Abstract
Glutathione S-transferases (GSTs) enzymes are critical antioxidant and detoxification system responsible for long-term existence of nematodes in host species. Hence, 16 phytochemicals predicted and reported to have potential nematicidal activity have been docked to GST enzyme of Meloidogyne incognita to assess their binding affinity and inhibitory activity. In vitro effects of these phytochemicals from in silico results have been done for validation of docking studies and efficacy in GST inhibition of following compounds such as alpha- pinene, alpha- terpineol, beta- caryophyllene, capsaicin, cinnamic acid, citronellol, curcumin, eugenol, geraniol, isoeugenol, linalool, myristicin, neral, NVA (N-vanillylnonanamide), piperine, vanillin have been revealed. Nematode inhibition in vitro bioassay for selected compounds could conclude that maximum mortality was observed with highest concentrations of beta- caryophyllene (78%) followed by eugenol (61.6%), cinnamic acid (55%) and N-vanillylnonanamide (49%). These findings thus suggest that the above phytochemicals could be potentially developed as nematicidal molecules against M. incognita infections.
Keywords: Meloidogyne incognita, Docking, Glutathione S-transferase(s), Phytochemicals
Background
Plant-parasitic nematodes, especially root-knot species, including Meloidogyne incognita, infect a wide range of cultivated plants, and are responsible for billions of dollars in crop losses annually [1, 2]. The genus Meloidogyne comprises more than 80 species [3] and has a crucial role in damaging most of the crop production economically. On a worldwide basis, it causes yield loss in a range of crops and cost growers worldwide >$75b each year, despite control measures [4]. The impact of these species is enhanced by their wide host ranges and is estimated to infect more than 5500 plant species [5]. Chemical control of nematodes requires relatively large amount of nematicides and there are tremendous environmental as well as health issues associated with the production and application of synthetic nematicides. The need for more sustainable and less toxic methods of nematode management has increased research on alternative control measures to synthetic chemical control. This led to the search for eco-friendly and cost effective control of nematodes by natural steps.
Glutathione S-transferases (GST) are a large family of multifunctional dimeric enzymes involved in the metabolization of a broad variety of xenobiotics and reactive endogenous compounds [6]. The complete machinery of glutathione (GSH) system such as GSTs (glutathione-Stransferase( s)) and GSHPx (glutathione peroxidases) are major defense systems of nematodes. The mechanism of action of GST(s) (E.C.2.5.1.18), enzymes includes defense against oxidative attack via conjugation of electrophiles to glutathione and reduction of lipid hydroperoxides [7, 8]. GST has been exploited as potential target for many chemotherapeutic agents. An active GST is a homodimer of a 208 residue long monomer consisting of two domains (smaller α/β domain and larger α domain). The N-terminal small domain (residues 1 to 74) is a α/β structure with the folding topology βαβαββα arranged in the order β2, β1, β3and β4with β3 anti-parallel to the others, forming a regular β-sheet with a right-handed twist surrounded by three α- helices. The C terminal, large domain 2 (82–208 residues) is α-helical. The residues that interface the two βαβ and ββα motifs are Trp38, Phe8, Val33, Cys47, Leu52 and Leu43 in human π GST [9]. The secondary structure of M. incognita GST is shown in (Figure 1).
Figure 1.
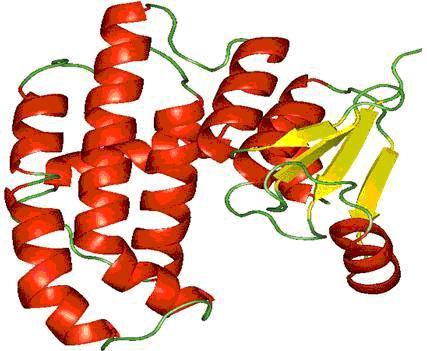
Secondary structure of M. incognita GST – Pymol visualization [17], helices is shown in red, sheets in yellow and loops in green.
Historically, herbs, shrubs and spices have enjoyed a rich tradition of use for their flavor enhancement characteristics and medicinal properties. The emphasis on medicinal plants, herbs and spices as herbal remedies are being lost due to lack of awareness and deforestation. As a result many valuable medicinal herbs as well as precious information are lost. Spices hold the promise of providing both significant clinical benefits and have potential for use as antibiotics. Spices are antimicrobial. This study was conducted with the objective of exploring the nematicidal activity of phytochemicals against M. incognita.
