveals a broad impact of c-Met-deficiency on the critical signaling pathways associated with fibrotic disease. (A) Unsupervised hierarchical cluster analyses of isolated hepatocytes at 1 w of injury phase. A total of 406 differentially expressed genes was identified using Bootstrap t-test (P-value < 0.01) (B) Top signaling pathways in Metfl/fl;Alb-Cre+/− hepatocytes as determined by GeneGo analysis. (C) Hierarchical cluster analysis during injury phase (1 d, 1 w, 2 w, 3 w). A total of 680 differentially expressed genes in Metfl/fl;Alb-Cre+/− livers were identified (FDR ≤ 0.05; fold change ≥2). Note a clear separation of both genotypes and different time points. (D) Top signaling pathways in Metfl/fl;Alb-Cre+/− livers as determined by GeneGo analysis.
Finally, to address broad transcriptional response to CCl4 exposure we isolated RNA from liver samples at various time-points during the injury phase (1 day, 1 wk, 2 wk, and 3 wk) and performed gene expression profiling using a 2-factor time course analysis taking into consideration both genotype-specific and time dependent transcriptional changes. A total of 680 genes including 147 upregulated and 533 down-regulated genes, were differentially expressed (FDR<0.05, ≥ 2-fold) in Metfl/fl;Alb-Cre+/− livers during injury phase (Supplemental Table S1). These genes were highly efficient in separating both genotypes as well as time points (Figure 5C). Major associated signaling pathways were related to tissue remodeling, inflammation, cell cycle, apoptosis as well as DNA damage and oxidative stress response (Figure 5D). Among the dominant networks and canonical pathways were profibrotic signaling, such as TGF-β1, as well as pathways related to inflammation, stress response and survival (Figures 4C and 5D; Supplemental Figure 4). We also found a compensatory up-regulation of EGFR signaling supporting our previous work on Metfl/fl;Alb-Cre+/− mice [28]. Consistent with c-Met knockout profibrotic phenotype, there was increased expression of markers for HSC activation (Tgfβ1, Tgfbr3, Thy1,) and phagocyte/macrophage function and survival (Cd68, C6, C1qc, Csf2ra, Fcer1g, Fpr2, Stat1, Timd4, Bcl6, Tyrobp, Fger1g, Egfr1, etc.). Conversely, pathways related to acute stress response (Alb, Apoh, Cfb, Hnrndk, Hrg, Il6st, Pik3r2, Ptpn11, Rbb4, Tab1, Traf6), survival (NF-kB, Irak1 and 3, K-Ras, Pik3r2, Tnfrsf11a), DNA damage and oxidative stress regulation (Rad9, Rad52, Ercc4, Gsta1 and 2, Jun, Gclc, Prdxd2, Fxn) were progressively down-regulated reflecting impaired regenerative capacity in the absence of c-Met signaling [9-10, 27, 38]. We also detected a significant reduction in the expression levels of several members of MAP-kinase family (Mapk9, Mapk12, Mapk1ip1, Mapk3k7ip1) responsive to growth factor and stress stimuli and genes which function to coordinate S phase and cell division (Tk1, Rfc5, Skp2, Cup4a, Ncaph, Ccng2, Csnk2a, Cdc6, cdk10, cdc123, cdc78, Chaf1a). In accordance with previous studies [23], we also found a prominent effect of c-Met deletion on genes involved in cytoskeleton organization (Actb, Actl6a, Tuba1a, Tuba8), chemotaxis and cell migration (Thy1, Cd207, Cd74, Il6st, Tyrobp, Ifngr2, Tspan8, Serpina1a), intercellular communications (Cdh5, Tjp1, Gjb2, Vezt) and adhesion (Vcam1, Cdh5, Cpxm1, Cul4a). Furthermore, loss of c-Met decreased transcript levels of genes encoding wide range of trans-membrane proteins which transport various molecules across extra- and intracellular membranes, including efflux transporters (Abca7, Abcd4, Abcc9, Abcf1, Atp2a3, Atp5a1, Atp5e), influx transporters of the solute carrier (SLC) family (Slc4a3, Slc7a4, Slc7a6os, Slc11a1, Slc15a3, Slc16a9, Slc23a2, Slc27a5, Slc37a, Slc39a13) as well as genes regulating traffic of membrane proteins and cargo sorting (Dync1li1, Snx14, Snx17, Exosc8, Tmed4, Vps26b, Vapa, Cav1).[39] Consistent with this, Metfl/fl;Alb-Cre+/− mice treated with CCl4 also displayed down-regulation of several members of DUB family of cytokine-regulated deubiquitinating enzymes (Usp1, Usp2, Usp10, Usp14), important regulators of the diverse cellular processes including receptor internalization and vesicle trafficking [40-41] well as a range of G-protein coupled receptors, the major sensors of outside molecules which function in intracellular signaling (Gpr1, Gpr65, Gpr108, Gpr124, Gpr137, Gpr146)[42]. Expression levels of S100a11, a member of the Ca(2+)-binding S100 protein family thought to facilitates membrane vesicle-fusion events such as endocytosis and membrane repair [43] as well as CAV1, the main structural protein of caveolae involved in cholesterol homoeostasis, transcytosis, and endocytosis [44] were also markedly decreased. Together these data reveal the massive changes in cellular metabolism and microenvironment caused by the deletion of c-Met receptor in hepatocytes.
