Abstract
The objective of this study was to elucidate the effect of partial reduction of the mitochondrial fission protein, dynamin-related protein 1 (Drp1) on mitochondrial activity and synaptic viability. Recent knockout studies of Drp1 revealed that homozygote Drp1 knockout mice are embryonic lethal due to reduced mitochondrial fission, and that this reduced fission leads to developmental defects in the brain. In contrast, heterozygote Drp1 knockout mice appear to be normal in terms of lifespan, fertility, and viability, and phenotypically these animals are not different from wild-type mice. However, the effects of partial Drp1 reduction on mitochondrial function and synaptic activity are not well understood. In the present study, we sought to characterize synaptic, dendritic and mitochondrial proteins, and mitochondrial function and GTPase enzymatic activity, in Drp1 heterozygote knockout mice. Interestingly, we found no significant changes in synaptic, dendritic, and mitochondrial proteins in the Drp1 heterozygote knockout mice compared to the wild-type mice. Further, mitochondrial function and GTPase enzymatic activity appeared to be normal. However, H2O2 and lipid peroxidation levels were significantly reduced in the Drp1 heterozygote knockout mice compared to the wild-type mice. These findings suggest that partial Drp1 reduction does not affect mitochondrial and synaptic viability and may have therapeutic use in treating patients with Alzheimer’s disease and Huntington’s disease.
Keywords: Dynamin-related protein 1, Mitochondria, Oxidative stress, Alzheimer’s disease, Huntington’s disease, Mitochondrial dynamics
Introduction
Structural abnormalities in mitochondria have been reported to be involved in mitochondrial dysfunction and oxidative stress in aging, cancer, diabetes, obesity, cardiovascular diseases and in age-related neurodegenerative diseases, such as Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s disease, and amyotrophic lateral sclerosis [1–17]. These structural abnormalities are primarily caused by an imbalance in highly conserved, GTPase genes that are essential for mitochondrial fission and fusion. Dynamin-related protein 1 (Drp1) is essential for mitochondrial fission, and mitofusins 1 and 2 (Mfn1, Mfn2), and optic atrophy 1 (Opa1) are involved in mitochondrial fusion [18–22].
In normal, healthy mammalian cells, such as neurons, fission and fusion cells balance equally, resulting in the maintenance of mitochondrial dynamics and distribution. However, recent research has found that neurons that express mutant proteins (e.g., amyloid beta in AD and mutant huntingtin in HD) show impaired fission and fusion balance [2,3,8–11,23,24]. This impaired balance is primarily due to the abnormal interaction between Drp1 (a fission protein) and mutant proteins. These abnormal interactions increase in disease progression, in AD and HD [8,9,11,24], ultimately creating structural abnormalities and consequent dysfunction in mitochondria. Mitochondrial dysfunction ultimately inhibits the production of adenosine triphosphate (ATP) and damages neurons [2,38–10,23,24]. Given this chain of responses triggered by the interaction of Drp1 and mutant huntingtin (in HD) and amyloid beta (in AD) proteins, researchers have suggested that a means to inhibit this interaction may provide a therapeutic strategy to prevent mitochondrial abnormalities and dysfunction in AD and HD [20,25].
Drp1 is a highly, evolutionary conserved, large GTPase protein that is critically involved in regulating structural features of mitochondria, including their size, shape, distribution, and remodeling, and in maintaining mitochondrial viability [18,20,22]. Further, Drp1 may also be involved in mitochondrial division, mitochondrial distribution, peroxisomal fragmentation, phosphorylation, ubiquitination, and SUMOylation [20,22]. Most importantly, Drp1 is critical in GTPase enzymatic activity, which in turn is critical for mitochondrial division.
In studies aimed at elucidating the normal function of Drp1, researchers found that Drp1 homozygote knockout mice (Drp1−/−) have developmental abnormalities, particularly in the forebrain, and they die, on average, shortly after embryonic day 11.5–12.5 [26,27]. Similarly, neural cell-specific (NS) Drp1−/− mice died shortly after birth, from brain hypoplasia. Primary culture of the NS-Drp1−/− mouse forebrain showed a decrease in neurites and the presence of defective synapse formation, the latter which was suggested to due to aggregated mitochondria that failed to distribute properly within cell processes [27]. Wakabayashi et al [26] found mitochondria formed extensive networks, and peroxisomes were elongated in Drp1−/− embryonic fibroblasts. Brain-specific Drp1 depletion resulted in the development of a few giant mitochondria instead of many short, tubular mitochondria observed in control cells [26]. Overall, these results suggest that Drp1 is critical for mitochondrial maintenance, division, and distribution in neurons.
In a diseased state, such as AD and HD, Drp1 has been found to interact with mutant huntingtin and amyloid beta proteins, leading to excessive mitochondrial fragmentation, mitochondrial dysfunction, oxidative stress, and neuronal damage [8,9,11]. A key question is how much Drp1 expression is sufficient to inhibit the interaction between Drp1 and the mutant proteins, in order to prevent excessive mitochondrial fragmentation – yet is sufficient to balance mitochondrial fission and fusion, and to maintain mitochondrial function and neuronal survival in a diseased state? To address this issue, we characterized Drp1+/− mice and Drp1+/+ mice for mitochondrial activity (mitochondrial proteins, mitochondrial function and GTPase Drp1 enzymatic activity), and synaptic viability (synaptic and dendritic proteins).