Methodology
Database screening and activity prediction:
Comprehensive reviews of plants with nematicidal phytochemicals were initially made and this led to the selection of nematicidal phytochemicals from different spices. The nematicidal phytochemicals were screened and collected from Dr. Duke's phytochemical and ethno-botanical databases (http://ars-grin.gov/duke/) and through literature search. PASS server [10] was used to predict nematicidal activity and GST substrate activity of the phytochemicals (http://195.178.207.233/PASS/AP.html). PreADMET server (http:/preadmet.bmcrd.org/) was used to predict the druglikeness and ADME-Tox (Absorption, Distribution, Metabolism Excretion and Toxicity) properties [11]. The ADME-Tox properties of a compound together with its pharmacological properties such as drug likeness are conventionally identified as part of drug development. Those compounds obeying the ADMET rules and druglikenes rules were short listed for docking studies. A total of 16 nematicidal compounds were selected and these 16 compounds were reported to have nematicidal activity in an earlier docking study and in vitro assay conducted in our laboratory in animal parasite Dirofilaria immitis, the canine filarial nematode [12].
Ligand Structure:
The canonical smiles notations of phytochemicals predicted to have nematicidal activity were collected from PubChem (http://pubchem.ncbi.nlm.nih.gov/),ChemSpider (http://chemspider.com) and DrugBank (http://www.drugbank.ca/). The 3D structures of compounds were developed by 3D Structure Generator CORINA [13, 14] using canonical smiles of the compound. Energy minimization and molecular optimization of all compounds were done using Arguslab 4.0.1 [15]. Geometry optimization was carried out using AM1 (Austin Model 1) semi-empirical quantum mechanics force field in Arguslab 4.0.1. The best conformer thus obtained was based on energy minimization and geometry optimization. The final structures exhibiting lowest energy were saved in *.pdb format for input in to MVD environment.
Modelling of target protein structur:
Glutathione-S-transferase of M. incognita has been selected as the target for docking study. There were no experimental structures for GST of M. incognita; hence we modeled 3D structure of GST by using Modeller9v8 software [16]. An active GST is a homodimer of a 208 residue long monomer consisting of two domains, smaller α/β domain and larger α domain. In this study we modelled a 3D structure of GST by X-ray Crystal Structure of a major nematode C. elegans specific GST (CE01613) (PDB ID: 1ZL9, Chains A) used as the template. The modeled structure revealed that GST predominantly consists of α helix. The secondary structure of modeled protein was viewed by PyMolv1.4.1 software [17]. The target and template was superimposed by DaliLite pairwise comparison of protein structure [18] and the backbone RMSD value was found to be 3.9 A°.
Target structure validation:
The Ramachandran plot is probably the best indicator for assessing the quality of experimentally and theoretically designed determination of three dimensional protein coordinates [19]. Ramachandran plot was identified by Procheck program [20] of Structural Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/) and Ramachandran Z-score was determined through Structure validation server of WHAT IF Web Interface [21]. Further, modeled protein is validated by molecular dynamics and mechanics by using various empirical forcefields such as ANOLEA [22], GROMOS [23], VERIFY-3D [24] and Errat protein structure verification algorithm [25] to identify overall quality factor of the model.
