Abstract
In this study, chemical feature based pharmacophore models of MMP-1, MMP-8 and MMP-13 inhibitors have been developed with the aid of HypoGen module within Catalyst program package. In MMP-1 and MMP-13, all the compounds in the training set mapped HBA and RA, while in MMP-8, the training set mapped HBA and HY. These features revealed responsibility for the high molecular bioactivity, and this is further used as a three dimensional query to screen the knowledge based designed molecules. These pharmacophore models for collagenases picked up some potent and novel inhibitors. Subsequently, docking studies were performed for the potent molecules and novel hits were suggested for further studies based on the docking score and active site interactions in MMP-1, MMP-8 and MMP-13.
Keywords: Collagenases, Osteoarthritis, S1' loop, Pharmacophore, Induced Fit, HypoGen
Background
Variety of biological processes such as embryonic development, tissue remodeling and tissue repair involve controlled degradation of extra cellular matrix (ECM). This feature is a fundamental part of growth, invasion, and metastasis of malignant tumors [1]. Matrix metalloproteinases (MMPs), a family of extracellular zinc-dependent neutral endopeptidases, are collectively capable of degrading essentially all ECM components. They are the prime factors indulged in breaking down the extracellular matrix contributing to disease states such as arthritis, atherosclerosis, tumor cell invasion and metastasis [2–4]. They are classified according to their domain structure into collagenases, gelatinases, stromelysins, matrilysin and membrane type MMPs (MT-MMPs) [5].
Among MMPs, collagenases are intimately involved in collagen homeostasis by post-translational proteolytic degradation. They principally comprise MMP-1 (collagenase-1), MMP-8 (collagenase-2) and MMP-13 (collagenase-3) [6]. Collagenases are the only endogenous enzymes that can readily cleave the triple helical domain of fibrillar collagens I, II and III. Collagen degradation is commenced by collagenases by making a sitespecific cleavage about three-quarter of the distance from Nterminus, followed by spontaneous collagen denaturation [7]. These interstitial collagenases degrade type I, II and III collagen in cartilage; this is a committed step in the development of rheumatoid arthritis as well as osteoarthritis and is revealed by elevated levels of these collagenases [8, 9].
Collagenases show interesting differences in the crystal structures, despite being highly homologous to one another. X ray analyses of the enzyme–inhibitor complex of collagenases suggested that the S1' subunit is a selectivity pocket for collagenase inhibitors [10–13]. The S1' subsite, also called the S1'-specificity pocket, is the most prominent pocket within the catalytic domain of collagenases. Differences in the relative size and shape of the S1' pockets in MMP-1, MMP-8 and MMP-13 suggest that this pocket is a critical determinant of MMP inhibitor selectivity [1]. The quite flexible loop forms a major portion of the S1' pocket and it undergoes a conformational change on inhibitor binding [14, 15]. The loop is of the same length in MMP-8 and MMP-13 and two residues are shorter in MMP-1[16]. A comparison of the available 3D structure of MMP-1, MMP-8 and MMP-13 shows the variability of amino acid residues in the S1' loop. This variability of the amino acid residues in the S1' loop causes difference in the shape of loops [16]. The structural features of these enzymes are most decisive in determining MMP substrate specificity and thereby inhibitor specificity which is enclosed within the catalytic domain [17]. Synthetic inhibitors specifically targeting MMP-1, MMP-8 and MMP-13 are unclear. Selectivity is more vital in minimizing the detrimental effects during long term medical treatment [18]. It has been reported that side effects were observed in the clinical studies of collagenase inhibitors, because they showed broadspectrum inhibition. Therefore, specific inhibition of MMP-1, MMP-8 and MMP-13 are considered to be an attractive target in drug discovery research [19, 20].