4. Discussion
In this study, we used Metfl/fl;Alb-Cre+/− mice and a model of chronic CCl4-induced liver injury to investigate the molecular and cellular mechanisms underlying the anti-fibrotic activities of HGF/c-Met. We provide evidence that genetic loss of c-Met in hepatocytes induces a state of “organ stress” characterized by multi-factorial changes both in parenchymal and non-parenchymal cell compartments in the liver. These include reduced hepatocyte proliferation, rapid pathological calcification of necrotic areas with impact on the ability of macrophages to remove or sequester necrotic material, and excessive activation of hepatic stellate cells, thereby aggravating inflammation and fibrosis. Transcritpome analysis revealed that loss of c-Met receptor in hepatocytes caused a widespread deregulation of critical signaling pathways associated with immune cell trafficking, oxidative stress and DNA damage response, in addition to proliferation and cell death. These findings establish a broad protective role for c-Met signaling in hepatocytes against toxic liver damage leading to development of fibrotic disease.
There is extensive evidence for a mitogenic role of HGF/c-Met [15, 24, 25, 37]. We and others have confirmed the central role of c-Met in cell cycle initiation and progression in gene knockout studies in rodents [9, 38]. The extent of mito-inhibition varied in c-Met conditional knockout mice expressing Cre-recombinase under the control of a ubiquitously expressed interferon-inducible or hepatocyte-specific Alb-Cre promoter [9, 29, 38] varied and was less severe in Metfl/fl;Alb-Cre mice most likely due to the adaptive up-regulation of alternative survival and proliferating signaling [21, 23, 28]. Nevertheless, in both models a conditional c-met inactivation caused an inhibition of Erk1/2 activation and a subsequent suppression of cell cycle regulators in a model of two-thirds partial hepatectomy. Consistent with this, in Metfl/fl;Alb-Cre fibrotic livers we have detected a significant reduction in the expression levels of several members of MAP kinase family (Mapk9, Mapk12, Mapk1ip1, Mapk3k7ip1) responsive to growth factor and stress stimuli and genes which function to coordinate S phase and cell division (Tk1, Rfc5, Skp2, Cup4a, Ncaph, Ccng2, Csnk2a, Cdc6, cdk10, cdc123, cdc78, Chaf1a). A significant number of down-regulated genes encoded proteins controlling proper chromosome segregation (Sma3, Smc1a, Cep78), spindle integrity and spindle checkpoint signaling (Spc24, Cspp1, Mad2l1bp, Anapc5) in line with our earlier work which established a critical role for c-Met in control of G2/M transcriptional program [38].