Molecular Docking:
Molecular Docking study was carried out by using Molegro Virtual Docker [26]. MVD performs flexible ligand docking, so the optimal geometry of the ligand will be determined during the docking. MVD includes MolDock Score [26] and PLANTS Score [27] for evaluating docking solutions. MVD returns multiple poses representing different potential binding modes. Here, clustering has been used to reduce the number of poses found during the docking run and only the most promising ones was reported. During docking, both protein and ligands were prepared for which bonds, bond orders, explicit hydrogens, charges, flexible torsions, were assigned at the missing region by the MVD program for both the protein and ligands. From the docking wizard ligands were selected and Moldock score scoring function has been used. The intact protein structure was loaded on to MVD platform for docking process. Potential binding sites (also referred to as cavities or active sites) has been identified using the built-in cavity detection algorithm of MVD. To reduce overall computing time, Ignore distant atoms option is used to ignore atoms far away from the binding site. The Enforce hydrogen bond directionality option is used to check if bonding between potential hydrogen bond donors and acceptors can occur. If hydrogen bonding is possible, the hydrogen bond energy contribution to the docking score is assigned a penalty based on the deviations from the ideal bonding angle. Using this option can significantly reduce the number of unlikely hydrogen bonds reported also Moldock score, Rerank score, total interactions and number of hydrogen bonds were identified from the pose by enabling the ligand evaluation terms. The search algorithm is taken as Moldock SE and numbers of runs are taken 10 and max iterations were 2000 with population size 50 with an energy threshold of 100. At each step least ‘min’ torsions/translations/rotations were tested and the one giving lowest energy was chosen. After the docking simulation was over, the poses which were generated were sorted by rerank score. The Rerank Score uses a weighted combination of the terms used by the MolDock score mixed with a few addition terms (the Rerank Score includes the Steric (by LJ12-6) terms which are Lennard-Jones approximations to the steric energy – the MolDock score uses a piecewise linear potential to approximate the steric energy) [26]. The reranking score function is computationally more expensive than the scoring function used during the docking simulation but it is generally better than the docking score function at determining the best pose among several poses originating from the same ligand [26]. While the rerank-score in MVD provides an estimate of the strength of the interaction, it is not calibrated in chemical units and it does not take complex contributions (such as entropy) into account. The scoring function used by MolDock is derived from the piecewise linear potential (PLP) scoring functions [28]. The scoring function used by MolDock further improves these scoring functions with a new hydrogen bonding term and new charge schemes [26]. Based on evolutionary algorithms (EAs) classification moldock algorithm may be classified as an Evolutionary simulator (ES), since it employs direct ranking of the solutions and the crossover operators. MolDock showed better overall performance in docking simulations when compared with other software.
In vitro GST Assay:
Based on the availability of compounds, those compounds with least dock score and high interaction with active-site was taken for in vitro studies. The phytochemicals β-caryophyllene, capsaicin, cinnamic acid, citronellol, curcumin, eugenol, geraniol, isoeugenol, linalool, myristicin, neral, α-pinene, piperine, terpineol, vanillin and strychnine were purchased in the pure form from Sigma Chemicals, USA; glutathione (GSH) and 1-chloro-2, 4-dinitrobenzene (CDNB) were purchased from Sisco Research Laboratories Pvt. Ltd., (Mumbai, India). M. incognita; used for the in vitro study was cultured in the Indian Institute of Spices Research, Nematology laboratory, Calicut.
GST crude enzyme was obtained by centrifuging an aliquot containing ~2000 nematodes at 1000 rpm for 2 min, and washing twice with phosphate buffered saline (PBS) at pH 7.4. The nematodes were ground with micro pestle and glass powder. The solution was centrifuged at 10000 rpm at 4 °C for 30 min. Supernatant was dialyzed against PBS overnight and made up to 2 ml. The following phytochemicals were used to study their GST inhibitory activity, at a concentration of 0.001 mg/ml in ethanol: β-caryophyllene, capsaicin, cinnamic acid, citronellol, curcumin, eugenol, geraniol, isoeugenol, linalool, myristicin, neral, α-pinene, piperine, terpineol, vanillin and strychnine (dissolved in water).
The dialyzed enzyme fraction (0.1 ml) was incubated in the presence of 1 ml of 0.001 mg/ml concentration of the phytochemicals listed above, in the presence of 1 mM glutathione reduced (GSH), and 0.1 M phosphate buffer, pH 6.5, for 1 hour at room temperature. A control containing ethanol was also maintained. GST activity was measured using the method of Habig et al. [29], by initiating the reaction with the addition of 1 mM 1-chloro-2, 4-dinitrobenzene (CDNB) and following the change in absorbance at 340 nm, in a Shimadzu 1601 UV-Visible spectrophotometer. The GST activity was expressed as change in absorbance at 340 nm per minute per ml crude enzyme extract. Two replicates of each treatment were maintained.