In the present study, we have generated pharmacophore models using Catalyst [21, 22] software for a diverse set of collagenase inhibitors (MMP-1, MMP-8 and MMP-13) with an aim to obtain pharmacophore model that would provide the chemical features responsible for activity. These pharmacophore features were used to screen the databases to find novel inhibitors. Further induced fit docking was performed to validate these inhibitors against MMP-1, MMP-8 and MMP-13. This in turn would be able to provide useful knowledge for developing specific new and active drug candidates targeting collagenases (MMP-1, MMP-8 and MMP-13).
Methodology
Pharmacophore modeling using Catalyst:
A set of 337 MMP-1 inhibitors with activity ranging from 0.4 nM to 100000 nM, 148 MMP-8 inhibitors with activity ranging from 0.13 nM to 78000 nM and 371 MMP-13 inhibitors with activity ranging from 0.16 nM to 100000 nM were selected from GOSTAR (gostardb.com). The molecules were divided into training and test set for the development and validation of pharmacophore models. The selection of training set is the most crucial part as it determines the quality of generated pharmacophore models. In this study, 21 of 337, 22 of 148 and 21 of 371 compounds were chosen for training set based on the diversity observed in their chemical structures and experimental activities for MMP-1, MMP-8 and MMP-13 respectively. The remaining compounds were used as test set for pharmacophore validation process. All the molecules were exported and then minimized using modified CHARMM force field in catalyst package [21, 22]. For each molecule, a maximum of 250 conformations were generated using the ‘best quality’ conformational search option within Catalyst's ConFirm module. It generates the conformations using ‘Poling’ algorithm. The molecules were then submitted to the catalyst hypothesis generation.
Model validation and Knowledge based screening:
The best pharmacophore hypothesis was used initially to screen 316 MMP-1, 126 MMP-8 and 350 MMP-13 test set molecules. The same model has also been used to select potent molecules from 10,000 library molecules designed using Scaffold Hoping (Knowledge based screening). Library molecules were generated based on the knowledge of binding interaction of known ligands reported with MMP-1, MMP-8 & MMP-13 and also the common features necessary for biological activity of molecule [24–26]. These molecules were built using Cerius2 software [27] and conformations for each compound were generated using best conformational analysis. These molecules were further screened for their activities using the developed pharmacophore models.
Ligand preparation:
Ligand structures were built using Maestro v9.1 and geometrically minimized using OPLS_2005 force field by ligprep module of Maestro 9.1 (Schrödinger suite, LLC) [28]. Ligprep produces a single, low energy, 3D structure for each input structure with various ring conformations, ionization states and tautomers using various criteria including molecular weight or specified numbers and types of functional groups present.
Protein preparation:
Protein preparation and refinement studies were performed on MMP-1 (PDB ID: 1HFC), MMP-8 (PDB ID: 3DPE) and MMP-13 (PDB ID: 1XUC) using protein preparation module (Schrödinger suite, LLC) [28] in which the water molecules were removed, hydrogen atoms were added, bond orders were assigned and orientation of hydroxyl groups were optimized. Finally, energy minimization was carried out using default constraint of 0.3 Å RMSD and OPLS 2005 force field.
Induced fit docking:
Induced fit docking method for protein structures of MMP-1, MMP-8 and MMP-13 was performed using Induced fit docking of Schrodinger package [28]. During docking process, the ligands were optimized using OPLS or MMFF force field, thus changing its conformation to find the best fit that can closely fit to the S1' pocket of MMP-1,MMP-8 and MMP-13. The binding affinity of each protein and ligand complex was reported as Glide Score [29]. All graphic images were picturised using PyMol program (www.pymol.org) [30]. Non-bonded interactions like hydrophobic was observed using LigPlot program [31] and these interactions can increase the binding affinity between target drug interfaces.