It is well known that the hepato-protective effects of HGF/c-Met signaling can dramatically attenuate the process of liver fibrosis by direct inhibition of pro-fibrotic molecules (e.g. TGF-β) as well as promotion of ECM resolution [12-13, 45]. In agreement with these data, our results demonstrate that the absence of hepatocyte c-Met signaling caused increased expression of pro-fibrotic molecules, such as TGF-β (Figure 4C, Supplemental Figure 4B), in parallel with excessive HSCs activation and ECM accumulation in Metfl/fl;Alb-Cre+/− livers challenged with CCl4 (Figure 2). The TGF-β upregulation could contribute at least in part to suppression of hepatocyte mitogenesis in Metfl/fl;Alb-Cre+/− fibrotic livers.
Pathophysiologically, the outcome of liver injury depends on the interactions between inflammatory cells (e.g. macrophages), non-inflammatory cells (e.g. HSCs) and hepatocytes [1]. The primary deficiencies in c-Met deficient livers involve repair impediment associated with hepatocyte damage, inflammation, and activation of matrix producing HSCs (Figures 1-3). Remarkably, DNA microarray analysis of isolated hepatocytes during early fibrotic response revealed that loss of c-Met receptor caused a strong upregulation of chemotactic cytokines and adhesion molecules (Figure 5, A and B) suggesting a direct parenchymal involvement in the production of factors associated with immunological functions.
Consistent with c-Met knockout phenotype, among the overexpressed genes were several members of “CC” (CCL2, CCL4, and CCL5) and “CXC” (CXCL10) families of chemokines as well as cell adhesion molecules (e.g. ICAM-1) implicated in inflammatory cell trafficking into sites of injury and initiation of immunologic and inflammatory reactions. Thus, CCL2, also known as monocyte chemoattractant protein 1 (MCP-1), expressed by both hepatocytes and macrophages, has been shown to participate in macrophage fusion and foreign body giant cell formation [46-47]. Noteworthy, the increased expression of proinflammatory CCL5, known as RANTES, has been identified in hepatocytes in response to inflammatory conditions including hepatocellular steatosis [48]. Upregulation of RANTES was also found in renal tubular epithelial cells during obstructive nephropathy although no or little RANTES protein was observed in normal kidney [49]. Significantly, HGF gene therapy was capable of suppressing epithelial RANTES expression in parallel with inhibition of inflammatory cells infiltration [49-50].
Exposure to CCl4 also increased hepatic expression of CXCL10, another important cytokine implicated in progression of liver fibrosis [51-52]. It has been shown that CXCL10 promoted liver fibrosis by preventing hepatic stellate cell inactivation by natural killer cells [53]. These findings are in line with a recent study which related the pro-fibrotic effects of c-Met deficiency in hepatocytes to increased survival of HSCs in a model of liver fibrosis induced by bile duct ligation [27].
Genotype and time-dependent transcriptome analysis of CCl4-exposed livers confirmed the wide-ranging impact of c-Met deficiency on critical signaling pathways associated with fibrotic disease (Figure 5, C and D). Among the top canonical pathways affected by the loss of hepatocyte c-Met there were acute phase response signaling, production of nitric oxide and ROS in macrophages, mitochondrial dysfunction, PDGF signaling, interferon signaling, HGMB1 signaling, toll-like receptor signaling along with NF-kB, IL-6, EGF, and TGF-β signaling (Figures 4 and 5, Supplementary Figure S4). Functional analysis of differentially expressed genes in Metfl/fl;Alb-Cre+/− livers treated with CCl4 also revealed modulation of several members of G-protein coupled receptors (GPCRs) implicated in regulation of immune system and inflammation, reorganization of cytoskeletal network and intercellular communication between cells (e.g. Actb, Tjp1, Gjb2), deregulation of a range of receptors (e.g. GPCRs, Tnfrs8, Tnfrs10b, Tnfrsf11a, Ifngr2, Il3ra, Fcgr4, Ppargc1a, etc.) and membrane transporters, including members of ATP-binding cassette and solute carrier families. The loss of c-Met also affected the genes involved in ubiquitin-mediated signaling (e.g. Usp1, 2, 10 and 14, Sumo1) known to act in transcriptional regulation, nuclear transport, and protein stability [40-41].