Bioassay of selected compounds for nematode inhibition:
In vitro bioassay has been conducted to determine differences in the ability of M. incognita populations to survive 24 hours in exposure to different phytochemical concentrations. Phytochemicals with good dock score and those with good percentage of inhibition in in vitro GST assay have been taken for in vitro bioassay based on the availability of compounds. Bioassay was conducted with 4 phytochemicals namely beta-caryophyllene, eugenol, norvalinamide and cinnamic acid. Compounds were taken in four different concentrations such as 200 µg/ml, 100 µg/ml, 50 µg/ml and 25 µg/ml, three replicates for each phytochemical were maintained. M. incognita, 100 in numbers were taken in each well containing 1 ml of phytochemical in specified concentrations. Phytochemical were dissolved in 0.3% Dimethyl sulfoxide (DMSO) in sterile distilled water (SDW), with control of 0.3% DMSO in SDW. After 24 hours of incubation, the mortality rates of nematodes were noted.
Result & Discussion
Target structure Modeling and validation:
Target structure was modeled by Modeller9v8 and 3D structure validation was done to identify overall quality factor of modeled protein. The secondary structure of M. incognita GST is shown in (Figure 1) and the electro negative and electro positive regions are given in (Figure 2), respectively. Ramachandran Z-score was identified which express how well the backbone conformations of all residues in the model correspond to the known allowed areas in the Ramachandran plot (Figure 3) is within expected ranges for modeled structures. Stereochemical validation shows that residues in allowed region of Ramachandran plot is 98.8%, disallowed region in Ramachandran plot is 1.1%, Ramachandran Z-score is 1.906. The overall quality factor of the model was identified to be 69.43% by further validation of modeled protein using molecular dynamics and mechanics methods such as various empirical force fields such as ANOLEA, GROMOS, VERIFY3D, and Errat protein structure verification algorithm.
Figure 2.
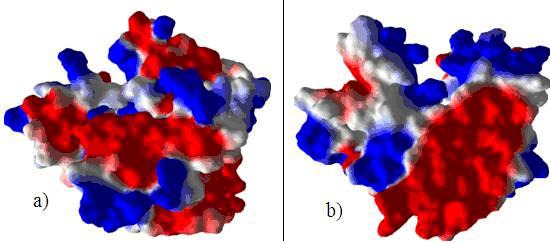
(a) and (b) Argus lab [15] representation of the electrostatic surface potentials of M. incognita GST showing the highly acidic and basic regions of the protein (a) and a view of the same molecule turned 180 about the x axis (b). The negative potential (colored red) and the positive potential (colored blue) respectively.
Figure 3.
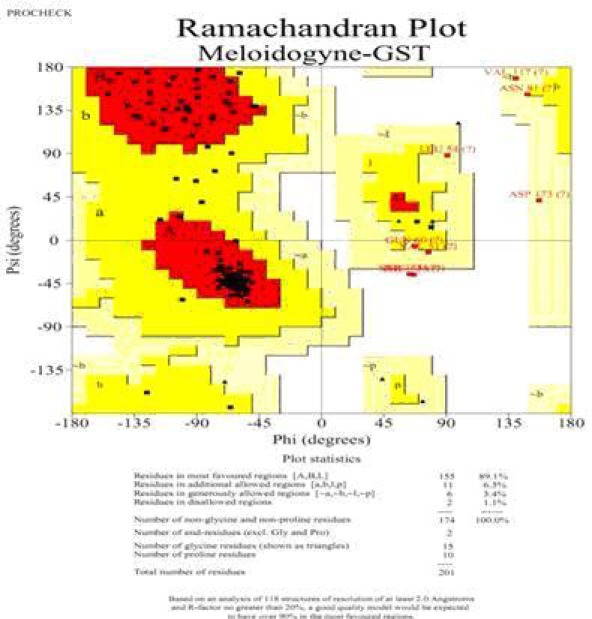
Ramachandran plot for modeled M. incognita GST
Molecular docking:
The entire protein structure was loaded on to MVD platform for finding potential active sites or cavities. A total of three cavities (Figure 4) were detected in modelled GST enzyme by using Molegro Virtual Docker cavity prediction and were named cav1, cav2 and cav3 the volume and surface area details are given in Table 1 (see supplementary material).
Figure 4.