Materials and Methods
Drp1 mice and brain tissues
We used brain tissues Drp1 heterozygote knockout (Drp1+/−) mice and control, wild-type (WT) Drp1+/+ [26] to determine the effects of partial Drp1 deficiency on mitochondrial dynamics and proteins, and on synaptic processes. The Drp1+/+ and Drp1+/− mice were housed at The Johns Hopkins University. The Johns Hopkins Institutional Animal Care and Use Committee approved all procedures for animal care. We genotyped all the mice for the Drp1 gene, using DNA prepared from tail biopsies of 2–3 week old mice, following the protocol described in Wakabayashi et al. [26].
Immunoblotting analysis
We performed immunoblotting analyses of protein lysates from 3-month-old Drp1+/+ mice (n=3) and Drp1+/− mice (n=3). Twenty μg protein lysates were resolved on a 4–12% Nu-PAGE gel (Invitrogen, Grand Island, NY). These resolved proteins were transferred to PVDF membranes (Novax Inc, San Diego, CA) and then incubated with a blocking buffer (5% dry milk dissolved in a TBST buffer) for 1 h at room temperature. The nylon membranes were incubated overnight with primary Drp1 antibodies (1:200 rabbit polyclonal Novus Biological Inc, Littleton, CO), Fis1 (1:200 dilution, Protein Tech Group Inc, Chicago, IL), Mfn1 (1:300 rabbit polyclonal, Santa Cruz Biotechnology Inc, Santa Cruz, CA), Mfn2 (1:300 rabbit polyclonal, Abcam, Cambridge, MA), Opa1 (1:500 mouse-monoclonal, BD Biosciences, San Jose, CA), CypD (1:500 mouse monoclonal, EMD Calbiochem, Gibbstown, NJ), synaptophysin (1:400 mouse monoclonal, Millipore, Temecula, CA), PSD95 (1:400 rabbit polyclonal, Abcam), MAP2 (1:500 rabbit polyclonal, Millipore) and β-actin (1:500 mouse monoclonal Chemicon, Millipore).
The membranes were washed with a TBST buffer 3 times at 10-min intervals and then incubated for 2 h with appropriate secondary antibodies, followed by 3 additional washes at 10-min intervals. Details of secondary antibodies and their dilutions are given Table 1. Mitochondrial, synaptic and dendritic proteins were detected with the Supersignal West Pico chemiluminescent reagent (Thermo Scientific). Scanned images of the exposed X-ray film were analyzed with ImageJ to determine relative band intensity. Quantification was performed on western blots of cerebral cortex and cerebellum protein lysates from Drp1+/+ mice and Drp1+/− mice.
Table 1.
Summary of antibody dilutions and conditions used in the immunoblotting analysis of mitochondrial, synaptic and dendritic proteins.
| Marker | Primary antibody – species and dilution | Purchased from Company, State | Secondary antibody, dilution | Purchased from Company, City & State |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Fis1 | Rabbit Polyclonal 1:400 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn1 | Rabbit Polyclonal 1:400 | Santa Cruz Biotechnology, INC, Santa Cruz, CA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn2 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals INC, Gibbstown, NJ | Sheep anti-mouse HRP 1:8,000 | GE Healthcare Amersham, Piscataway, NJ |
| Synaptophysin | Mouse Monoclonal 1:400 | Millipore, Temecula, CA | Sheep anti-mouse HRP 1:8,000 | GE Healthcare Amersham, Piscataway, NJ |
| PSD95 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| MAP2 | Rabbit Polyclonal 1:500 | Millipore, Temecula, CA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| B-actin | Mouse Monoclonal 1:500 | Sigma-Aldrich, St Luis, MO | Sheep anti-mouse HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
Immunohistochemistry and immunofluorescence analyses
Using immunofluorescence techniques, we performed immunostaining for synaptophysin and PSD95 (synaptic proteins), MAP2 (dendritic protein), Drp1, Fis1 (fission proteins), Mfn1 (fusion protein), and CypD (matrix protein) in brain sections we excised from Drp1+/+ mice and Drp1+/− mice. The brain specimens were embedded paraffin, and sections were cut into widths of 15 μm. The sections were deparaffinized by a 10-min wash with xylene, followed by a 5-min wash in each of 3 serial dilutions of alcohol (at 95, 70, and 50%). The sections were again washed once for 10 min, with double-distilled H2O, and then for 6 more times with phosphate-buffered saline (PBS) at pH 7.4, at 5 min each. To block the endogenous peroxidase, sections were treated for 15 min with 3% H2O2 and then with 0.5% Triton dissolved in PBS at pH 7.4. The sections were blocked for 1 h with a solution of 0.5% Triton in PBS+10% goat serum+1% bovine serum albumin.
The sections were incubated overnight at room temperature with the following antibodies: anti-Drp1 (1:200 rabbit polyclonal dilution, Santa Cruz Biotechnology), Fis1 (1:200 dilution, Protein Tech Group Inc), Mfn1 (1:200 rabbit polyclonal Santa Cruz Biotechnology Inc), Mfn2 (1:400 rabbit polyclonal, Abcam), Opa1 (1:500 mouse-monoclonal BD Biosciences), CypD (1:500 mouse monoclonal, EMD Calbiochem), pyruvate dehyrogenase (1:1000, Mitosciences, Eugene, OR), synaptophysin (1:400 mouse monoclonal, Millipore), PSD95 (1:400 rabbit polyclonal, Abcam) and MAP2 (1:500, rabbit polyclonal, Millipore).