In the setting of c-Met deficiency, the mechanistic significance of prooxidant-mediated hepatic fibrosis may be particularly relevant in potentially initiating a cascade of pro-fibrotic events. Previously, we have demonstrated that unchallenged c-Met conditional livers exhibit increased oxidative damage and reduced levels of GSH which were compensated by a sustain activation of key transcription factors protecting against oxidative and xenobiotic stress, including Nrf2 and NF-κB [21, 28]. However, when subjected to a chronic treatment with CCl4, Metfl/fl;Alb-Cre+/− livers were not capable of maintaining an adequate pro-survival signaling and displayed a strong down-regulation of the NF-κB and Nrf2 dependent genes (Figure 4C, Supplemental Figure 4C). Consistent with these findings, CCl4-exposed c-Met knockout livers displayed a more significant elevation of lipid peroxidation products, known as strong inducers of HSC activation [54], during early fibrotic response relative to control mice (Figures 4A). These findings show that increased oxidative stress is one of the hallmark features of the hepatocyte c-Met deficiency which may aggravate CCl4 induced liver fibrosis. However, the loss of c-Met signaling in the context of chronic CCl4 liver injury caused a plethora of diverse cellular and molecular changes, including aberrant tissue remodeling, disruption of a coordinated wound-healing response and reduced stress resistance, exceeding the sole secondary effects ascribed to oxidative stress.
Collectively, these findings provide mechanistic explanation for the high susceptibility of Metfl/fl;Alb-Cre+/− mice to chemically induced hepatic fibrosis. The profibrotic effects of c-Met deficiency were mediated by activation of the major proinflammatory genes in addition to increased oxidative stress and lack of compensatory regeneration. Although more work is required to determine the exact contribution of each of these factors, the study establishes a key protective role for intact c-Met signaling in the maintenance of structural integrity and adaptive plasticity of the liver under adverse conditions and strongly supports current efforts directed at modulating the HGF/c-Met signaling pathway in therapeutic settings [55].
Supplementary Material
Highlights.
Loss of c-Met signaling in hepatocytes aggravates hepatic fibrogenesis caused by CCl4 exposure
Loss of c-Met signaling in hepatocytes initiates a cascade of pathological events affecting both parenchymal and nonparenchymal cells
Transcritpome analysis revealed that the profibrotic effects of c-Met deficiency were mediated by activation of the major proinflammatory gene sets in addition to increased oxidative stress and lack of compensatory regeneration
These findings establish a broad protective role for c-Met signaling in hepatocytes against toxic liver damage leading to development of fibrotic disease
Acknowledgements
This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research. We thank Tanya Hoang for technical assistance.
Support: This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research
List of Abbreviations
- HGF
hepatocyte growth factor
- NAFLD/NASH
non-alcoholic fatty liver disease/steatohepatitis
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
- CCl4
carbon tetrachloride
- TGF-β
transforming growth factor β
- PDGF-β
platelet derived growth factor β
- BrdU
bromodeoxyuridine
- IHC
immunohistochemistry
- FDR
false discovery rate
- HSC
hepatic stellate cells
- GPCR
G-protein-coupled receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brunt EM. Nonalcoholic steatohepatitis: definition and pathology. Semin Liver Dis. 2001;21:3–16. doi: 10.1055/s-2001-12925. [DOI] [PubMed] [Google Scholar]