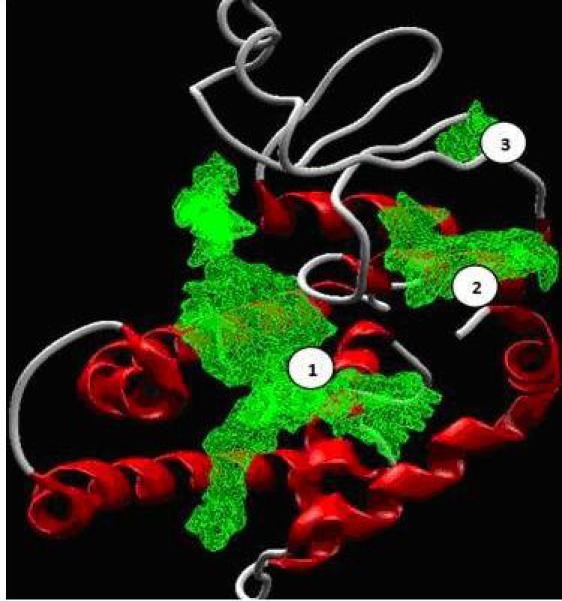
Three cavities detected in Modelled GST of M. incognita
Docking study has been conducted with entire protein structure loaded on to MVD platform. All the three cavities were selected in the MolDock GRID radius 35 A° for docking simulation. Out of the three cavities, all 16 compounds were docked in to Cav1 of the modelled GST protein. Selected compounds exhibited good dock scores and hydrogen bonds interactions in docking studies. alpha pinene (PubChem CID: 6654, MW: 136.234040 g/mol), alpha terpineol (PubChem CID: 17100, MW: 154.249320 g/mol, beta caryophyllene (PubChem CID: 5281515, MW: 204.351060 g/mol), capsaicin (PubChem CID: 1548943, MW: 305.411880 g/mol), cinnamic acid (PubChem CID: 444539, MW: 148.158620 g/mol), citronellol (PubChem CID: 8842, MW: 156.265200 g/mol), curcumin (PubChem CID: 969516; MW: 368.380 g/mol), eugenol (PubChem CID: 3314 , MW: 164.201080 g/mol), geraniol (PubChem CID: 637566, MW: 154.249320 g/mol), isoeugenol (PubChem CID: 853433, MW: 164.201080 g/mol), linalool (PubChem CID: 6549; MW: 154.249 g/mol), myristicin (PubChem CID: 4276, MW: 192.211180 g/mol), neral (PubChem CID: 638011, MW: 152.233440 g/mol ), piperine (PubChem CID: 638024, MW: 285.337660 g/mol), Nvanillylnonanamide (PubChem CID: 2998, MW: 293.401180g/mol) and vanillin (PubChem CID: 1183; MW: 152.147 g/mol) are promising hits as GST inhibitors of natural origin. All of the selected 16 phytochemicals showed good dock score with the target. Docking results showed that all the selected compounds docked satisfactorily to the GST enzyme with good docking scores. Hence these phytochemicals, with less docking energy and greater number of hydrogen bond interactions, were selected as promising nematicidal compounds against M. incognita after docking studies; Table 2 (see supplementary material). Shows the number of H-bond interactions, MolDock score and rerank score between target and phytochemicals.
Docking view showing hydrogen bond interaction of ligands with the GST enzyme is shown in (Figure 5). Pictures were generated using Molegro Virtual Docker; hydrogen bond interaction of these nematicidal compounds with target is shown as green dashed lines in the figure. Ligands are shown in stick model and the target is displayed as secondary structure backbone.
Figure 5.