On the day after the primary antibody incubation, sections were washed once with 0.1% Triton in PBS and were then incubated with appropriate biotinylated secondary antibodies for 1 h at room temperature. Details of secondary antibodies and their dilutions are given in Table 2. The sections were washed with PBS 3 times for 10 min each and then incubated for 1 h with labeled streptavidin, an HRP solution (Molecular Probes, Eugene, OR). The sections were each washed 3 more times for 10 min each, with PBS at pH 7.4, and then were treated with Tyramide Alexa 594 (red) or Alexa 488 (green) (Molecular Probes) for 10 min at room temperature. They were cover-slipped with Prolong Gold (Invitrogen) and photographed with a confocal microscope. Randomly selected immunofluorescence images from cerebral cortex sections of Drp1+/+ mice and Drp1+/− mice at 40X magnification were analyzed with ImageJ to determine relative immunofluorescence intensity between Drp1+/+ mice and Drp1+/− mice for mitochondrial, synaptic and dendritic proteins. To determine mitochondrial morphology, we photographed the hippocampal and cerebral cortex sections from Drp1+/+ (WT) and Drp1+/− mice that were immunostained with mitochondrial matrix marker, pyruvate dehydrogenase.
Table 2.
Summary of antibody dilutions and conditions used in the immunohistochemistry/immunofluorescence analysis of mitochondrial and synaptic proteins using brain sections from DRP+/+ and DRP+/− mouse.
| Marker | Primary antibody – species and dilution | Purchased from Company, State | Secondary antibody, dilution, Alexa dye | Purchased from Company, City & State |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, INC, Santa Cruz, CA | Goat anti - rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| Fis1 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, INC, Santa Cruz, CA | Goat anti-rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, M VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| Mfn1 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, INC, Santa Cruz, CA | Goat anti-rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| Mfn2 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Goat anti - rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals INC, Gibbstown, NJ | Goat anti-mouse Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| Pyruvate dehydrogenase | Mouse Monoclonal 1:1000 | Mitosciences Eugene, OR | Donkey anti-mouse Alexa 488 | Molecular Probes, Eugene, OR |
| Synaptophysin | Mouse Monoclonal 1:300 | Millipore, Temecula, CA | Goat anti-mouse Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| PSD95 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Goat anti - rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
| MAP2 | Rabbit Polyclonal 1:400 | Millipore, Temecula, CA | Goat anti rabbit Biotin 1:300, ABC, TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories INC, Burlingame, CA Molecular Probes, Eugene, OR |
Mitochondrial Function
H2O2 production
Using an Amplex® Red H2O2 Assay Kit (Molecular Probes), we measured the production of H2O2 in the tissues from the cerebral cortex and the cerebellum of Drp1+/− mice (n=3) and Drp1+/+ mice (n=3), as previously described [28]. Briefly, the production of H2O2 was measured in mitochondria isolated from cerebral cortex and the cerebellum of Drp1+/− mice and Drp1+/+ mice. A BCA Protein Assay Kit (Pierce Biotechnology) was used to measure protein concentration in a reaction mixture that contained mitochondrial proteins (μg/μl), Amplex Red reagents (50 μM), horseradish peroxidase (0.1 U/ml), and a reaction buffer (1X). The mixture was incubated at room temperature for 30 min, followed by spectrophotometer readings of fluorescence (570 nm). Finally, H2O2 production was determined, using a standard curve equation expressed in nmol/μg mitochondrial protein.
Cytochrome oxidase activity
Cytochrome oxidase activity was measured in mitochondria isolated from the cerebral cortex and cerebellum of Drp1+/− mice (n=3) and Drp1+/+ mice (n=3), as described in Manczak et al. [23]. Enzyme activity was assayed spectrophotometrically with a Sigma Kit (Sigma-Aldrich) following manufacturer’s instructions. Briefly, 2 μg mitochondrial protein was added to 1.1 ml of a reaction solution containing 50 μl 0.22 mM ferricytochrome c fully reduced by DTT, Tris-HCl at pH 7.0, and 120 mM potassium chloride. The absorbance of wavelength at 550 mM (or its decrease) was recorded in 1-min reactions, at 10-sec intervals. Cytochrome c oxidase activity was measured according to the following formula: mU/mg total mitochondrial protein = (A/min sample − (A/min blank) × 1.1 mL × 21.84). The protein concentrations were determined, following the BCA method.
ATP levels
The levels of ATP were measured in mitochondria that were isolated from cerebral cortex and cerebellum tissues of Drp1+/− mice (n=3) and Drp1+/+ mice (n=3), with a ATP determination kit (Molecular Probes) [29]. This bioluminescence assay is based on the reaction of ATP with recombinant firefly luciferase and its substract luciferin. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to the ATP concentration, which is measured using a luminometer. We measured ATP from mitochondrial pellets, using a standard curve method.
Lipid peroxidation assay
Lipid peroxidates are unstable indicators of oxidative stress in neurons [30]. 4-hydroxy-2-nonenol (HNE) is the final product of lipid peroxidation that was measured in cerebral cortex and cerebellum tissues of Drp1+/− mice (n=3) and Drp1+/+ mice (n=3), with an HNE-His ELISA Kit (Cell BioLabs, Inc., San Diego, CA). Briefly, freshly prepared protein was added to a 96-well protein binding plate and incubated overnight at 4°C. It was then washed 3 times with a wash buffer. The washed protein and the anti-HNE-His antibody were then added to wells and incubated for 2 h at room temperature, and then washed 3 times. Next, the samples were incubated with a secondary antibody that was conjugated with peroxidase for 2 h at room temperature. They were then incubated with an enzyme substrate. Optical density was measured to quantify the level of HNE (lipid peroxidation).