- [3].Poynard T, Bedossa P, Opolon P, The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups Natural history of liver fibrosis progression in patients with chronic hepatitis C. Lancet. 1997;349:825–832. doi: 10.1016/s0140-6736(96)07642-8. [DOI] [PubMed] [Google Scholar]
- [4].Brunt EM. Nonalcoholic steatohepatitis. Semin Liver Dis. 2004;24:3–20. doi: 10.1055/s-2004-823098. [DOI] [PubMed] [Google Scholar]
- [5].Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- [7].Friedman SL, Rockey DC, Bissell DM. Hepatic fibrosis 2006: report of the Third AASLD Single Topic Conference. Hepatology. 2007;45:242–249. doi: 10.1002/hep.21459. [DOI] [PubMed] [Google Scholar]
- [8].Jiao J, Friedman SL, Aloman C. Hepatic fibrosis. Curr Opin Gastroenterol. 2009;25:223–229. doi: 10.1097/mog.0b013e3283279668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huh CG, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sato M, Kakubari M, Kawamura M, Sugimoto J, Matsumoto K, Ishii T. The decrease in total collagen fibers in the liver by hepatocyte growth factor after formation of cirrhosis induced by thioacetamide. Biochem Pharmacol. 2000;59:681–690. doi: 10.1016/s0006-2952(99)00359-7. [DOI] [PubMed] [Google Scholar]
- [12].Ueki T, Kaneda Y, Tsutsui H, Nakanishi K, Sawa Y, Morishita R, Matsumoto K, Nakamura T, Takahashi H, Okamoto E, Fujimoto J. Hepatocyte growth factor gene therapy of liver cirrhosis in rats. Nat Med. 1999;5:226–230. doi: 10.1038/5593. [DOI] [PubMed] [Google Scholar]
- [13].Inagaki Y, Higashi K, Kushida M, Hong YY, Nakao S, Higashiyama R, Moro T, Itoh J, Mikami T, Kimura T, Shiota G, Kuwabara I, Okazaki I. Hepatocyte growth factor suppresses profibrogenic signal transduction via nuclear export of Smad3 with galectin-7. Gastroenterology. 2008;134:1180–1190. doi: 10.1053/j.gastro.2008.01.014. [DOI] [PubMed] [Google Scholar]
- [14].Kim WH, Matsumoto K, Bessho K, Nakamura T. Growth inhibition and apoptosis in liver myofibroblasts promoted by hepatocyte growth factor leads to resolution from liver cirrhosis. Am J Pathol. 2005;166:1017–1028. doi: 10.1016/S0002-9440(10)62323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Matsuda Y, Matsumoto K, Ichida T, Nakamura T. Hepatocyte growth factor suppresses the onset of liver cirrhosis and abrogates lethal hepatic dysfunction in rats. J Biochem. 1995;118:643–649. doi: 10.1093/oxfordjournals.jbchem.a124958. [DOI] [PubMed] [Google Scholar]
- [16].Ozaki I, Zhao G, Mizuta T, Ogawa Y, Hara T, Kajihara S, Hisatomi A, Sakai T, Yamamoto K. Hepatocyte growth factor induces collagenase (matrix metalloproteinase-1) via the transcription factor Ets-1 in human hepatic stellate cell line. J Hepatol. 2002;36:169–178. doi: 10.1016/s0168-8278(01)00245-8. [DOI] [PubMed] [Google Scholar]
- [17].Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- [18].Jamall IS, Finelli VN, Que Hee SS. A simple method to determine nanogram levels of 4-hydroxyproline in biological tissues. Anal Biochem. 1981;112:70–75. doi: 10.1016/0003-2697(81)90261-x. [DOI] [PubMed] [Google Scholar]
- [19].Scheuer PJ. Classification of chronic viral hepatitis: a need for reassessment. J Hepatol. 1991;13:372–374. doi: 10.1016/0168-8278(91)90084-o. [DOI] [PubMed] [Google Scholar]
- [20].Buege JA, Aust SD. Microsomal lipid peroxidation. Methods Enzymol. 1978;52:302–310. doi: 10.1016/s0076-6879(78)52032-6. [DOI] [PubMed] [Google Scholar]
