Abstract
The mechanism of light-triggered conformational change and signaling in light-oxygen-voltage (LOV) domains remains elusive in spite of extensive investigation and their use in optogenetic studies. The LOV2 domain of Avena Sativa phototropin1 (AsLOV2), a member of the Per-Arnt-Sim (PAS) family, contains an FMN chromophore that forms a covalent bond with a cysteine upon illumination. This event leads to the release of the carboxy terminal Jα helix, the biological output signal. Using mutational analysis, circular dichroism and NMR, we find that the largely ignored amino terminal helix is a control element in AsLOV2’s light-activated conformational change. We further identify a direct amino-to-carboxy terminal “input-output” signaling pathway. These findings provide a framework to rationalize the LOV domain architecture, as well as the signaling mechanisms in both isolated and tandem arrangements of PAS domains. This knowledge can be applied in engineering LOV-based photoswitches, opening up new design strategies and improving existing ones.
All kingdoms of life contain Per-Arnt-Sim (PAS) domains as part of signaling networks that respond to a diverse array of environmental stimuli and modulate a variety of output responses.1–4 These domains are usually part of larger proteins, some of which can contain multiple PAS domains. Many PAS domains are attached to signaling proteins such as kinases and phosphodiesterases3, others to membrane proteins5 and some function as single domain proteins.6 The PAS domains have very diverse sequences but all have a highly conserved 100–120 residue α/β fold termed the PAS core.7 Generally, the input sensor is a ligand contained in a binding pocket located on one side of the five stranded β sheet. The output is believed to be mediated by amino and/or carboxy termini, typically helices, which reside on the other side of the sheet3,8–13 (Fig. 1).
Fig. 1.
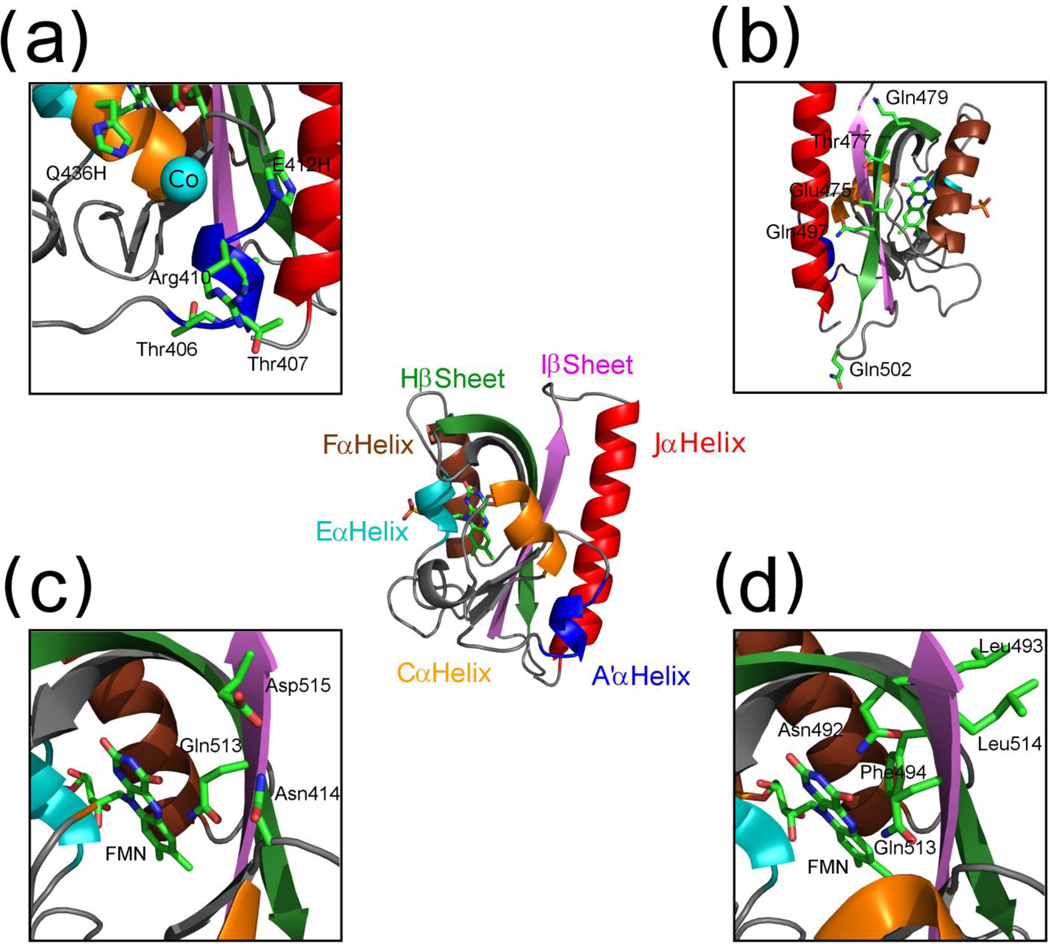
AsLOV2 structure. (a) A’α helix mutations and the biHis site with Co2+ atom (turquoise). (b) The location of residues hypothesized to stabilize the Jα helix through electrostatic interactions. (c) Hypothesized N414, Q513 and D515 network (Cα helix is truncated for clarity). (d) Residues hypothesized to transmit signal directly through Hβ and Iβ strands (Cα helix is truncated for clarity). Structural images were created using Pymol (Schrödinger, LLC).
In response to blue light, algae and plants use PAS-containing phototropins to activate signaling cascades that end in phototropism or chloroplast rearrangement.14–16 Phototropins utilize a subclass of PAS domains termed the Light-Oxygen-Voltage (LOV) domain. In phototropins, two LOV domains occur in series followed by a kinase domain. In Arabidopsis thaliana, either of the LOV domains can activate phosphorylation through the attached carboxy terminal kinase domain in vitro.17,18
The LOV domain is an ideal model to study PAS domain signaling because the signaling state is photoexcitable and reversible. Blue light activation of A. sativa LOV2 (AsLOV2) occurs when the non-covalently bound flavin mononucleotide (FMN) chromophore absorbs a photon and forms a metastable covalent bond between its C4a atom and the sulfur of the neighboring C450.19 This cysteine adduct decays in seconds-hours depending on the local environment20 and the covalent bond destabilization appears to be caused by a base catalyzed deprotonation of the FMN.20,21 After covalent bond formation, the protein undergoes a conformational change that results in the undocking of the carboxy terminal Jα helix, an event that triggers kinase activity.22,23 In particular, the Jα helix docking equilibrium shifts by ~100-fold from a mostly docked state in the dark to a mostly undocked condition in the lit state. 24–27 This ~ 3 kcal mol−1 shift in equilibrium free energy can be used to regulate effector activity.
Although output signals are known for AsLOV2 at a gross level, the molecular level mechanism by which the FMN-cysteinyl adduct formation results in undocking of the Jα helix located over 15 Å away remains largely unknown. Many studies suggest that signaling occurs through the β sheet separating the chromophore and the Jα helix3,22,24,26,28,29, potentially due to an increase in dynamics in the lit state.26 Recently, temperature dependent FTIR studies observed changes in both the α-helical and β-sheet region prior to the undocking of the Jα helix. This fast response suggests the presence of a S390-I2 state, although the origin is unclear of the early α-helical signal change.30 In crystallographic studies illumination of LOV2 crystals results in the backbone of the entire protein moving by only ~0.2 Å RMSD.31 Nevertheless, some side chain displacements and changes in hydrogen bonding patterns are observed in the region near the chromophore and the amino terminal A’α helix, which is interpreted as part of a structural signal.