GTPase enzymatic activity
Using a Novus Biological calorimetric kit (Littleton, CO), we measured GTPase enzymatic activity in cerebral cortex and cerebellum tissues from Drp1+/+ mice (n=3) and Drp1+/− mice (n=3), following GTPase assay methods described in Shirendeb et al. [11], based on GTP hydrolyzing to GDP and to inorganic phosphorous (Pi). We measured GTPase activity, based on the amount of Pi that the GTP produces. By adding the ColorLock Gold (orange) substrate to the Pi generated from GTP, we assessed GTP activity, based on the inorganic complex solution (green). Calorimetric measurements (green) were read in the wavelength range of 650 nm. We compared GTPase activity in cereberal cortex and cerebellum tissues from the Drp1+/− mice and the Drp1+/+ mice.
Results
Immunoblotting analysis
To determine the effects of partial Drp1 deficiency on mitochondrial proteins (Drp1, Fis1, Mfn1, Mfn2, Opa1, and CypD), on synaptic proteins (synaptophysin and PSD95), and on dendritic proteins (MAP2), mitochondrial, synaptic, and dendritic proteins were measured in cerebral cortex and cerebellum tissues from Drp1 +/− mice and Drp1+/+ mice.
Mitochondrial proteins
As shown in Fig. 1A and 1B, we found reduced levels of Drp1 in the cerebral cortex (P=0.2) and cerebellum (P=0.06) tissues from the Drp1+/− mice and the Drp1+/+ mice, respectively, but the levels were not significantly different, indicating only partial Drp1 reduction in the brain tissues from the Drp1+/− mice. However, we did not find reduced levels of Fis1 in the cerebral cortex (P=0.9) and cerebellum (P=0.1) tissues from the Drp1+/− mice and the Drp1+/+ mice, respectively (Fig. 1A and 1B). We also did not find any significant reduction in the Mfn1 levels in the cerebral cortex (P=0.8) and cerebellum (P=0.1) tissues from the Drp1+/− mice and the Drp1+/+ mice, respectively. Similar results were found for Mfn2 and CypD (Fig. 1A and 1B).
Figure 1.
Immunoblotting analysis of mitochondrial proteins. A represents representative immunoblots of mitochondrial proteins in Drp1+/+ mice and Drp1+/− mice. Fifty μg of protein lysate was used from cerebral cortex and cerebellum tissues from the Drp1+/+ and Drp1+/− mice. Immunoblotting analyses were performed, using Drp1, Fis1, Mfn1, Mfn2, CypD, and beta actin, as a control for equal loading protein. B represents quantitative densitometric analysis of mitochondrial proteins.
Synaptic and dendritic proteins
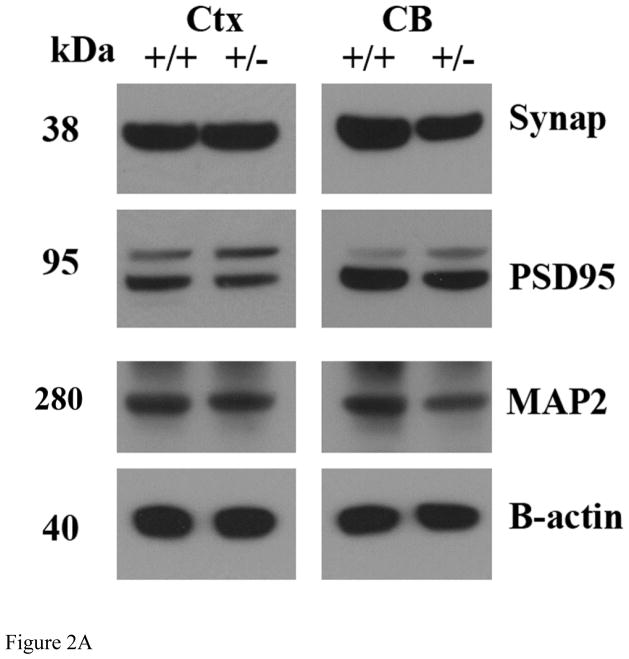
Our quantitative immunoblotting analysis of synaptophysin and PSD95 revealed no significant changes in the cerebral cortex (P=0.9 for synaptophysin and P=0.7 for PSD95) and cerebellum (P=0.1 for synaptophysin and P=0.9 for PSD95) tissues from in Drp1+/− mice and Drp1+/+ mice (Fig. 2A and Fig. 2B). Dendritic protein, MAP2 was reduced in the cerebellum of Drp1+/− mice relative to Drp1+/+ mice but not significant (P=0.06). However, MAP2 levels were unchanged between Drp1+/+ mice and Drp1+/− mice.
Figure 2.
Immunoblotting analysis of synaptic and dendritic proteins. A represents representative immunoblots of synaptic and dendritic proteins in Drp1+/+ mice and Drp1+/− mice. Fifty μg of protein lysate was used from cerebral cortex and cerebellum tissues from the Drp1+/+ and Drp1+/− mice. Immunoblotting analyses were performed, using synaptophysin, PSD95, MAP2, and beta actin, as a control for equal loading protein. B represents quantitative densitometric analysis of synaptic and dendritic proteins.