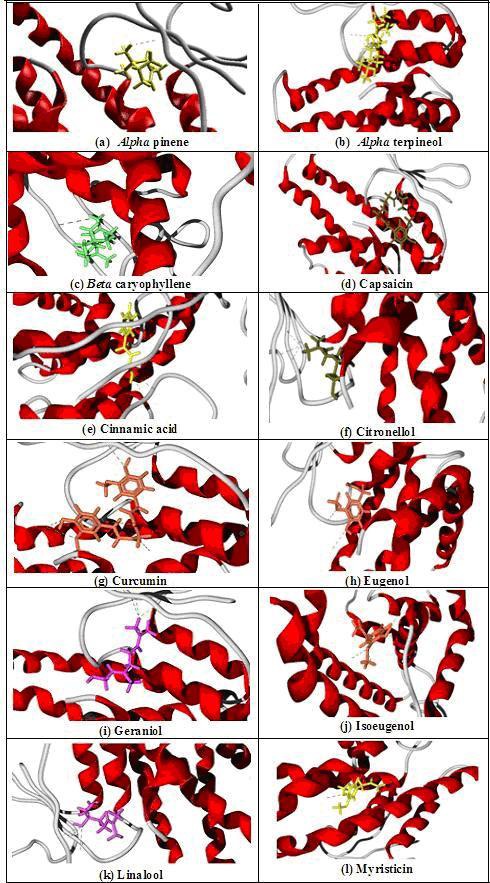
Docking view showing Hydrogen bond interaction of ligands with GST enzyme.(a) Alpha pinene, (b) Alpha terpineol, (c) Beta caryophyllene, (d) Capsaicin, (e) Cinnamic acid, (f) Citronellol, (g) Curcumin, (h) Eugenol, (i) Geraniol, (j) Isoeugenol, (k) Linalool, (l) Myristicin.
In vitro GST assay:
Compounds with the lowest dock score and high interaction were taken for in vitro studies based on the availability of the compound. All of the selected 16 phytochemicals showed good dock score. Hence all of them were taken for in vitro studies.
In vitro studies indicate that beta caryophyllene (99.860%), myristicin (99.442%), eugenol (98.607%), capsaicin (94.707%), alpha terpineol (94.150%), geraniol (92.757%), curcumin (92.479%), neral (92.201%), cinnamic acid (92.201%), Nvanillylnonanamide (89.415%), piperine (89.136%), linalool (89.136%), isoeugenol (88.301%), vanillin (87.465%), alpha pinene (85.515%) and citronellol (74.930%) have good potential as nematicidal compounds against M. incognita GST Table 3 (see supplementary material). The compounds which are found to be quite effective inhibitors of GST in in silico had been validated by alternate methods such as in vitro GST assay and in vitro bioassay experiments. The reason why there is a slight variation in in-silico docking and in vitro activities could be because, inside a living system, and in experimental conditions binding of target protein and compound tend to deviate from their in silico behavior in an unpredicted way. However, we cannot detect whether all the residues in the target to which the compound is making bonds in in silico environment is accessible to the compound in vitro. Moreover, in silico computational tools such as in silico docking provides visual inspection of the binding modes of target and protein. The slight difference in the correlation between the in silico and in vitro results could be attributed to chemical conditions and environmental differences in each case.
The results for in vitro studies proved that sixteen lead molecules selected by virtual screening and docking studies were able to inhibit the GST enzyme isolated from Meloidogyne incognita, in vitro. Thus the above in vitro study largely validates the results obtained from in silico docking studies. Hence these compounds beta caryophyllene, myristicin, eugenol , capsaicin , alpha terpineol , geraniol, curcumin , neral, cinnamic acid, norvalinamide, piperine, linalool, isoeugenol, vanillin, alpha pinene and citronellol can be considered for novel nematicidal compounds through inhibition of GST enzyme. This has been validated by in vitro studies. Based on the availability of compounds and live nematodes, those compounds showed good activity against M. incognita GST has been taken for in vitro bioassay studies.
Bioassay of selected compounds for nematode inhibition:
Bioassay of selected compounds for nematode inhibition by four of the selected compounds based on their availability viz. Beta- caryophyllene, cinnamic acid, eugenol and (N vanillylnonanamide) NVA at four different concentrations was evaluated in an in vitro bioassay on M. incognita. After 24 hours of incubation, the mortality rates of nematodes were noted in numbers. Among the four compounds the maximum mortality was observed with the highest concentrations (200 µgml−1) of Beta- caryophyllene (78%) followed by eugenol (61.6%), cinnamic acid (55%) and N-vanillylnonanamide (49%) in Table 4 (see supplementary material). The mortality of nematodes was directly proportional to the concentration of the compounds.