Result & Discussion
Synthetic inhibitors taken for this study include hydroxamate, non-hydroxamate, carboxylate, phosphinate, aminocarboxylates, thiol, sulphonates, pyrrolidine, diazepine, etc., which tends to have a greater inhibition towards MMPs [28]. Hydroxamate diazepine and phenyl sulfonyl acetamide inhibitors are potent inhibitors of MMP-9 and MMP-13 both in vitro and in vivo, for osteoarthritis in rabbit model [32, 33]. Since many of them have been quite potent and in some cases fairly selective against collagenases, these molecules were further taken as a basement to design the target specific inhibitors for collagenases (MMP-1, MMP-8 and MMP-13) using pharmacophore modeling.
Pharmacophore generation and validation studies using HypoGen:
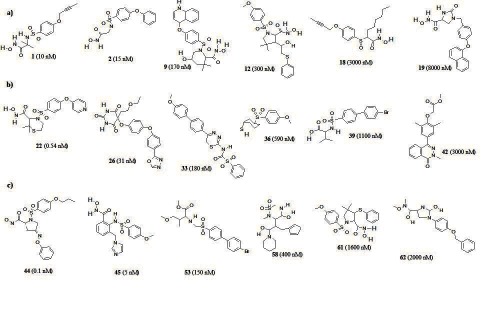
Ten hypotheses were generated using 21 diverse training set molecules for MMP-1 and MMP-13, and 22 molecules for MMP- 8 in HypoGen. (Figure 1 a, b & c) show some of the molecules selected as the training set for MMP-1, MMP-8 and MMP-13. The best hypothesis for MMP-1 and MMP-13 consists of 1) one hydrogen bond acceptor, 2) one hydrogen bond donor and ring aromatic whereas MMP-8 consists of 1) two hydrogen bond acceptor, 2) one hydrogen bond donor and one hydrophobic.
Figure 1.
Structures of some of the training set molecules for (a) MMP-1, (b) MMP-8 & (c) MMP-13 (experimental IC50 values, in parentheses).
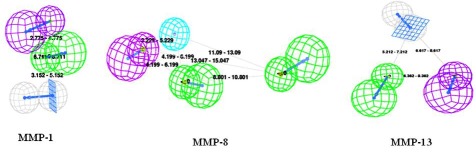
The values of ten hypotheses such as cost, correlation (r), and root-mean-square deviations (RMSD) are statistically significant Table 1A, 1B & 1C (see supplementary material). The pharmacophore (Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP-13 respectively) having high correlation coefficient (r), lowest total cost, and lower RMSD value was chosen to estimate the activity of test set. The best models Hypo-1,11 and 21 for MMP-1, MMP-8 and MMP-13 respectively has been given in (Figure 2) and the parameters that describe Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP-13 respectively are shown in Table 1A, 1B & 1C (see supplementary material).
Figure 2.
Hypogen feature with its distance constraints, features are color coded with green: hydrogen bond acceptor, magenta: hydrogen bond donor, white: ring aromatic and blue: hydrophobic for MMP-1, MMP-8 and MMP-13.