Immunostaining analysis
To determine whether partial Drp1 deficiency affects the localization and distribution of mitochondrial, synaptic, and dendritic proteins, we conducted immunohistochemistry and immunofluorescence analyses of Drp1, Fis1, Mfn1, CypD, synaptic (synaptophysin and PSD95), and dendritic (MAP2) proteins in brain tissues from Drp1+/− mice and Drp1+/+ mice. We quantified the immunoreactivities of all the proteins.
Mitochondrial proteins
We found Drp1 to be ubiquitous in the hippocampus and cerebral cortex tissues from both the Drp1+/− mice and Drp1+/+ mice, and we found Drp1 to be reduced in the cerebral cortex sections from Drp1+/− mice relative to Drp1+/+ mice, but not significant (P=0.06) (Fig. 3). As shown in Fig. 4, similar to Drp1, Fis1 was uniformly expressed in the brain. However, immunoreactivity of Fis1 was not significantly decreased in the cerebral cortex of the Drp1+/− mice relative to Drp1+/+ mice (P=0.06).
Figure 3.
Immunoreactivity of Drp1 in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) represents the hippocampus (10X the original magnification); (B), the cerebral cortex (10X the original magnification); (C), the cerebral cortex (40X the original magnification) and (D) quantification of Drp1 immunoreactivity in the cerebral cortex.
Figure 4.
Immunoreactivity of Fis1 in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) represents the hippocampus (10X the original magnification); (B), the cerebral cortex (10X the original magnification); (C), the cerebral cortex (40X the original magnification) and (D) quantification of Fis1 immunoreactivity in the cerebral cortex.
We did not find any change in the immunoreactivity of mitochondrial fusion protein, Mfn1 (P=0.6) in the cerebral cortex brain sections from the Drp1+/− mice relative to Drp1+/+ mice, suggesting that reduced expression Drp1 did not affect mitochondrial fusion proteins in the brain (Fig. 5).
Figure 5.
Immunoreactivity of Mfn1 in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) the cerebral cortex (10X the original magnification); (B), the cerebral cortex (40X the original magnification) and (C) quantification of Mfn1 immunoreactivity in the cerebral cortex.
Similar to the fusion proteins, matrix protein, CypD immunoreactivity was unchanged in the brain sections from the Drp1+/− and Drp1+/+ mice (Fig. 6) (P=0.4).
Figure 6.
Immunoreactivity of cyclophilin D in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) represents the hippocampus (10X the original magnification); (B), the cerebral cortex (10X the original magnification); (C), the cerebral cortex (40X the original magnification) and (D) quantification of CypD immunoreactivity in the cerebral cortex.
Synaptic and dendritic proteins
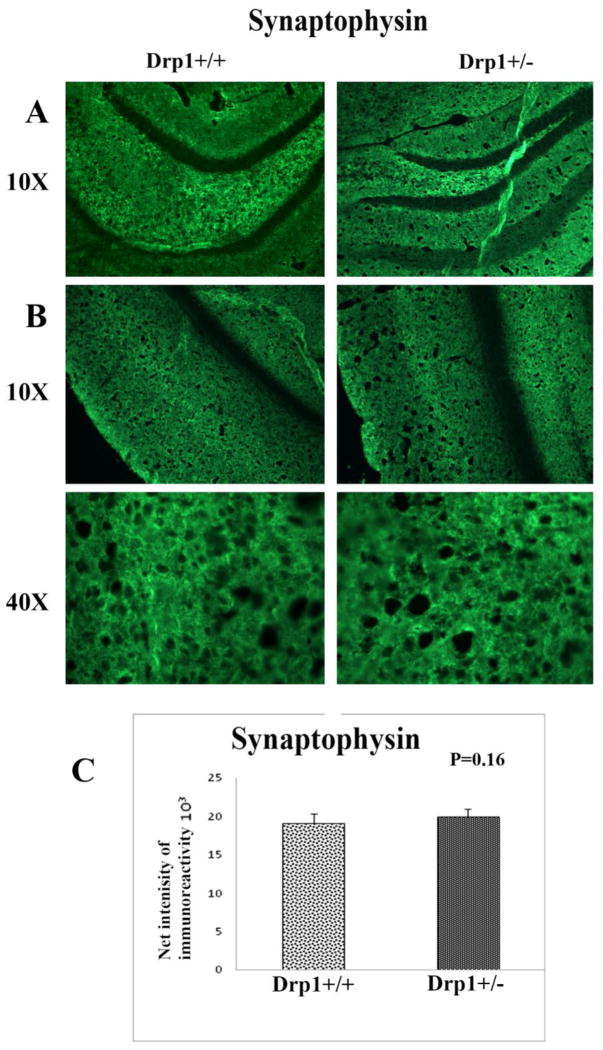
Our quantitative immunohistochemistry analyses of synaptic proteins revealed that immunoreactivities of synaptophysin (P<0.16) (Fig. 7) and PSD95 (P<0.29) (Fig. 8) were unchanged in the brain sections from the Drp1+/− and Drp1+/+ mice. These results indicate that a partial reduction in Drp1 does affect the expression of synaptic proteins. Similar to the immunoreactivity of synaptic proteins, the immunoreactivity of MAP2 did not change between the Drp1+/− and Drp1+/+ mice (Fig. 9) (P=0.11)
Figure 7.