Here, we find that adduct formation triggers unfolding of the A’α helix which in turn promotes the undocking of the adjoining Jα helix. We also investigate a number of previous hypotheses related to changes in β dynamics upon illumination. The role of the amino terminus in AsLOV2 signaling is consistent with other LOV domain studies, suggesting that this behavior is a widely used mechanism for the entire family. We also suggest that the direct communication between the spatially adjacent termini in phototropins provides a simple mechanism for propagating a signal through a tandem series of LOV domains to a protein’s output effector domain.
RESULTS
Mutational Analysis of AsLOV2
To identify how adduct formation triggers the undocking of the Jα helix, we examined the effects of mutations throughout the PAS core of the AsLOV2 domain, measuring the FMN photorecovery rate and Jα docking equilibrium (Fig. 1). Blue light illumination (λ~400–450 nm) and adduct formation resulted in a decrease in absorbance in the region near λmax = 448 nm. We measured the photorecovery rate by tracking absorbance at 448 nm for 29 mutations. Recovery times were within 10% of the WT (Asphot1 residues 404–560) protein for ten mutations (time constant τ = 81 ± 2s, Table 1). Five mutations located in or near the FMN binding site produced longer recovery times. Another ten mutations dispersed throughout the protein resulted in faster recovery times. Our results are consistent with prior studies which observed that the changes in the local environment or steric interactions near the FMN chromophore are responsible for the slowing of photorecovery, while faster recovery can occur upon changes in solvent accessibility resulting from structural perturbations distal to the chromophore.20
Table 1.
Secondary structure changes and photorecovery times
| Construct | δ222a | FMN τ (s) |
|---|---|---|
| WT | 0.30 | 81 |
| WT(404–546) W491Y/L408W | 0.30 | 78 |
| ΔJα | 0.15 | 82 |
| ΔJα W491Y/L408W | 0.16 | 79 |
| Δ408–411 ΔJα | 0.10 | 83 |
| T406A/T407A | 0.38 | 56 |
| T407P | 0.27 | 81 |
| T407W | 0.29 | 82 |
| Δ408–411 | 0.11 | 71 |
| L408D | 0.18 | 96 |
| R410P | 0.20 | 108 |
| R410G | 0.35 | 78 |
| Δ410–412 | 0.34 | 72 |
| K413A | 0.22 | 59 |
| N414V | 0.25 | ≥ 12 hrsb |
| F434L | 0.35 | 12 |
| E475A | 0.20 | 67 |
| Q479L | 0.30 | 49 |
| N492A | 0.18 | 54 |
| L493A | 0.11 | 121 |
| F494L | 0.15 | 206 |
| H495L | 0.15 | 85 |
| Q497A | 0.38 | 74 |
| R500A | 0.39 | 57 |
| Q502A | 0.30 | 78 |
| Q513A | 0.18 | 261 |
| L514A | 0.42 | 83 |
| D515V | 0.25 | 54 |
| H519A | 0.30 | 80 |
| E412H/Q436H | 0.27 | 39 |
δ222 = (θ222,lit − θ222,dark)/θ222,dark
Aggregation prone.
We also tracked the loss of secondary structure upon blue light photoexcitation using far-UV circular dichroism (CD) amplitude change at 222 nm (Table 1). In AsLOV2, photoexcitation resulted in a reduction in the magnitude of the (negative) CD signal at 222 nm (θ222). This decrease, which primarily reflects the result of a loss of helical structure, is attributed to the unfolding in the Jα helix, although other structural changes could be occurring as well. No obvious correlation existed between photorecovery times and light activated conformational changes.
The fractional change in CD signal at 222 nm of WT AsLOV2 in the dark state relative to lit state, δ222 = (θ 222,lit-θ 222,dark)/θ222,dark, was 0.30 ± 0.01. Notably, a construct lacking the Jα helix entirely (AsLOV2ΔJα, residues 404–520) still had a significant CD change, δ222 = 0.15 (Fig. 2a and Table 1). This result indicated that structures other than Jα helix undergo a conformation change upon illumination, although some portion of the residual CD change could be attributable to the FMN during light activation.
Fig. 2.
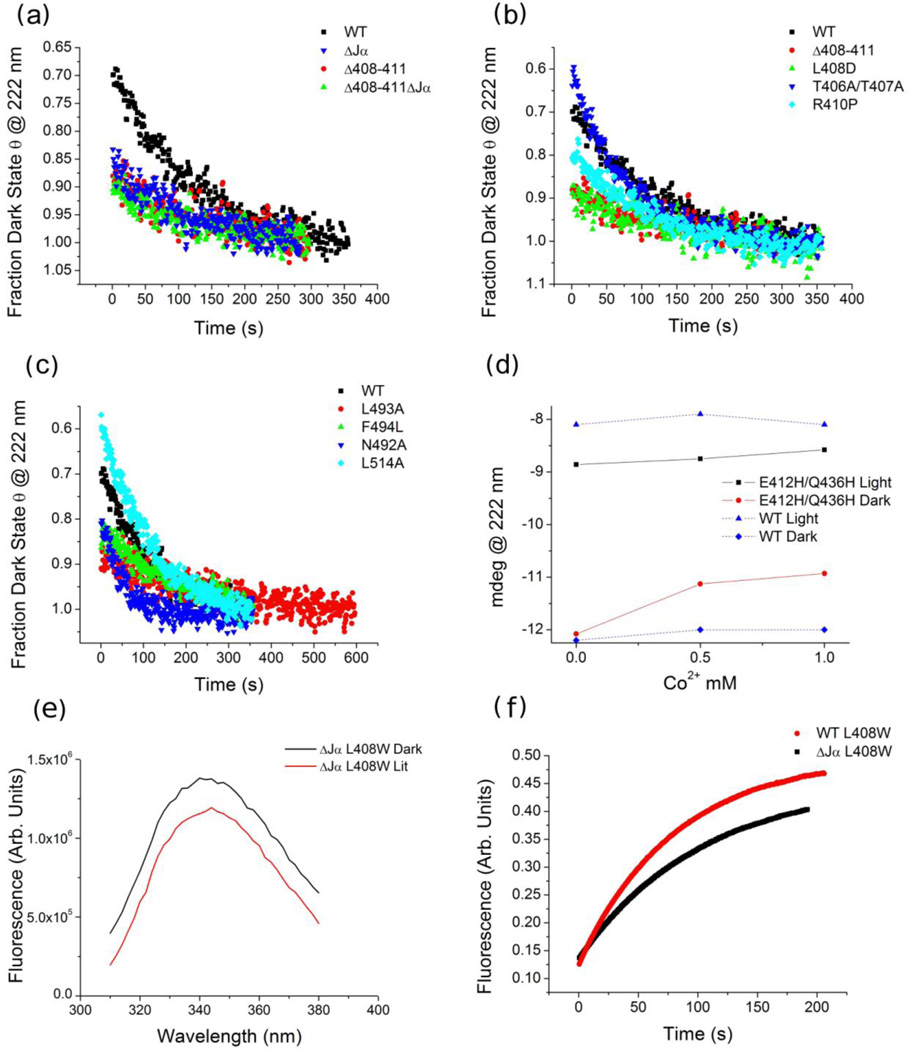
Photorecovery following CD and fluorescence. (a–c) The normalized dark state recovery of WT AsLOV2 and variants. (d) The effect of Co2+ on the lit (black) and dark state (red) ellipticity values of the WT and biHis mutant (E412H/Q436H). (e) Fluorescence traces during photo-recovery of AsLOV2 WT and ΔJα variants with a single tryptophan L408W on the A’α helix (λexcite = 280 ± 5 nm). (f) Fluorescence emission spectrum with λexcite = 295 ± 2 nm to minimize the contribution of the three tyrosines.