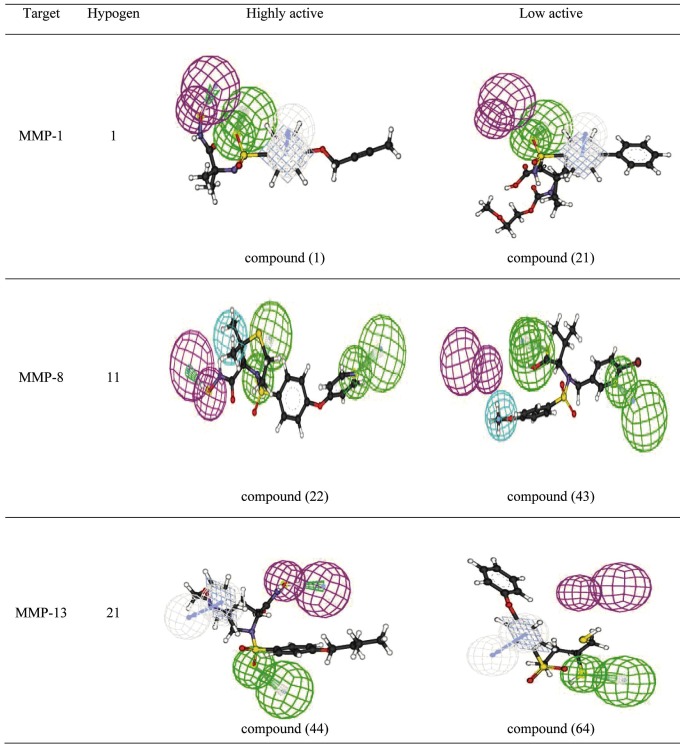
Two statistical methods were employed to rank the ten resultant hypotheses. In the first method, all the ten hypotheses were evaluated using a test set of known MMP-1, MMP-8 and MMP-13 inhibitors, which are not included in the training set. Predicted activities of the test set were calculated using all ten hypotheses and correlated with the experimental activities. Of the ten hypotheses, the best hypothesis Hypo-1 is characterized by the highest cost difference (58.88), lowest RMSD value error (0.87) and with correlation (0.89) for MMP-1. Hypo-11 is best for MMP-8, with highest cost difference (64.95), lowest RMSD value error (1.04) and correlation (0.85). Hypo-21 is the best hypothesis for MMP-13 with highest cost difference (63.90), lowest RMSD value error (1.15) and correlation (0.87). These results conclude that Hypo-1, 11 and 21 are best ranking pharmacophore for MMP-1, MMP-8 and MMP-13 respectively, among the 10 hypotheses obtained. In MMP-1 and MMP-13, all the compounds in the training set map hydrogen bond acceptor (HBA) and ring aromatic (RA), while in MMP-8, the training set map HBA and hydrophobic (HY) and these features revealed that they should be mainly responsible for the high molecular bioactivity. Thus this should be taken into account in discovering or designing novel inhibitors. The most active compounds 1, 22 and 44, has a highest fitness score of 6.80, 8.01 and 8.20 sequentially, when mapped Hypo-1, 11 and 21 to MMP-1, MMP-8 and MMP-13 respectively (Figure 3) whereas the least active compounds 21, 43 and 64 maps to a lowest value of 4.8, 4.13 and 5.73 (Figure 3). It is evident that as error, weight and configuration components are very low and not deterministic to the model, the total pharmacophore cost is also low and close to the fixed cost. Also, as total cost is less than the null cost, this model accounts for all the pharmacophore features and has a good predictability power. A second statistical test includes calculation of false positives, false negatives, enrichment and goodness of hit to determine the robustness of hypotheses. Under all validation conditions, Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP-13 respectively performed superior as compared to the other hypotheses and demonstrated excellent prediction of MMP-1, MMP-8 and MMP-13 inhibitory activities of the training set compounds Table 2A, 2B & 2C (see supplementary material).
Figure 3.