Immunoreactivity of synaptophysin in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) represents the hippocampus (10X the original magnification); (B), the cerebral cortex (10X the original magnification); (C), the cerebral cortex (40X the original magnification) and quantification of synaptophysin immunoreactivity in the cerebral cortex.
Figure 8.
Immunoreactivity of PSD95 in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) represents the hippocampus (10X the original magnification); (B), the cerebral cortex (10X the original magnification); (C), the cerebral cortex (40X the original magnification) and (D) quantification of MAP2 immunoreactivity in the cerebral cortex.
Figure 9.
Immunoreactivity of MAP2 in brain sections from Drp1+/+ mice and Drp1+/− mice. (A) the cerebral cortex (10X the original magnification); (B), the cerebral cortex (40X the original magnification) and (C) quantification of MAP2 immunoreactivity in the cerebral cortex.
Mitochondrial Network
To determine mitochondrial network, we immunostained the hippocampal and cortical sections from Drp1+/+ and Drp1+/− mice using mitochondrial matrix marker, pyruvate dehydrogenase. As shown in Figure 10, we did not find increased mitochondrial network in cerebral cortex regions from Drp1+/− mice relative to Drp1+/+ mice. However, we observed slightly increased mitochondrial network in hippocampal neurons of Drp1+/− mice relative to Drp1+/+ mice.
Figure 10.
Immunoreactivity of mitochondrial matrix marker, pyruvate dehydrogenase from Drp1+/+ and Drp1+/− mice. (A) represents hippocampal section 9100X the orginal magnification) and (B) represents cerebral cortex (100X the original magnification. White arrows indicate elongated mitochondria.
Mitochondrial function
To determine whether partial deficiency of Drp1 expression affects mitochondrial function in Drp1/− mice, we characterized mitochondrial function by measuring H2O2 production, cytochrome oxidase activity, lipid peroxidation, and ATP production in cerebral cortex and cerebellum tissues from Drp1+/− mice and Drp1+/+ mice.
H2O2 production
Significantly decreased levels of H2O2 were found in cerebral cortex tissues from Drp1+/− mice relative to Drp1 +/+ mice (P<0.04) (Fig. 11). However, H2O2 levels were unchanged in the cerebellum of Drp1+/− mice relative to Drp1 +/+ mice. These findings suggest that partial reduction of Drp1 expression influences H2O2 levels in the cortex but not in the cerebellum.
Figure 11.
Mitochondrial functional parameters in Drp1+/+ mice and Drp1+/− mice. (A) represents hydrogen peroxide in cerebral cortex and cerebellum tissues from Drp1+/− mice and Drp1+/+ mice. Significantly reduced hydrogen peroxide production was found in the cerebral cortex of Drp1+/− mice compared to Drp1+/+ mice (P<0.05). (B) represents cytochrome oxidase activity. Cytochrome oxidase activity was unchanged in the cerebral cortex and cerebellum tissues from Drp1+/− mice compared to Drp1+/+ mice. (C) represents lipid peroxidation in cerebral cortex and cerebellum tissues from Drp1+/− mice and Drp1+/+ mice. Significantly reduced lipid peroxidation levels were found in the cerebral cortex of Drp1+/− mice compared to Drp1+/+ mice (P<0.05). (D) represents synaptosomal mitochondrial ATP. Synaptosomal mitochondrial ATP was unchanged in the cerebral cortex and cerebellum tissues from Drp1+/− mice compared to Drp1+/+ mice.
Cytochrome oxidase activity
Cytochrome oxidase activity, an indicator of complex IV of oxidative phosphorylation, was unchanged in the cerebral cortex (P=0.2) and cerebellum (P=0.07) from Drp1+/− mice relative to Drp1 +/+ mice (Fig. 11), indicating that a partial reduction of Drp1 did not alter cytochrome c oxidase activity.
ATP production
Similar to cytochrome oxidase activity, we did not observe any change in the level of ATP level for the cerebral cortex (P=0.6) and cerebellum (P=0.8) tissues from Drp1+/− and Drp1+/+ mice, respectively (Fig. 11).
Lipid peroxidation
We found significantly reduced levels of lipid peroxidation in the cerebral cortex tissues from the Drp1+/− mice relative to the Drp1+/+ mice (P<0.03) (Fig. 11). However, the levels were unchanged in the cerebellum tissues from the Drp1+/− and Drp1+/+ mice (P=0.51).
GTPase enzymatic activity
As shown in Fig. 12, GTPase Drp1 enzymatic activity was reduced in cerebral cortex tissue from both the Drp1+/− and Drp1+/+ mice, but not significantly reduced (P=0.10) (Fig. 12). GTPase Drp1 enzymatic activity was unchanged in cerebellum tissue from the Drp1+/− and Drp1+/+ mice (P=0.9). These findings suggest that partial reduction of Drp1 does not affect GTPase Drp1 enzymatic activity.
Figure 12.
GTPase enzymatic activity in Drp1+/+ mice and Drp1+/− mice. GTPase enzymatic activity was unchanged in the cerebral cortex and cerebellum tissues from Drp1+/− mice compared to Drp1+/+ mice.