Potential nematicidal value of phytochemicals has been studied in order to find out natural nematicidal compounds and their efficacy. Phytochemicals have been initially tested by in silico docking studies with GST and then carried on to in vitro GST assay and finally validated by in vitro bioassay using live nematodes. Mode of action of selected phytochemicals on nematodes and percentage of inhibition of nematode GST by each phytochemical is understood by the study. The beneficial effects of natural compounds on nematode control and management have been revealed. This study offers a promising area of non-chemical control and management against plant parasitic nematode. Further work in this area may result in the development of a potent nematicidal molecule from spices.
Supplementary material
Acknowledgments
The authors would like to thank the facilities provided by Director, Indian Institute of Spices Research (IISR), Calicut and DBT, New Delhi for funding.
Footnotes
Citation:Babu et al, Bioinformation 8(7): 319-325 (2012)
References
- 1.GN Agrios, et al. In Plant Pathology. 1997;565:597. [Google Scholar]
- 2.JN Sasser, DW Freckman. Society of Nematology. 1987:7. [Google Scholar]
- 3.G Karssen, et al. Plant parasitic nematode genus Meloidogyne Goldi. U.K: Brill Academic Press; 1892. p. 9. [Google Scholar]
- 4.JN Sasser, DW Freckman. Vistas on Nematology, Society of Nematologist. 1987:7–14. [Google Scholar]
- 5.DL Trudgill, Blok VC. Annual Review of Phytopathology. 2001;39:53. doi: 10.1146/annurev.phyto.39.1.53. [DOI] [PubMed] [Google Scholar]
- 6.Peter Reinemer, et al. Journal of Molecular Biology. 1996;255:289. doi: 10.1006/jmbi.1996.0024. [DOI] [PubMed] [Google Scholar]
- 7.PM Brophy, DI Pritchard. Experimental Parasitology. 1994;79:89. [Google Scholar]
- 8.R Ahmad, AK Srivastava. Parasitol Res. 2008;102:805. doi: 10.1007/s00436-007-0836-9. [DOI] [PubMed] [Google Scholar]
- 9.R Bhargavi, S Vishwakarma. Bioinformation. 2005;1:25. [Google Scholar]
- 10.DA Filimonov, VV Poroikov. BIOS Scientific Publishers. 1996:47. [PubMed] [Google Scholar]
- 11.Kwang Lee Sung, et al. AbstrConf Comb ChemJpn. 2005;21:22. [Google Scholar]
- 12.Azeez Shamina, et al. J Mol Model. 2012;18:151. doi: 10.1007/s00894-011-1035-2. [DOI] [PubMed] [Google Scholar]
- 13.H Schönberger, CH Schwab. J Mol Model. 2000;6:379. [Google Scholar]
- 14.J Sadowski, et al. J Chem INF Comput Sci. 1994;34:1000. [Google Scholar]
- 15.CJ Peng, et al. Comp Chem. 1995;16:49. [Google Scholar]
- 16.N Eswar, et al. Current Protocols in Bioinformatics. 2006;15:30. [Google Scholar]
- 17.MA Lill, ML Danielson. J Comput Aided Mol Des. 2011;25:13. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- 18.L Holm, J Park. Bioinformatics. 2000;16:566. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 19.WW Rob, et al. Comput. Appl Biosci. 1997;13:425. doi: 10.1093/bioinformatics/13.4.425. [DOI] [PubMed] [Google Scholar]
- 20.RA Laskowski, et al. Crystallography of Biological Macromolecules. 2001:722. [Google Scholar]
- 21.K Wilson, et al. J Mol Biol. 1998;276:417. doi: 10.1006/jmbi.1997.1526. [DOI] [PubMed] [Google Scholar]
- 22.F Melo, et al. ProcIntConfIntellSyst Mol Biol. 1997;5:187. [Google Scholar]
- 23.RP Walter, et al. JmPhys Chem. 1999;103:3596. [Google Scholar]
- 24.R Lüthy, et al. Nature. 1992;356:83. [Google Scholar]
- 25.C Colovos, TO Yeates. Protein Sci. 1993;2:1511. [Google Scholar]
- 26.R Thomsen, MH Christensen. J Med Chem. 2006;49:3315. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 27.O Korb, et al. Chem Inf Model. 2009;49:84. [Google Scholar]
- 28.JM Yang, CC Chen. Proteins. 2004;55:288. [Google Scholar]
- 29.WH Habig, MJ Pabst. J BiolChem. 1974;249:7130. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