Conformational Changes Associated with Jα undocking
We sought to identify the light-sensitive conformational changes in the AsLOV2ΔJα construct by performing NMR measurements employing a Carr-Purcell-Meiboom-Gill (CPMG) refocusing component.32,33 For protein ensembles with µs-ms interconversion times between different states, CPMG measurements can provide quantitative information on weakly populated conformations.27,34,35 Using these methods, well-to-well kinetics and docking equilibrium of the Jα helix in the dark state were measured.27 A subsequent study found that mutations that altered the Jα helix docking equilibrium in the dark state, as determined by CPMG measurements, also changed the δ222 value.25
Here we employed a 15N-1H HSQC sequence which incorporates a CPMG pulse train within the INEPT transfer period. This pulse train suppresses the loss of signal due to multi-site exchange so that an increase in peak intensity reflects an increase in µs-ms dynamics. This protocol allows for the facile measurement of changes in conformational fluctuations across the entire protein for a large number of Jα containing AsLOV2 variants, as defined in terms of the ratio ICPMG/ INHSQC.
Using this pulse sequence, we probed multi-site exchange processes for WT AsLOV2 and seven variants (K413A, F434L, F452L, E475A, F494L, R500A and H519A). These variants had a range of values for dark-to-lit state changes in δ222 from 0.20 to 0.39. To quantify the change for each mutant, the CPMG values were averaged for the assignable residues in each of AsLOV2’s eleven secondary structural elements and three loops (Fig. 1). These values are compared to those observed in the WT AsLOV2. This averaging procedure across secondary structure elements helps remove possible bias from the selection of residues in regions that are prone to fraying or rigidity.
As previously demonstrated25, conformational fluctuations of the Jα helix in the dark state reflects a docking-undocking process that can be perturbed by mutations. In the present study, we hypothesized that if a mutation perturbed a structural element that was related to Jα helix undocking, the change in fluctuations of the structural element should be correlated with the changes of the Jα helix. Accordingly, we looked for structural elements which had changes in µs-ms fluctuations that were correlated with the changes in dark-to-lit state δ222 CD signal. We anticipated that this comparison would identify structures whose fluctuations were related to Jα undocking.
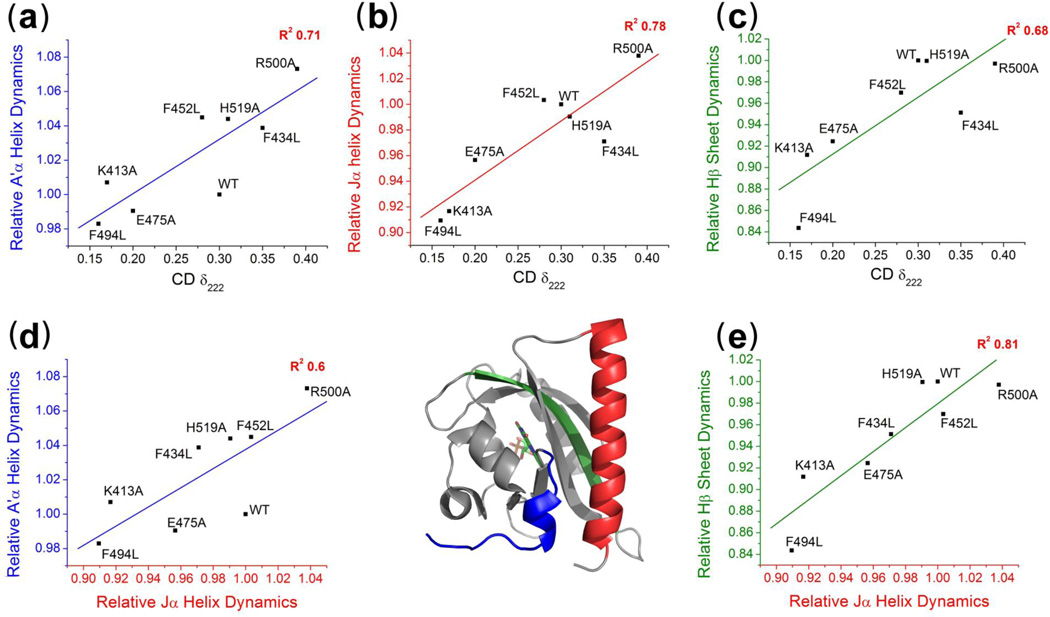
AsLOV2 has fourteen structural elements. We were able to measure dynamics for four or more residues in ten of these regions (Fig. 3). For three elements, the shift in the CPMG values upon mutations correlated with the shift in the dark-to-lit CD change, δ222, with a correlation coefficient stronger than R2 > 0.6 (Table S1), the Jα helix (R2 = 0.78), the Hβ strand (R2 = 0.68) and the A’α helix (R2 = 0.71). The strong correlation between δ222 and the Jα helix CPMG values indicates the change in Jα docking affinity in the lit state tracks with changes in its fluctuations in the dark state. This result reaffirms that the changes in dark state fluctuations of Jα helix probed by the CPMG measurement are largely attributable to changes in the Jα docking rates and equilibrium, consistent with earlier findings.25,27
Fig. 3.
Correlated dynamics across AsLOV2 structural elements identified through mutations. (a–c) Correlations of the CPMG values for secondary structure elements with the light-activated secondary structure change as measured by CD parameter δ222 for the A’α helix (blue), Jα helix (red) and Hβ strand (green). (d,e) Correlations of the CPMG values between the Jα helix and the A’α helix and Hβ strand.
This explanation can be extended to the other two strongly correlated regions, the Hβ strand and A’α helix. The Jα helix undocking is known to alter the NH chemical shifts of adjacent structural elements.36 As a result, a strong correlation is expected between the CPMG values for Jα helix and any structural element that senses the Jα helix undocking event. This situation applies to both the Hβ strand and A’α helix because the shifts in their CPMG values upon mutation correlate well with the shift for the Jα (R2Hβ = 0.81 and R2A’α = 0.6) and they are both in direct contact with the Jα helix (the Hβ strand has the most contacts with the Jα) (Fig. 3d,e). The CPMG values for the Gβ and Iβ strands correlate to lessor degree with the Jα helix dynamics (R2Gβ = 0.55 and R2Iβ = 0.47) and they are not well correlated with light activated conformational change as measured by δ222 (R2Gβ = 0.23 and R2Iβ = 0.26). These results suggest that the Gβ and Iβ strands experience only a mild conformational change upon Jα helix undocking or adduct formation. (Table S2). Overall, the correlations of the shift upon mutation of the CPMG values for the A’α helix, the Hβ strand and the Jα helix both with each other and with changes in δ222 can be attributable to the same process, Jα helix undocking.
Furthermore, any change in fluctuations identified by the CPMG measurements cannot be attributable to photoexcitation because the NMR measurements are conducted in the dark. Hence, the observed changes in Hβ and A’α helix fluctuations identified in the CPMG measurement in the dark do not result from changes in FMN’s photo-state. Rather, the CPMG data indicate that the observed increase in dark state fluctuations of these elements is a consequence of Jα undocking rather than the underlying cause of the light-induced undocking. These results along with the significant change in δ222 in the construct lacking the Jα helix provide a strong motivation for a reexamination of the mechanism of photo-induced undocking of the Jα helix.