Discussion
The objective of this study is to characterize mitochondrial, synaptic and dendritic proteins and mitochondrial function in Drp1 heterozygote knockout (Drp1+/−) mice. The Drp1+/− mice that express partial reduction of Drp1 are normal in terms of lifespan, fertility, and viability [26], and phenotypically these mice are similar to WT (Drp1+/+) mice. Characterization of Drp1+/− mice is critical because homozygote Drp1 knockout (or Drp1−/−) mice are embryonic lethal with reduced mitochondrial fission, leading to developmental and synaptic defects in the brain, and however, it was unclear, if synaptic and mitochondrial deficiencies are present in Drp1+/− mice. In the present study, we found no significant changes in mitochondrial, synaptic, and dendritic proteins in the Drp1+/− mice. Further, mitochondrial function and GTPase Drp1 enzymatic activity appeared to be normal in these mice. However, H2O2 and lipid peroxidation levels were significantly reduced in Drp1+/− mice compared to control, WT mice. Since synaptic and mitochondrial proteins were normal and lipid peroxidation and H2O2 were reduced, these findings implicate that partial reduction of Drp1 is expected to reduce excessive mitochondrial fragmentation, maintain mitochondrial, and synaptic functions in neurons, in diseased states, such as AD and HD.
Mitochondrial function in Drp1 heterozygote knockout mice
Mitochondrial cytochrome oxidase activity and ATP production were normal in the brain tissues from the Drp1+/− mice that we studied. However, lipid peroxidation and H2O2 levels were significantly reduced, compared to WT mice. The reduction in H2O2 may be related to partial deficiency of Drp1 since reduced Drp1 expression has been associated with a reduction in mitochondrial fragmentation, in turn possibly leading to decreased H2O2 production and lower lipid peroxidation that we found in the brain tissues from the Drp1+/− mice.
In AD and HD (and other neurodegenerative diseases), increased production of free radicals and lipid peroxidation have been identified as early changes in the diseased state, followed by altered mitochondrial enzyme activities and reduced ATP levels and neuronal damage. These abnormalities in mitochondrial function have been associated with mitochondrial structural changes [2,3,8,11,24], and these changes may occur primarily due to the abnormal interaction between mutant huntingtin and amyloid beta protein(s) and Drp1. Several recent reports support this notion in AD and HD [8,9,11,24]. In studies of mitochondria and amyloid beta in AD, researchers have found that amyloid beta interacts with Drp1 in an age and disease progression-dependent manner, induces elevated production of GTPase Drp1 enzymatic activity, and causes excessive mitochondrial fragmentation and mitochondrial dysfunction [8,24]. In investigations of mitochondria and mutant huntingtin in HD, two recent studies found that mutant huntingtin interacts with Drp1 mainly in disease-affected neurons [9,11], and this interaction increased as the disease progressed. They also found Drp1 interaction is polyQ repeat number in disease progression. These studies suggest that mutant huntingtin and amyloid beta proteins interact with Drp1, causing both structural and functional changes in mitochondria in AD and HD. These studies also implicate the reduction of amyloid beta and mutant huntingtin may reduce Drp1 interaction and decrease mitochondrial fragmentation that reduced production of either mutant proteins or Drp1 or both mutant proteins and Drp1 may protect affected neurons from AD and HD. The present findings of reduced H2O2 and lipid peroxidation in Drp1+/− mice support the implication that partial deficiency of Drp1 may reduce mitochondrial structural and functional effects in a disease state.
Mitochondrial, synaptic, and dendritic proteins in heterozygote Drp1 knockout mice
Possible factors for the embryonic lethality of homozygote Drp1 knockout mice are developmental and synaptic defects that may be primarily caused by reduced mitochondrial fission and elevated mitochondrial fusion in the axonal tracts of embryonic cells [26,27]. In contrast to the homozygote knockouts, the heterozygote Drp1 knockouts (Drp1+/− mice) are normal and viable. However, there have been no published reports of investigations into synapses and mitochondria. Our study is the first to investigate mitochondria activity and synaptic viability, and we have found no significant alterations in mitochondrial, synaptic, or dendritic proteins in the cerebral cortex and cerebellum tissues from the Drp1+/− mice. Partial reduction of Drp1 did not alter mitochondrial fission (Drp1, FIs1), fusion (Mfn1, Mfn2), or matrix proteins (CypD). Further, quantitative analyses of key synaptic and dendritic proteins – synaptophysin, PSD95, and MAP2 – were intact in the Drp1+/− mice, confirming that partial reduction of Drp1 may not be harmful to nondiseased neurons. Further research into the role of partial Drp1 deficiency in a diseased state, such as AD and HD, is still needed, to evaluate the extent of reduction that Drp1 can protect synaptic or neuronal damage in the presence of amyloid beta and mutant huntingtin. Therefore, Drp1+/− mice remain good candidates to study partial Drp1 deficiency in a diseased state.
Mitochondrial and synaptic deficiencies are reported to be associated with synaptic damage, neurodegeneration and clinical symptoms found in AD and HD patients [31–36]. Other neurodegenerative diseases, such as ALS, PD, and ataxias, may also be associated with synaptic damage and neurodegeneration. The increased production of defective mitochondria following the interaction of Drp1 and particular mutant proteins (e.g., amyloid beta, mutant huntingtin, mutant SOD1, mutant PD proteins, and mutant ataxins) may lead to the inability of mitochondria to transport to axons, dendrites, and terminals of neurons and may lead to the inability of mitochondria to produce sufficient quantities of ATP at synapses. These mitochondrial impairments may induce synaptic damage, and neurodegeneration, and may ultimately lead to clinical symptoms seen in AD and PD patients.