Shows Hypo-1, 11&21 mapping to highly active and low active compounds for MMP-1, MMP-8 & MMP-13
Analyzing the results, in MMP-1 out of 7 highly active molecules, 5 were predicted correctly as highly active, and the rest were predicted as moderately active. Among the 9 moderately active molecules, 6 molecules were predicted as moderately active, 2 were predicted as highly active and 1 was predicted as low active molecule. Out of 5 low active molecules, one was predicted as moderately active and remaining was predicted as low active. In MMP-8 out of 10 highly active molecules, 8 were predicted correctly as highly active, and the rest were predicted as moderately active. Among the 7 moderately active molecules, 2 molecules were predicted as highly active 4 were predicted as moderately active and 1 was predicted as low active molecule. Out of 5 low active molecules, 2 were predicted as moderately active and rest was predicted as low active. In MMP-13, out of 8 highly active molecules, 6 were predicted correctly as highly active, and the rest were predicted as moderately active. Among the 9 moderately active molecules, all molecules were predicted as moderately active. Out of 4 low active molecules, 1 was predicted as moderately active and rest was predicted as low active. Activities of the compounds were correctly predicted and fit values also confer a good measure of how well the pharmacophoric features of Hypo-1, Hypo-11 and Hypo-21 for MMP-1, MMP-8 and MMP-13 respectively were mapped onto the chemical features of the compounds. The best models Hypo-1,11 and 21 for MMP-1, MMP-8 and MMP-13 respectively has been given in (Figure 2) and the parameters that describe Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP-13 respectively are given in Table 1A, 1B & 1C (see supplementary material). Figure 3 shows all the features of Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP-13 respectively (acceptor, donor, hydrophobic and ring aromatic) that were mapped onto the highly active compounds of training set (1, 22, and 44) and onto the inactive compound of training set 21, 43 and 64 for MMP-1, MMP-8 and MMP-13 respectively. The correlation values along with above predictions make the pharmacophore suitable to predict molecular properties well. Hypo-1, 11 and 21 was used to search the test set of known MMP-1, MMP-8 and MMP-13 inhibitors respectively. Database mining was performed using the BEST flexible searching technique. The results were analyzed using a set of parameters such as hit list (Ht), number of active percent of yields (%Y), percent ratio of actives in the hit list (%A), enrichment factor of 2.91, 2.82 and 2.96 (E), false negatives, false positives, and goodness of hit score of 0.75, 0.70 and 0.80 (GH). Hypo-1, 11 and 21 (for MMP-1, MMP-8 and MMP-13 respectively) succeeded in the retrieval of 80% of the active compounds. An enrichment factor of 2.91, 2.82, 2.96 and a GH score of 0.75, 0.70, 0.80 indicates that the quality of the model is acceptable.
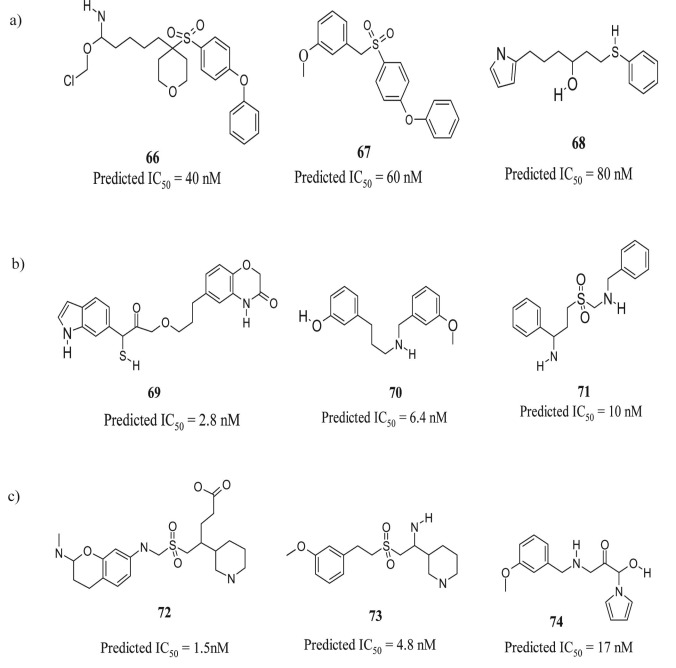
Overall, a strong correlation was observed between the predicted Hypo-1, 11, 21 and the experimental activity for MMP-1, MMP-8, and MMP-13 inhibitory (IC50) of the training and test compounds. However, Hypo-1, 11 and 21 models has a greater tendency to show false positives. This could be attributed to high structural similarity in the active and inactive MMP-1, MMP-8 and MMP-13 inhibitors, resulting in inability to discriminate this pattern by pharmacophore models. We have selected Hypo-1, 11 and 21 for MMP-1, MMP-8 and MMP -13 respectively as a 3D query to search a subset of knowledge based designed database of 10,000 compounds to retrieve compounds with novel structural scaffolds and desired features. The initial screening of Hypo-1, 11 and 21 yielded 3000 compounds and further cluster analysis of these hits corresponded to 220 unique cluster representatives. We further extended this study to structure-based design to limit the number of false positive hits and to further understand the binding of inhibitors to the active site of all three MMPs. (Figure 4 a, b & c) shows some of the identified and optimized potent lead molecules through virtual library screening for MMP-1, MMP-8 and MMP-13.