The N-terminal A’α helix unfolding controls Jα undocking
In the crystal structure31, the small amino terminal A’α helix is in contact with the Jα helix (Fig. 1d). Although the protein backbone does not appreciably move in the crystal structure upon illumination, side chain rearrangements and electron density differences are observed in the region between the A’α helix and the chromophore.31 Accordingly, we created an AsLOV2 variant lacking both the Jα and A’α helices (AsLOV2ΔJαΔA'α, lacking residues 408–411 and 520–560). The fractional change in CD upon illumination is reduced in AsLOV2ΔJαΔA'α to δ222 = 0.10, as compared to the 0.15 and 0.30 observed in AsLOV2ΔJα and WT AsLOV2, respectively (Fig. 2a and Table 1). These results are particularly significant as they suggest that even in the absence of the Jα helix, the A’α helix is folded in the dark state and unfolds upon illumination. This photosensitive event could be the critical event that shifts the docking equilibrium of the Jα helix from favorable to unfavorable (Fig. 5a).
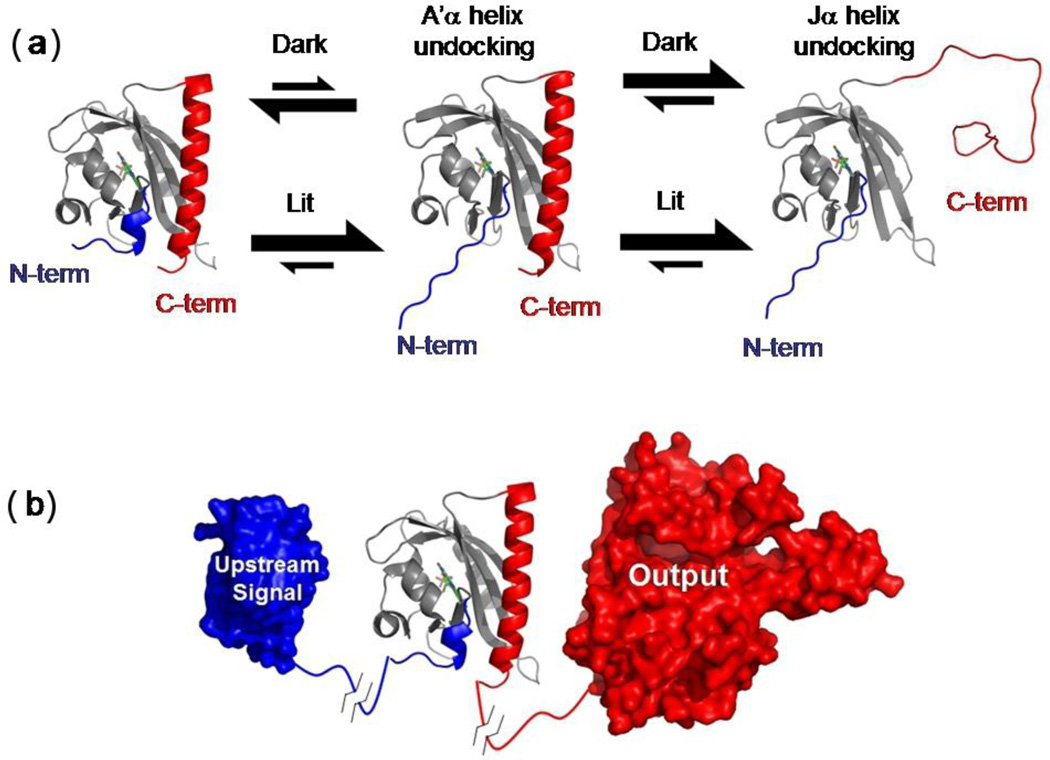
Fig. 5.
LOV docking equilibria, architecture and signaling. (a) Docking reactions of the A’α and Jα helices in the lit and dark states. For simplicity, only the route where the A’α undocking precedes the Jα undocking is shown. (b) In phototropins containing LOV2, the amino-to-carboxy terminal communication within each LOV domain may influence the other LOV domain via a carboxy-to-amino communication allowing for a greater output effect as compared to a single LOV domain.
To obtain additional, site resolved information about the photo-induced conformational changes related to the A’α helix, we replaced L408 with a tryptophan. In the crystal structure, this tryptophan would be partially buried by both the Jα and A’α helices. To focus on this tryptophan only, we removed the remaining tryptophan (W491Y) in the AsLOV2ΔJα and in a truncated WT construct (404–546), which lacks the putative unstructured carboxy region that contains another tryptophan.
Conformational changes were probed with fluorescence taking advantage of the sensitivity of tryptophan’s fluorescence to its environment. The fluorescence quantum yield of the WT and AsLOV2ΔJα constructs decreased by nearly the same amount upon excitation (Fig. 2e). Further, the emission spectrum of AsLOV2ΔJα underwent an ~ 4 nm red shift (Fig. 2f) upon excitation. Both features were to be expected if the tryptophan becomes more solvent exposed in the lit state, e.g. upon unfolding of the A'α helix, consistent with similar studies on LOV domains.37,38 While it may be considered that the decrease in quantum yield could also be explained by an electron transfer reaction from the tryptophan to the lit state FMN chromophore,39 the red shift was indicative of the tryptophan undergoing a change in its environment. These results support our hypothesis that the A’α helix is folded in the dark even upon truncation of the Jα helix, and that it undergoes photo-induced conformational change, which in turn promotes the unfolding the Jα helix in the WT construct.
To further examine this possibility, we created additional constructs that contain the Jα helix but a modified A'α helix. We either truncated a large portion (AsLOV2ΔA'α), or destabilized it using a helix-breaking proline substitution (R410P), or substituted an inward facing core residue with a carboxylic acid (L408D). In the L408D construct, we presumed that the burial of the charged acidic group would constitutively unfold the A’α helix, as the other core acidic substitutions had done for the Jα helix in LOV2 (e.g. I539E40) and various secondary structures in ubiquitin.41 We also strengthened the helix, replacing the two solvent exposed β-branched residues with helix stabilizing alanines (T406A/T407A).
For all three A’α helix-challenged variants, the change in CD signal upon illumination was greatly reduced. For the deletion (AsLOV2ΔA'α) and the charge buried variant (L408D), δ222 = 0.11 and 0.18, respectively (Fig. 2a and Table 1). The helix-stabilized variant, T406A/T407A, had δ222 = 0.37, a value even greater than the 0.30 observed in the WT AsLOV2 (Fig. 2b and Table 1).
For the AsLOV2ΔA'α and charge buried variants, the extent of undocking of the Jα helix was reduced compared to wild-type (i.e. lower δ222 value). This reduction could be due to the Jα being largely unfolded in the dark state or to a smaller fraction undocking in the lit state. To determine which situation was applicable, we conducted NMR 15N-1H HSQC measurements (Fig. S1). In contrast to the well dispersed wild type AsLOV2’s spectrum, the A’α-destabilized mutants had poorly dispersed, exchanged broadened and depleted spectra. These spectra were similar to that observed both for AsLOV2 in the light23 as well as for AsLOV2ΔJα indicating that the Jα helix is largely undocked in the dark. Conversely, the A’α-stabilized T406A/T407A variant had a wild-type 15N-1H HSQC spectrum and the CPMG measurements show a decrease in Jα helix dynamics (Table S2).
To further demonstrate that the docking equilibrium of the Jα helix can be controlled by interactions involving the A’α helix, we introduced a bihistidine (biHis) metal ion binding site between the Cα and A’α helices using substitutions E412H and Q436H (Fig. 1d). BiHis sites have been used in protein folding studies to identify regions that have come together at the rate limiting step by examining the effects of metal binding on folding rates.42–44 In spite of the biHis site being located over 10 Å away from the Jα helix, an increase in [Co2+] both decreased the θ222 amplitude in the dark as well as the magnitude of the light-induced change, δ222 (Fig. 2d). These two results can be explained by the A’α helix being pulled towards the Cα helix upon titration of Co2+ which results in the helix being farther away from the Jα helix weakening its interactions with the Jα helix. As a result, the Jα is docked less often in the dark state, and hence the fraction that can become undocked upon illumination is reduced (Fig. 5a).