In summary, we found that Drp1+/− mice (Drp1 heterozygote mice) do not have mitochondrial and synaptic deficiencies and, further, that these mice show reduced oxidative stress (levels of H2O2 and lipid peroxidation). Thus, Drp1+/− mice are excellent candidates for further studies of partial Drp1 deficiency in mitochondrial structure and mitochondrial function, and in deficiencies in synaptic viability in AD and HD.
Research highlights.
We investigated mitochondrial activity and synaptic viability in Drp1+/− and Drp1+/+ mice.
No significant changes were found in synaptic, dendritic, and mitochondrial proteins in Drp1+/− mice compared to Drp1+/+ mice.
Mitochondrial function and GTPase Drp1 enzymatic activity appeared to be normal.
Free radicals and lipid peroxidation levels were significantly reduced in the Drp1+/− mice compared to Drp1+/+ mice.
Acknowledgments
This research was supported by NIH grants AG028072, RR000163 (to PHR) and GM089853 (HS) and Alzheimer Association grant IIRG-09-92429 (to PHR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Swerdlow RH. The neurodegenerative mitochondriopathies. J Alzheimers Dis. 2009;17:737–751. doi: 10.3233/JAD-2009-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du H, Guo L, Fang F, Chen D, Sosunov AA, McKhann GM, Yan Y, Wang C, Zhang H, Molkentin JD, Gunn-Moore FJ, Vonsattel JP, Arancio O, Chen JX, Yan SD. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer’s disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci USA. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 7.Chang DT, Reynolds IJ. Mitochondrial trafficking and morphology in healthy and injured neurons. Prog Neurobiol. 2006;80:241–268. doi: 10.1016/j.pneurobio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle DA, Reddy PH. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirendeb UP, Calkins MJ, Manczak M, Anekonda V, Dufour B, McBride JL, Mao P, Reddy PH. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzmicic J, Del Campo A, Lopez-Crisosto C, Morales PE, Pennanen C, Bravo-Sagua R, Hechenleitner J, Zepeda R, Castro PF, Verdejo HE, Parra V, Chiong M, Lavandero S. Mitochondrial Dynamics: a Potential New Therapeutic Target for Heart Failure. Rev Esp Cardiol. 2011;64:916–923. doi: 10.1016/j.recesp.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Krzywanski DM, Moellering DR, Fetterman JL, Dunham-Snary KJ, Sammy MJ, Ballinger SW. The mitochondrial paradigm for cardiovascular disease susceptibility and cellular function: a complementary concept to Mendelian genetics. Lab Invest. 2011;91:1122–1135. doi: 10.1038/labinvest.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camara AK, Bienengraeber M, Stowe DF. Mitochondrial approaches to protect against cardiac ischemia and reperfusion injury. Front Physiol. 2011;2:13. doi: 10.3389/fphys.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC. Bioenergetic Origins of Complexity and Disease. Cold Spring Harb Symp Quant Biol. 2011 Dec 22; doi: 10.1101/sqb.2011.76.010462. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook CC, Higuchi M. The awakening of an advanced malignant cancer: An insult to the mitochondrial genome. Biochim Biophys Acta. 2011 Sep 2; doi: 10.1016/j.bbagen.2011.08.017. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy PH, Reddy TP. Mitochondria as a therapeutic target for aging and neurodegenerative diseases. Curr Alzheimer Res. 2011;8:393–409. doi: 10.2174/156720511795745401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev. 2011;67:103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer CS, Osellame LD, Stojanovski D, Ryan MT. The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell Signal. 2011;23:1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama Y, Zhang Z, Sesaki H. Mitochondrial division: molecular machinery and physiological functions. Curr Opin Cell Biol. 2011;23:427–434. doi: 10.1016/j.ceb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manczak M, Mao P, Calkins MJ, Cornea A, Reddy AP, Murphy MP, Szeto HH, Park B, Reddy PH. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer’s disease neurons. J Alzheimers Dis. 2010;20(Suppl 2):S609–S631. doi: 10.3233/JAD-2010-100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2011;20:4515–4529. doi: 10.1093/hmg/ddr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy PH, Shirendeb UP. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington’s disease. Biochim Biophys Acta. 2012;1822:101–110. doi: 10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 28.Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 29.Drew B, Leeuwenburgh C. Method for measuring ATP production in isolated mitochondria: ATP production in brain and liver mitochondria of Fischer-344 rats with age and caloric restriction. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1259–R1267. doi: 10.1152/ajpregu.00264.2003. [DOI] [PubMed] [Google Scholar]
- 30.Hall ED, Bosken JM. Measurement of oxygen radicals and lipid peroxidation in neural tissues. Curr Protoc Neurosci. 2009;Chapter 7(Unit 7.17.1–51) doi: 10.1002/0471142301.ns0717s48. [DOI] [PubMed] [Google Scholar]
- 31.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–64. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 32.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 33.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 34.Folstein SE. Huntington’s Disease. Johns Hopkins University Press; 1990. [Google Scholar]
- 35.Montoya A, Price BH, Menear M, Lepage M. Brain imaging an cognitive dysfunctions in Huntington’s disease. J Psychiatry Neurosci. 2006;31:21–29. [PMC free article] [PubMed] [Google Scholar]
- 36.Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]