Figure 4.
Some of the newly identified potent lead molecules for (a) MMP-1, (b) MMP-8 & (c) MMP-13.
Docking studies for MMP-1, MMP-8 and MMP-13:
For furhter validation, docking is performed for 220 hits (MMP- 1, MMP-8 & MMP-13) using Induced fit docking mode of Maestro. Most of the compounds show hydrogen bonding interactions with the S1 pocket residues. Docking results shows that known (yellow green) and newly identified compounds (purple) occupy the S1' loop region with the almost similar conformations. Especially, highly active compounds are forming at least two specific or unique hydrogen bond interactions in the S1' loop and shows high glide score with collagenases (MMP-1, MMP-8 and MMP-13). The docking scores and the hydrogen bonding interaction for newly identified molecules with MMP-1, MMP-8 and MMP-13 were shown in Table 3 (see supplementary material).. To investigate the ligand binding affinity at the hydrophobic S1' pocket, we further estimated the hydrophobic interactions using LIGPLOT software (Figure 5). The hydrogen and hydrophobic interactions of highly active ligands for MMP-1, MMP-8 and MMP-13 are discussed below.
Figure 5.
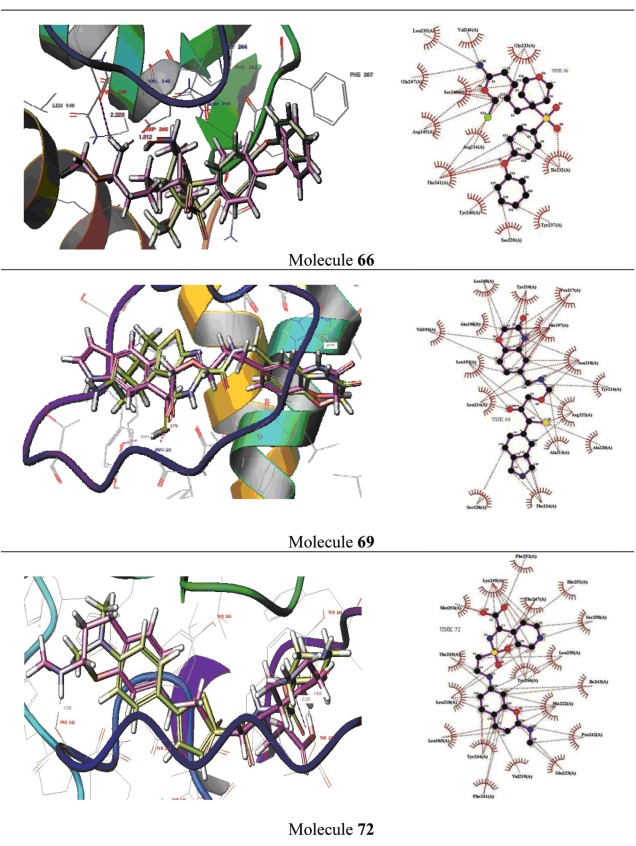
Shows docked conformations of molecule 66, 69 & 72 in the S1 loop of MMP-1, MMP-8 & MMP-13 respectively. Dotted lines represent hydrogen bonds. Hydrophobic interactions are represented as arcs. Ligand represented as ball and sticks.