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations have been performed on AsLOV2 in both the lit and dark states for hundreds of nanoseconds but they do not shown any of the major conformational changes seen during in vitro experimentation.4,28,45 This difference can be accounted for by the conformational changes occurring in vitro on micro to millisecond timescales. To overcome this issue, we created two less stable constructs in silico, AsLOV2ΔA’α and AsLOV2ΔJα to promote conformational changes (Fig. 4a,b). Our corresponding in vitro experiments indicated that when the A’α helix unfolds, the Jα helix should unfold but the opposite should not be true. To investigate this possibility in silico, we performed explicit solvent simulations using the CHARMM22 force field. Each simulation was run for at least 50 ns and repeated at least twice with different initial velocity distributions to test reproducibility.
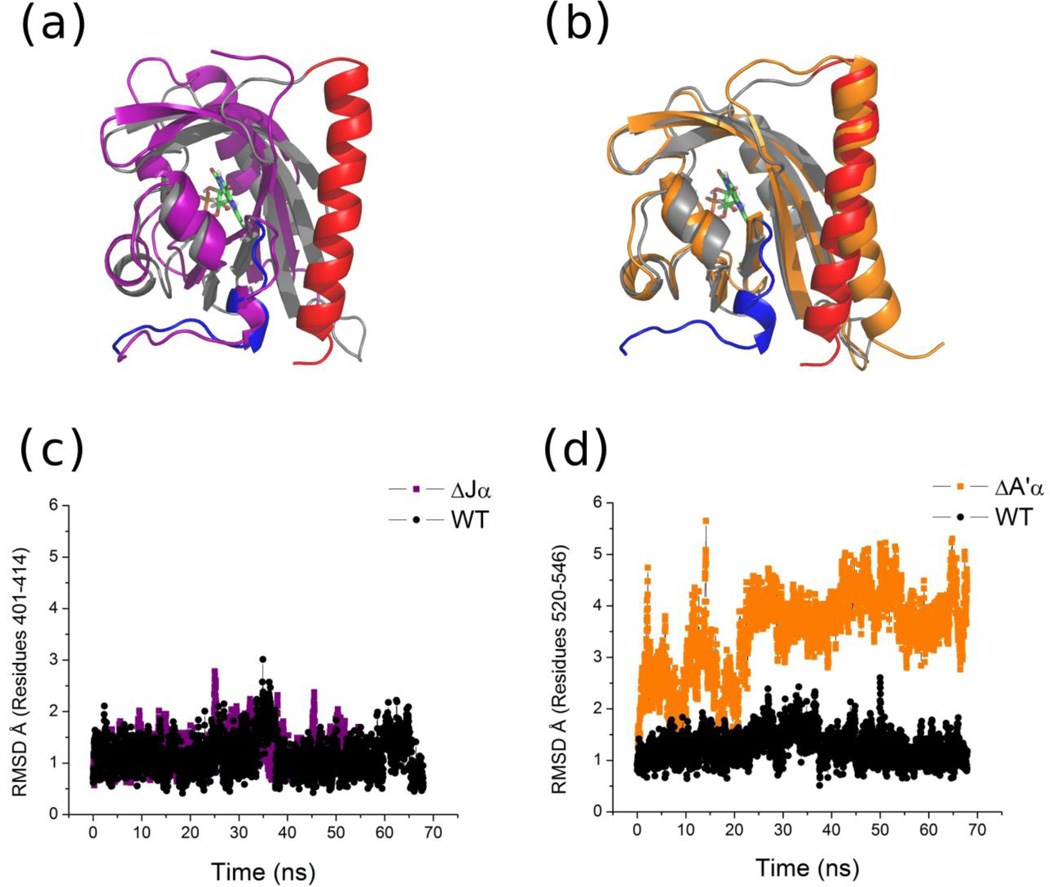
Fig. 4.
MD simulations of AsLOV2 constructs in the dark state.(a) Structural alignment of the final frame for WT AsLOV2 (grey) and AsLOV2ΔJα (purple). (b) Structural alignment of the final frame for WT AsLOV2 (grey) and AsLOV2ΔA’α (orange). (c) RMSD (Å) for the amino terminus (401–414) from the starting state for the WT AsLOV2 and AsLOV2ΔJα. (d) RMSD (Å) for the Jα helix (520–546) from the starting state for WT AsLOV2 and AsLOV2ΔA’α.
In the AsLOV2ΔJα trajectories, the A’α helix did not undergo any significant structural change. The RMSD of the A’α helix for both WT AsLOV2 and the AsLOV2ΔJα construct stayed at 1.3 Å over the course of the simulations (Fig. 4a,c). This small variance indicates that even without the Jα helix, the A’α helix has sufficient interactions to stay folded and docked on the PAS core over the length of the simulation. In contrast, the AsLOV2ΔA’α construct shows unwinding and unfolding of the Jα helix at the carboxy terminus (Fig. 4b). For the AsLOV2ΔA’α construct, the RMSD fluctuated up to 5 Å for Jα helix while it remained below 2 Å for the wild type protein (Fig. 4d). This difference indicates that the amino terminal region and the A’α helix are required to maintain the Jα helix’s native conformation. These simulations match our experimental results in that they find the docking of the Jα helix is more sensitive to the presence of the A’α helix than vice-versa.
In summary, our critical findings are i) disruption or stabilization of the A’α helix markedly affects the fraction of Jα helix that goes from a docked to undocked conformation upon illumination; ii) A’α helix conformation is light dependent; iii) this light-induce change does not depend on the presence of the Jα helix, but the conformational status of the Jα helix depends on the presence of the A’α helix, iv) the interactions between the two helices modulates the docking status of the Jα helix. Based on these results, we believe that the photo-controlled unfolding of the A’α helix shifts the equilibrium of the Jα helix from docked to undocked (Fig. 5a), although other factors involving the β sheet may contribute to the undocking equilibrium as well, as discussed below.
Role of the β sheet
In this section, we examine the contribution of the β sheet in the photo-induced Jα undocking event. As discussed in the NMR section, β sheet fluctuations increase in response to Jα undocking in both the dark and lit state. This result motivated us to test a number of hypotheses about the role of β sheet in Jα helix undocking. MD simulations observed that a reorientation of Q513 transmits stress through the Iβ strand, which results in the breakage of the Q497-D540 and Q479-E518 bonds, followed by undocking of Jα helix.28 To test this proposal, we made mutations Q479L and Q497A but still observed a δ222 value consistent with Jα helix undergoing light activated conformational change (Fig. 1b and Table 1). We then proceeded to make mutations to other residues that could form interactions with the Jα helix, T477V and Q502A, and still observed no change in δ222 as compared to the WT value. These two results do not support the proposal that changes upon illumination in the hydrogen bonding or electrostatic interactions between the β sheet and the Jα helix are responsible for undocking.