- [21].Gomez-Quiroz LE, Factor VM, Kaposi-Novak P, Coulouarn C, Conner EA, Thorgeirsson SS. Hepatocyte-specific c-Met deletion disrupts redox homeostasis and sensitizes to Fas-mediated apoptosis. J Biol Chem. 2008;283:14581–14589. doi: 10.1074/jbc.M707733200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Andersen JB, Loi R, Perra A, Factor VM, Ledda-Columbano GM, Columbano A, Thorgeirsson SS. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51:1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Burr AW, Toole K, Chapman C, Hines JE, Burt AD. Anti-hepatocyte growth factor antibody inhibits hepatocyte proliferation during liver regeneration. J Pathol. 1998;185:298–302. doi: 10.1002/(SICI)1096-9896(199807)185:3<298::AID-PATH88>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [25].Phaneuf D, Moscioni AD, LeClair C, Raper SE, Wilson JM. Generation of a mouse expressing a conditional knockout of the hepatocyte growth factor gene: demonstration of impaired liver regeneration. DNA Cell Biol. 2004;23:592–603. doi: 10.1089/dna.2004.23.592. [DOI] [PubMed] [Google Scholar]
- [26].Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [27].Giebeler A, Boekschoten MV, Klein C, Borowiak M, Birchmeier C, Gassler N, Wasmuth HE, Muller M, Trautwein C, Streetz KL. c-Met confers protection against chronic liver tissue damage and fibrosis progression after bile duct ligation in mice. Gastroenterology. 2009;137:297–308. 308, e291–294. doi: 10.1053/j.gastro.2009.01.068. [DOI] [PubMed] [Google Scholar]
- [28].Takami T, Kaposi-Novak P, Uchida K, Gomez-Quiroz LE, Conner EA, Factor VM, Thorgeirsson SS. Loss of hepatocyte growth factor/c-Met signaling pathway accelerates early stages of N-nitrosodiethylamine induced hepatocarcinogenesis. Cancer Res. 2007;67:9844–9851. doi: 10.1158/0008-5472.CAN-07-1905. [DOI] [PubMed] [Google Scholar]
- [29].Ishikawa T, Factor VM, Marquardt JU, Raggi C, Seo D, Kitade M, Conner EA, Thorgeirsson SS. Hepatocyte growth factor (HGF)/c-Met signaling is required for stem cell mediated liver regeneration. Hepatology. 2011 doi: 10.1002/hep.24796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moreira RK. Hepatic stellate cells and liver fibrosis. Arch Pathol Lab Med. 2007;131:1728–1734. doi: 10.5858/2007-131-1728-HSCALF. [DOI] [PubMed] [Google Scholar]
- [31].Han X, Sterling H, Chen Y, Saginario C, Brown EJ, Frazier WA, Lindberg FP, Vignery A. CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J Biol Chem. 2000;275:37984–37992. doi: 10.1074/jbc.M002334200. [DOI] [PubMed] [Google Scholar]
- [32].De Minicis S, Bataller R, Brenner DA. NADPH oxidase in the liver: defensive, offensive, or fibrogenic? Gastroenterology. 2006;131:272–275. doi: 10.1053/j.gastro.2006.05.048. [DOI] [PubMed] [Google Scholar]
- [33].Kisseleva T, Brenner DA. Fibrogenesis of parenchymal organs. Proc Am Thorac Soc. 2008;5:338–342. doi: 10.1513/pats.200711-168DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Novo E, Parola M. Redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair. 2008;1:5. doi: 10.1186/1755-1536-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanchez A, Alvarez AM, Benito M, Fabregat I. Transforming growth factor beta modulates growth and differentiation of fetal hepatocytes in primary culture. J Cell Physiol. 1995;165:398–405. doi: 10.1002/jcp.1041650221. [DOI] [PubMed] [Google Scholar]
- [36].Ramadori G, Neubauer K, Odenthal M, Nakamura T, Knittel T, Schwogler S, Meyer zum Buschenfelde KH. The gene of hepatocyte growth factor is expressed in fat-storing cells of rat liver and is downregulated during cell growth and by transforming growth factor-beta. Biochem Biophys Res Commun. 1992;183:739–742. doi: 10.1016/0006-291x(92)90545-v. [DOI] [PubMed] [Google Scholar]
- [37].Nakamura T, Tomita Y, Hirai R, Yamaoka K, Kaji K, Ichihara A. Inhibitory effect of transforming growth factor-beta on DNA synthesis of adult rat hepatocytes in primary culture. Biochem Biophys Res Commun. 1985;133:1042–1050. doi: 10.1016/0006-291x(85)91241-0. [DOI] [PubMed] [Google Scholar]