Binding mode of molecule - 66 into the S1' loop of MMP-1:
Docking results shows that both the ligands occupy the S1' loop region with the almost similar conformation with a glide score - 7.816 and -7.089 Kcal/mol. LigPlot software was used to understand the in-depth interaction pattern between both the ligands and MMP-1. Hydrophobic interaction was identified with amino acid residues Leu 140, Phe 207, Arg 214, Ile 232, Gly 233, Leu 235, Tyr 237, Ser 239, Tyr 240, Phe 242, Ser 243, Asp 245 and Gln 247. Both the ligands form hydrogen bonding interaction with the Val 246 and specific amino acid residue Asp 245 in the S1' loop region of MMP-1 (Figure 5).
Binding mode of molecule - 69 into the S1' loop of MMP-8:
Docking results shows that both the ligands occupy the S1' loop region with the almost similar conformation with a glide score - 7.240 and -7.216 Kcal/mol. For the above two ligands hydrophobic interaction was seen with amino acid residues Leu 160, Leu193, Val 194, His 197, Glu 198, Ala 213, Leu 214, Pro 217, Asn 218, Tyr 219, Ala 220, Arg 222, Thr 224 and Ser 228 using LigPlot software. Phenyl group in both the ligands occupy the solvent exposed region including His 207, Ser 228 and Thr 224. Both the ligands form hydrogen bonding interaction with the Pro 217 which is unique among MMP-8 and also form pi-pi stacking with the Arg 222 in the S1' loop region of MMP-8 (Figure 5).
Binding mode of molecule -72 into the S1' loop of MMP-13:
Docking results shows that both the ligands occupy the S1' loop region with the almost similar conformation with a glide score - 10.892 and -9.203 Kcal/mol. Using LigPlot software, hydrophobic interaction was seen with amino acid residues Leu 185, Leu 218, Val 219, His 222, Leu 239, Phe 241, Pro 242, Ile 243, Thr 245, Tyr 246, Thr 247, Lys 249, Ser 250, His 251, Phe 252 and Met 253. Both the ligands form hydrogen bonding interaction with the specific amino residue with the Met 253. Molecule 72 additional ly forms interaction with the Thr 247 and Pro 242 and also forms pi-pi stacking with the Tyr 244 in the S1' loop region of MMP-13 (Figure 5).
Conclusion
Statistically validated pharmacophore models were generated for collagenase inhibitors to locate the spatial arrangement of features, which are necessary for biological activity. Out of them, Hypo-1, 11 and 21 performed superior for MMP-1, MMP- 8 and MMP-13 respectively and also showed excellent prediction for inhibitory activities of the test set compounds and well complemented with receptor active sites, compared to the other hypotheses. The best hypothesis for MMP-1 and MMP-13 consists of one hydrogen bond acceptor, one hydrogen bond donor and ring aromatic, whereas MMP-8 consists of two hydrogen bond acceptors, one hydrogen bond donor and one hydrophobic group. These pharmacophore models were used to retrieve the molecules from the databases which were further validated using docking studies. Finally, three structurally diverse compounds with high Glide scores and interactions with critical active site amino acids for MMP-1, MMP-8 and MMP-13 were identified. From this study, we suggest that these molecules can be used for further studies and also serve as potential leads against collagenase infections.
Supplementary material
Acknowledgments
I acknowledge my colleague Saranyah.K, S.R.M University, India and GVK Biosciences, India for their constant encouragement and support.