Next we set out to test the suggestion that N414, Q513 and D515 form a hydrogen bond network that changes upon light activation and modulates Jα helix undocking46 (Fig. 1c). For the mutations N414V, Q513A and D515V, only Q513A has a significant effect on δ222 (Table 1) suggesting a separate or non-unique role for these residues. Another suggestion is that a side chain rotamer of Q513 causes a bulge in the Iβ strand which could affect Jα helix docking affinity.4 Using CD and NMR, we analyzed mutations made on the Hβ and Iβ strands, on both the chromophore side (N492A, F494L and Q513A), and the Jα helix side (L493A and L514A) (Fig. 1a). All mutations reduced the δ222 value except L514A, which had an increased value (Fig. 2c and Table 1). The NMR spectra did not provide further information as all of the mutants are similar to WT except for L493A which was significantly different and indicative of structural disruption. This provides the most promising support for the involvement of the β sheet in photo-induced undocking. It is still unclear if mutations to the β sheet have an active role in light activation, e.g. specifically promoting the Jα undocking in the light state, or whether each mutation equally affects the dark and lit state Jα docking equilibrium. These data represent a basis for further analysis of the role of the β sheet.
DISCUSSION
Our major goal is to identify how absorption of a photon by the FMN chromophore in AsLOV2 results in the undocking of the Jα helix located over 15 Å away. We have found that the absence of A’α helix results in the undocking of the Jα helix even in the dark (Fig. 5a). Our CD and fluorescence measurements indicate that the A’α helix unfolds upon illumination even upon deletion the Jα helix. Hence, the docking affinity in the dark is favorable between the PAS core and the A’α helix but not between the core and the Jα helix. These observations argue that the photosensitivity of A’α helix modulates the undocking of the Jα helix. This result is supported by our MD simulations and the crystallographic studies where side chain movements near the A’α helix are interpreted to be part of the structural signal.31 In addition, conformational changes of the A’α helix can account for the FTIR-monitored decrease in helical content that occurs prior to the undocking of the Jα helix at low temperature in AsLOV2.30
Light-triggered conformation changes have been observed by NMR24,26 and FTIR22,29,30,47,48 in the β sheet that separates the chromophore and the Jα helix. These changes have been suggested to play a role in the undocking of the Jα helix. This hypothesis is supported by FTIR spectroscopy on Adantium phy3 LOV2 that observed conformational changes in Jα helix when mutations are made to residues homologous to F49448 and Q51326,47,49 in AsLOV2, located on the Hβ or Iβ strand respectively. Nevertheless, we note that in the absence of the A’α helix in AsLOV2, the docking equilibrium of the Jα helix is unfavorable even in the dark. Hence, the light sensitivity of the A’α helix, which is not located on the face of the β sheet, is sufficient to explain the photocycling of the Jα helix although changes in the β sheet could be a factor as well.
We have examined the behavior of 29 mutations in this study and have unpublished data on over 50 others located throughout AsLOV2. Nevertheless, we are unable to conclusively identify a residue that is essential for the light sensitivity of Jα helix docking except for residues in or near the A’α helix, or the adduct-forming C450. This result suggests that there is not a single signal propagating from the FMN to the Jα helix. Potentially, a structural rearrangement of multiple residues produces a state with weaker A’α docking affinity. The conformational adjustment, however, must be sufficiently minor that the protein’s hydrogen network remains because hydrogen exchange protection factors remain above 105 across most of the core of the protein in the lit state.23
Our CD studies of the double helix truncation, AsLOV2ΔA’αΔJα, indicate there is still a residual light activated conformational change, δ222 = 0.10. This residual may be due in part to changes in the Cα and Eα helices, the β sheet, as suggested by the FTIR studies,22,30,47,48, a shrinkage in the core50 or the FMN chromophore. Previous CPMG measurements on WT AsLOV2 found Rex > 4 s−1 for two residues in the Cα helix indicating dynamics on the same timescale as Jα helix unfolding.27 A conformational change in the Cα helix could be involved in the signaling mechanism from the chromophore to the A’α helix as it is located between the two.
The side chain dynamics are increased in the lit state according to 13C-1H HSQC measurements. 23 This result supports the notion that conformational entropy increases upon Jα undocking, ΔSundock, and this increase favors the undocking of the helix. However, for entropy to be a contributing factor in the light-triggered increase in Jα undocking, the conformational entropy in the lit state must be higher than in the dark undocked state (i.e. ΔSlitundock > ΔSdarkundock). We are unaware of any such demonstration.
Another unknown factor is whether the strength of the A’α helix- Jα helix interactions in the docked conformation are decreased upon illumination. Such a decrease, however, is unnecessary for photo-triggering; mutations which have the same effect in both states can still alter the magnitude of the dark-lit switching.25 Irrespective of whether the A’α helix interactions change, it does not affect our conclusion that the docking of the A’α helix itself is light sensitive and this sensitivity can control the docking status of the Jα helix.
A central question is why do organisms continually use the PAS fold in signaling networks? PAS domains have highly conserved structures even when there is little sequence similarity. A lack of understanding may be attributed to the fact that many PAS domain structures that have been solved contain only the α/β core, lacking either the amino terminal or carboxy terminal regions and sometimes both, which may be critical to signaling.8–13
Our finding that the AsLOV2 output signal, Jα release, is promoted by the A’α undocking has implications that may explain the widespread use of the PAS fold. An analysis of the structure and function of PAS domains suggests that the amino terminus either undergoes a conformational change (e.g., in Vivid13, AsLOV2, PYP51 and hERG5,52) or is positioned at a dimer interface in PAS domains that experience a change in quaternary structure (e.g., in YtvA53,54, CitA55 and Vivid56). This observation along with the diversity of sequences flanking the α/β core suggests that the region occupied by the A’α helix is an allosterically sensitive but variable region of the PAS domain that provides each domain with its unique function.
In this light, the PAS domain architecture can be viewed as a large β sheet which provides a central scaffold. Attached on one side is the input sensor which is held in place by variable regions, for example, the Eα and Fα helices and the turn between the Gβ and Hβ strands in AsLOV2. Along the edge and continuing on to the other side of the β sheet are the variable amino- and carboxy termini (Fig. 5b). These elements are involved in protein-specific signaling mechanisms.
In crystallographic studies on Vivid, Zoltowski et al.13 identified a network of salt bridges and hydrogen bonds as the signaling conduit between the chromophore and its amino terminus. They suggest that a conformational change of the large amino terminus in Vivid occurs through changes in side chain interactions around the helix. This helix is positioned where the Jα helix is in AsLOV2 rather than where the A’α helix is located. Potentially, the 36 residues upstream of the amino terminal helix which are unresolved in the Vivid structure are positioned where the A’α helix is in AsLOV2, and they undergo a conformational change upon adduct formation for similar reasons.
Given our finding of a coupling between the neighboring A’α and Jα helices in AsLOV2, we suggest that signaling in tandem LOV domains can occur through interactions involving their termini (Fig. 5b). This coupling is distinct from a signaling mechanism which goes through the entire body of protein, as often is implied by the simplistic linear arrangement of protein “boxes” illustrating tandem LOV domain architectures. The mechanism involving amino-to-carboxy transmission may explain how signals can be propagated in PAS domains which lack an input sensor (e.g. chromophore). In addition, a number of PAS domain proteins, including phototropins, contain more than one PAS domain in tandem. The A. thaliana phototropin 2 contains sequentially arranged LOV1, LOV2, and kinase domains17. When LOV2 is photoinactivated with a C450A mutation, LOV1 still can activate the kinase domain potentially by signaling through LOV2.17 Although there can be a considerable number of residues (50–100) between LOV1 and LOV2 in phototropins, the A’α helix in these LOV2 domains typically has 100% identity across species. Potentially the signal from LOV1 propagates to the kinase domain through the amino terminus of LOV2.