- [38].Factor VM, Seo D, Ishikawa T, Kaposi-Novak P, Marquardt JU, Andersen JB, Conner EA, Thorgeirsson SS. Loss of c-Met disrupts gene expression program required for G2/M progression during liver regeneration in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- [41].Ramakrishna S, Suresh B, Baek KH. The role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci. 2011;68:15–26. doi: 10.1007/s00018-010-0504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- [43].Fan C, Fu Z, Su Q, Angelini DJ, Van Eyk J, Johns RA. S100A11 mediates hypoxia-induced mitogenic factor (HIMF)-induced smooth muscle cell migration, vesicular exocytosis, and nuclear activation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000901. M110 000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mastrodonato M, Calamita G, Rossi R, Mentino D, Bonfrate L, Portincasa P, Ferri D, Liquori GE. Altered distribution of caveolin-1 in early liver steatosis. Eur J Clin Invest. 2011;41:642–651. doi: 10.1111/j.1365-2362.2010.02459.x. [DOI] [PubMed] [Google Scholar]
- [45].Xia JL, Dai C, Michalopoulos GK, Liu Y. Hepatocyte growth factor attenuates liver fibrosis induced by bile duct ligation. Am J Pathol. 2006;168:1500–1512. doi: 10.2353/ajpath.2006.050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dambach DM, Watson LM, Gray KR, Durham SK, Laskin DL. Role of CCR2 in macrophage migration into the liver during acetaminophen-induced hepatotoxicity in the mouse. Hepatology. 2002;35:1093–1103. doi: 10.1053/jhep.2002.33162. [DOI] [PubMed] [Google Scholar]
- [47].Kyriakides TR, Foster MJ, Keeney GE, Tsai A, Giachelli CM, Clark-Lewis I, Rollins BJ, Bornstein P. The CC chemokine ligand, CCL2/MCP1, participates in macrophage fusion and foreign body giant cell formation. Am J Pathol. 2004;165:2157–2166. doi: 10.1016/S0002-9440(10)63265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kirovski G, Gabele E, Dorn C, Moleda L, Niessen C, Weiss TS, Wobser H, Schacherer D, Buechler C, Wasmuth HE, Hellerbrand C. Hepatic steatosis causes induction of the chemokine RANTES in the absence of significant hepatic inflammation. Int J Clin Exp Pathol. 2010;3:675–680. [PMC free article] [PubMed] [Google Scholar]
- [49].Giannopoulou M, Dai C, Tan X, Wen X, Michalopoulos GK, Liu Y. Hepatocyte growth factor exerts its anti-inflammatory action by disrupting nuclear factor-kappaB signaling. Am J Pathol. 2008;173:30–41. doi: 10.2353/ajpath.2008.070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gong R, Rifai A, Tolbert EM, Biswas P, Centracchio JN, Dworkin LD. Hepatocyte growth factor ameliorates renal interstitial inflammation in rat remnant kidney by modulating tubular expression of macrophage chemoattractant protein-1 and RANTES. J Am Soc Nephrol. 2004;15:2868–2881. doi: 10.1097/01.ASN.0000141962.44300.3A. [DOI] [PubMed] [Google Scholar]
- [51].Berres ML, Trautwein C, Schmeding M, Eurich D, Tacke F, Bahra M, Neuhaus P, Neumann UP, Wasmuth HE. Serum chemokine CXC ligand 10 (CXCL10) predicts fibrosis progression after liver transplantation for hepatitis C infection. Hepatology. 2011;53:596–603. doi: 10.1002/hep.24098. [DOI] [PubMed] [Google Scholar]
- [52].Harvey CE, Post JJ, Palladinetti P, Freeman AJ, Ffrench RA, Kumar RK, Marinos G, Lloyd AR. Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J Leukoc Biol. 2003;74:360–369. doi: 10.1189/jlb.0303093. [DOI] [PubMed] [Google Scholar]
- [53].Hintermann E, Bayer M, Pfeilschifter JM, Luster AD, Christen U. CXCL10 promotes liver fibrosis by prevention of NK cell mediated hepatic stellate cell inactivation. J Autoimmun. 2010;35:424–435. doi: 10.1016/j.jaut.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hui AY, Friedman SL. Molecular basis of hepatic fibrosis. Expert Rev Mol Med. 2003;5:1–23. doi: 10.1017/S1462399403005684. [DOI] [PubMed] [Google Scholar]
- [55].Ghiassi-Nejad Z, Friedman SL. Advances in antifibrotic therapy. Expert Rev Gastroenterol Hepatol. 2008;2:803–816. doi: 10.1586/17474124.2.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.