Footnotes
Citation:Kalva et al, 8(7): 301-308 (2012)
References
- 1.H Nagase, et al. Cardiovasc Res. 2006;69:562. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 2.CJ Malemud. Front Biosci. 2006;11:1696. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- 3.C Amalinei, et al. Rom J Morphol Embryol. 2010;51:215. [PubMed] [Google Scholar]
- 4.S Chakraborti, et al. Mol Cell Biochem. 2003;253:269. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 5.H Nagase, JF Woessner. J Biol Chem. 1999;274:21491. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 6.T Donahue, et al. Hernia. 2006;10:478. [Google Scholar]
- 7.H Nagase, et al. Biol Chem. 1997;378:151. [Google Scholar]
- 8.V Mattot, et al. J Cell Sci. 1995;108:529. doi: 10.1242/jcs.108.2.529. [DOI] [PubMed] [Google Scholar]
- 9.J Martel-Pelletier, et al. Lab Invest. 1994;70:807. [Google Scholar]
- 10.M Sawa, et al. Bioorg Med Chem Lett. 2002;12:581. doi: 10.1016/s0960-894x(01)00796-x. [DOI] [PubMed] [Google Scholar]
- 11.JC Spurlino, et al. Proteins. 1994;19:98. [Google Scholar]
- 12.G Pochetti, et al. J Med Chem. 2009;52:1040. doi: 10.1021/jm801166j. [DOI] [PubMed] [Google Scholar]
- 13.CK Engel, et al. Chem Biol. 2005;12:181. [Google Scholar]
- 14.B Lovejoy, et al. Nat Struct Biol. 1999;6:217. doi: 10.1038/6657. [DOI] [PubMed] [Google Scholar]
- 15.GE Terp, et al. J Med Chem. 2002;45:2675. doi: 10.1021/jm0109053. [DOI] [PubMed] [Google Scholar]
- 16.BG Rao. Curr Pharm Des. 2005;11:295. doi: 10.2174/1381612053382115. [DOI] [PubMed] [Google Scholar]
- 17.S Rowsell, et al. J Mol Biol. 2002;319:173. doi: 10.1016/S0022-2836(02)00262-0. [DOI] [PubMed] [Google Scholar]
- 18.D Georgiadis, A Yiotakis. Bioorg Med Chem. 2008;16:8781. doi: 10.1016/j.bmc.2008.08.058. [DOI] [PubMed] [Google Scholar]
- 19.K Maskos, et al. Biochimie. 2005;87:249. [Google Scholar]
- 20.M Sawa, et al. Bioorg Med Chem Lett. 2002;12:581. doi: 10.1016/s0960-894x(01)00796-x. [DOI] [PubMed] [Google Scholar]
- 21.Catalyst, Version 4.11, Accelry's Inc. San Diego, CA, USA: 2007. [Google Scholar]
- 22.EA Hecker, et al. J Chem Inf Comput Sci. 2002;42:1204. doi: 10.1021/ci020368a. [DOI] [PubMed] [Google Scholar]
- 23.AK Debnath. J Med Chem. 2002;45:41. doi: 10.1021/jm010360c. [DOI] [PubMed] [Google Scholar]
- 24.B Lovejoy, et al. Nat Struct Biol. 1999;6:217. doi: 10.1038/6657. [DOI] [PubMed] [Google Scholar]
- 25.G Pochetti, et al. J Med Chem. 2009;52:1040. doi: 10.1021/jm801166j. [DOI] [PubMed] [Google Scholar]
- 26.LG Monovich, et al. J Med Chem. 2009;52:3523. doi: 10.1021/jm801394m. [DOI] [PubMed] [Google Scholar]
- 27.Cerius2, version 4.8, Accelrys Inc., San Diego, USA: 2007. [Google Scholar]
- 28.RA Friesner, et al. J Med Chem. 2006;49:6177. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 29.PYMOL, version 1.2. Delano Scientific LLC. San Carlos, CA, USA.: [Google Scholar]
- 30.AC Wallace, et al. Protein Eng. 1995;8:127. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 31.JI Levin, et al. Bioorg Med Chem Lett. 1998;8:2657. doi: 10.1016/s0960-894x(98)00473-9. [DOI] [PubMed] [Google Scholar]
- 32.V Aranapakam, et al. J Med Chem. 2003;46:2376. doi: 10.1021/jm0205550. [DOI] [PubMed] [Google Scholar]
- 33.S Vadivelan, et al. Eur J Med Chem. 2009;44:2361. doi: 10.1016/j.ejmech.2008.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.