In prokaryotic organisms, most linkers between the PAS domains are 20–40 residues3, which generally is less than observed in phototropins. Möglich et al.46 identified that in prokaryotic tandem PAS structures, the PAS A and PAS B domains are linearly oriented in a head-to-tail arrangement. In Sinorhizobium meliloti DctB (PDB ID: 3E4P), we note that the helical interconnecting linker and the end of the long amino terminal helix of PAS A are positioned in the PAS B domain at the analogous positions occupied by the A’α and Jα helices in LOV2 domain, respectively. The interactions between these two helices in DctB and in other tandems, may help explain the required heptad phasing of the interconnecting helix. Furthermore, the flanking helices that are amino and carboxy terminal to the core contribute to the interface in many prokaryotic dimers.3
The AsLOV2 domain has been employed as the light-sensitive control element in optogenetic studies that have already much shown promise for addressing questions in cell biology.25,57–59 Our elucidation of the A’α helix-initiated undocking of the Jα helix can aid in the design of new synthetic AsLOV2-based photoswitches. Whereas all previous AsLOV2 fusions have focused on the Jα helix undocking, the realization that the A’α helix also undergoes a conformational change opens up the possibility of other design strategies involving the amino terminus.
METHODS
Cloning, Expression and Purification
A clone of A. sativa phot1 LOV2 (404–560) with a His6-Gβ1 fusion was generously provided by K. Gardner (University of Texas Southwestern Medical Center, Dallas). Mutations were made using the quickchange site directed mutagenesis strategy. All proteins were expressed in Escherichia coli BL21 (DE3) cells grown in M9 minimal medium supplemented with 15NH4Cl (1 g/L) at 37 °C to an OD 600 nm of 0.6 and induced with 1 mM IPTG. Cultures were then incubated for ~18 hours at 18 °C and pelleted and frozen. Frozen pellets were resuspended in 50 mM Tris 100 mM NaCl and 0.01% SDS and cells were lysed using sonication and clarified with centrifugation at 10000g for 45 min. The proteins were then purified using metal affinity chromatography and exchanged into 50 mM Tris, 1 mM EDTA, 5 mM DTT, pH 8. The His6-Gβ1 tag was removed by incubating overnight at 20 °C with His6-TEV protease. Metal affinity chromatography was then used to remove His6-Gβ1 and His6-TEV protease from the solution. The final protein contains residues GEF on the amino terminus and G on the carboxy terminus as cloning artifacts. Proteins were run on a Sephadex S100 size exclusion column (GE Healthcare) and if the UV-Vis 280/447 nm ratio differed greatly from ~2.6–2.9, proteins were further purified using anion exchange chromatography.
Circular Dichroism
Circular dichroism was performed using a Jasco J-715 spectrometer with ~1 µM protein in 50 mM NaH2PO4 200 mM NaCl pH 7 at 22 °C using a 1 cm pathlength cuvette. Light activating kinetic traces were collected by illuminating the sample using a 40 W white LED (BT DWNLT A, TheLEDlight.Com) for 30 seconds to photosaturate. Measurements were then made every second with a 2 nm bandwidth and fit with a single exponential decay using Origin 7 software (MicroCal Inc., Northampton, MA).
UV-Vis
UV-Vis spectra were acquired using an Olis HP 8452 Diode Array (Bogart, GA) with a 1 cm path length cuvette at 22 °C. Kinetic traces were acquired after illumination with photosaturation assayed by a severely diminished signal. Measurements were taken every 1 to 30 seconds depending on the recovery rate at the FMN central peak maximum, usually 448 nm, until complete recovery of the signal was achieved.
Fluorescence
Tryptophan fluorescence recovery was measured using a Jasco J-715 spectrometer with λexcitation = 275 ±5 nm and a Hanamatsu H5784-06 PMT with a 300–400 nm filter for emission detection. Experiments were performed at 22 °C by exposing the samples to a 40 W white LED. All experiments were performed in a 1 cm path length cuvette and normalized to 1 µM concentration using the FMN absorbance at 448 nm. Tryptophan λmax emission experiments were performed on a Horiba Fluorolog 3 fluorometer with excitation at 295 ± 2 nm with protein concentrations of 10 – 100 µM.
NMR Measurements
All NMR experiments were performed on a Varian Inova 600 MHz spectrometer with a cryoprobe at 25 °C. Proteins were concentrated to 100–500 µM and 10% D2O was added prior to experimentation. NMR data was processed using NMRPipe60 and analyzed using SPARKY61. NMR experiments were run using standard pulse sequences available in Varian BioPack, 15N-1H HSQC (gNhsqc) and CPMG 15N-1H HSQC (CPMGgNhsqc). The CPMG 15N-1H HSQC applies a CPMG pulse train during the standard INEPT transfer period thereby increasing the intensity of peaks by reducing the effect of conformational exchange (Rex) on the signal.33 The contribution of Rex to the 1HN transverse relaxation rates was determined by calculating the ratio of the peak intensities between the CPMG 15N-1H HSQC (ICPMG) and 15N-1H HSQC (INHSQC) spectra. Signal intensity gain due to the CPMG pulse train can be be approximated by ICPMG / INHSQC = eRext.62 To calculate the conformational dynamics of secondary structural elements the ICPMG / INHSQC ratio of assignable peaks for each element in each mutant were averaged and a ratio was calculated by comparing to the exact averaged assignable residues in WT AsLOV2. The NMR assignments of WT AsLOV2 were provided by K. Gardner (University of Texas Southwestern Medical Center, Dallas).
Molecular Dynamics
Simulations were run using standard procedures described elsewhere.4 Briefly, MD simulations were performed using the NAMD63 molecular dynamics package with the CHARMM22 force field on the dark state of AsLOV2 (PDB ID: 2V0U). All simulations were run on Teragrid using the Ranger resource. All simulations were performed by solvating the protein in TIP3P water box and neutralizing the charge of the system with six sodium ions. Parameters for FMN were taken from Freddolino et al. (2006).45 All simulations were performed using an NPT ensemble at a temperature of 300 K, a pressure of 1 atm, a 2-fs timestep and full electrostatics calculated every two steps using the PME method. The amino terminal truncations were made by removing every residue before 414 in the structure file. The Jα helix truncations were made by removing every residue after 520 in the structure. All simulations were performed at least twice with different initial velocity distributions to test reproducibility.
We investigate the mechanism of light-triggered conformational changes in AsLOV2
Spectroscopy indicates that the N-terminal helix unfolds upon illumination
This event triggers the unfolding of the C-terminal Jα helix
Results can be used in other LOV-based photoswitches and new designs strategies
An N- to C-terminal signaling mechanism is possible in other PAS domains.
Supplementary Material
Acknowledgements
We thank D. Strickland, S. Crosson, K. Moffat, M. Glotzer, and K. Gardner for helpful discussions and comments on the manuscript. This work was supported by research and training grants from the US National Institutes of Health (GM088668 to T.R.S. and M. Glotzer, 5T32GM007183-34 to B. Glick), and the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust (to T.R.S., M. Glotzer and E. Weiss). This research was supported in part by the National Science Foundation through TeraGrid resources provided by the Texas Advanced Computing Center under grant number TG-MCB090169.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim Proteins in Environmental Adaptation. Annual Review of Physiology. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 2.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 3.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song SH, Freddolino PL, Nash AI, Carroll EC, Schulten K, Gardner KH, Larsen DS. Modulating LOV Domain Photodynamics with a Residue Alteration outside the Chromophore Binding Site. Biochemistry. 2011 doi: 10.1021/bi200198x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 6.Borgstahl GE, Williams DR, Getzoff ED. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 7.Hefti MH, Francoijs KJ, de Vries SC, Dixon R, Vervoort J. The PAS fold - A redefinition of the PAS domain based upon structural prediction. European Journal of Biochemistry. 2004;271:1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x. [DOI] [PubMed] [Google Scholar]
- 8.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of Ligand Binding within a Completely Buried Cavity in HIF2 alpha PAS-B. Journal of the American Chemical Society. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada S, Sugimoto H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-Linked Histidine Kinase and the Response Regulator Complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Ayers RA, Moffat K. Changes in quaternary structure in the signaling mechanisms of PAS domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama Y, Nakasako M, Okajima K, Iwata A, Kashojiya S, Matsui Y, Tokutomi S. Light-Induced Movement of the LOV2 Domain in an Asp720Asn Mutant LOV2-Kinase Fragment of Arabidopsis Phototropin 2. Biochemistry. 50:1174–1183. doi: 10.1021/bi101689b. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Sayed N, Baskaran P, Beuve A, van den Akker F. PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J Biol Chem. 2008;283:1167–1178. doi: 10.1074/jbc.M706218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 15.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 16.Liscum E, Briggs WR. Mutations in the Nph1 Locus of Arabidopsis Disrupt the Perception of Phototropic Stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka D, Tokutomi S. Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13337–13342. doi: 10.1073/pnas.0506402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 19.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandre MTA, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JTM. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto A, Iwata T, Sato Y, Matsuoka D, Tokutomi S, Kandori H. Light signal transduction pathway from flavin chromophore to the J alpha helix of Arabidopsis phototropin1. Biophys J. 2009;96:2771–2778. doi: 10.1016/j.bpj.2008.12.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 24.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 25.Strickland D, Yao XL, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nature Methods. 2010;7 doi: 10.1038/nmeth.1473. 623-U18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash AI, Ko WH, Harper SM, Gardner KH. A Conserved Glutamine Plays a Central Role in LOV Domain Signal Transmission and Its Duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao XL, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J alpha photoswitch. Nature Chemical Biology. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peter E, Dick B, Baeurle SA. Mechanism of signal transduction of the LOV2-Jalpha photosensor from Avena sativa. Nat Commun. 2010;1:122. doi: 10.1038/ncomms1121. [DOI] [PubMed] [Google Scholar]
- 29.Koyama T, Iwata T, Yamamoto A, Sato Y, Matsuoka D, Tokutomi S, Kandori H. Different role of the Jalpha helix in the light-induced activation of the LOV2 domains in various phototropins. Biochemistry. 2009;48:7621–7628. doi: 10.1021/bi9009192. [DOI] [PubMed] [Google Scholar]
- 30.Alexandre MT, van Grondelle R, Hellingwerf KJ, Kennis JT. Conformational heterogeneity and propagation of structural changes in the LOV2/Jalpha domain from Avena sativa phototropin 1 as recorded by temperature-dependent FTIR spectroscopy. Biophys J. 2009;97:238–247. doi: 10.1016/j.bpj.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 32.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 33.Mulder FAA, Spronk CAEM, Slijper M, Kaptein R, Boelens R. Improved HSQC experiments for the observation of exchange broadened signals. Journal of Biomolecular Nmr. 1996;8:223–228. doi: 10.1007/BF00211169. [DOI] [PubMed] [Google Scholar]
- 34.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 35.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450 doi: 10.1038/nature06410. 838-U13. [DOI] [PubMed] [Google Scholar]
- 36.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J Am Chem Soc. 2004;126:3390–3391. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 37.Jentzsch K, Wirtz A, Circolone F, Drepper T, Losi A, Gartner W, Jaeger KE, Krauss U. Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry. 2009;48:10321–10333. doi: 10.1021/bi901115z. [DOI] [PubMed] [Google Scholar]
- 38.Losi A, Ternelli E, Gartner W. Tryptophan fluorescence in the Bacillus subtilis phototropin-related protein YtvA as a marker of interdomain interaction. Photochemistry and Photobiology. 2004;80:150–153. doi: 10.1562/2004-03-17-RC-116.1. [DOI] [PubMed] [Google Scholar]
- 39.Eisenreich W, Fischer M, Romisch-Margl W, Joshi M, Richter G, Bacher A, Weber S. Tryptophan (13)C nuclear-spin polarization generated by intraprotein electron transfer in a LOV2 domain of the blue-light receptor phototropin. Biochemical Society Transactions. 2009;37:382–386. doi: 10.1042/BST0370382. [DOI] [PubMed] [Google Scholar]
- 40.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-J alpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Z, Sosnick TR. Protein vivisection reveals elusive intermediates in folding. J Mol Biol. 397:777–788. doi: 10.1016/j.jmb.2010.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosnick TR, Krantz BA, Dothager RS, Baxa M. Characterizing the Protein Folding Transition State Using psi Analysis. Chem. Rev. 2006;106:1862–1876. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- 43.Baxa M, Freed KF, Sosnick TR. Quantifying the Structural Requirements of the Folding Transition State of Protein A and Other Systems. J. Mol. Biol. 2008;381:1362–1381. doi: 10.1016/j.jmb.2008.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krantz BA, Sosnick TR. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nature Struct. Biol. 2001;8:1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- 45.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophysical Journal. 2006;91:3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moglich A, Yang XJ, Ayers RA, Moffat K. Structure and Function of Plant Photoreceptors. Annual Review of Plant Biology, Vol 61. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 47.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto A, Iwata T, Tokutomi S, Kandori H. Role of phe1010 in light-induced structural changes of the neol-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- 49.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 50.Eitoku T, Nakasone Y, Matsuoka D, Tokutomi S, Terazima M. Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. Journal of the American Chemical Society. 2005;127:13238–13244. doi: 10.1021/ja052523i. [DOI] [PubMed] [Google Scholar]
- 51.Harigai M, Yasuda S, Imamoto Y, Yoshihara K, Tokunaga F, Kataoka M. Amino acids in the N-terminal region regulate the photocycle of photoactive yellow protein. J Biochem. 2001;130:51–56. doi: 10.1093/oxfordjournals.jbchem.a002961. [DOI] [PubMed] [Google Scholar]
- 52.Ng CA, Hunter MJ, Perry MD, Mobli M, Ke Y, Kuchel PW, King GF, Stock D, Vandenberg JI. The N-Terminal Tail of hERG Contains an Amphipathic alpha-Helix That Regulates Channel Deactivation. Plos One. 2011;6 doi: 10.1371/journal.pone.0016191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurk M, Dorn M, Kikhney A, Svergun D, Gartner W, Schmieder P. The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study. J Mol Biol. 403:78–87. doi: 10.1016/j.jmb.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 54.Moglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Vaidya AT, Chen C-H, Dunlap JC, Loros JJ, Crane BR. Structure of a Light-Activated LOV Protein Dimer That Regulates Transcription. Science Signaling. 2011;4 doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461 doi: 10.1038/nature08241. 104-U111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. Journal of Molecular Biology. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 61.Kneller DG, Kuntz ID. Ucsf Sparky - an Nmr Display, Annotation and Assignment Tool. Journal of Cellular Biochemistry. 1993:254–254. [Google Scholar]
- 62.van Tilborg PJA, Mulder FAA, de Backer MME, Nair M, van Heerde EC, Folkers G, van der Saag PT, Karimi-Nejad Y, Boelens R, Kaptein R. Millisecond to microsecond time scale dynamics of the retinoid X and retinoic acid receptor DNA-binding domains and dimeric complex formation. Biochemistry. 1999;38:1951–1956. doi: 10.1021/bi982526q. [DOI] [PubMed] [Google Scholar]
- 63.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.