Abstract
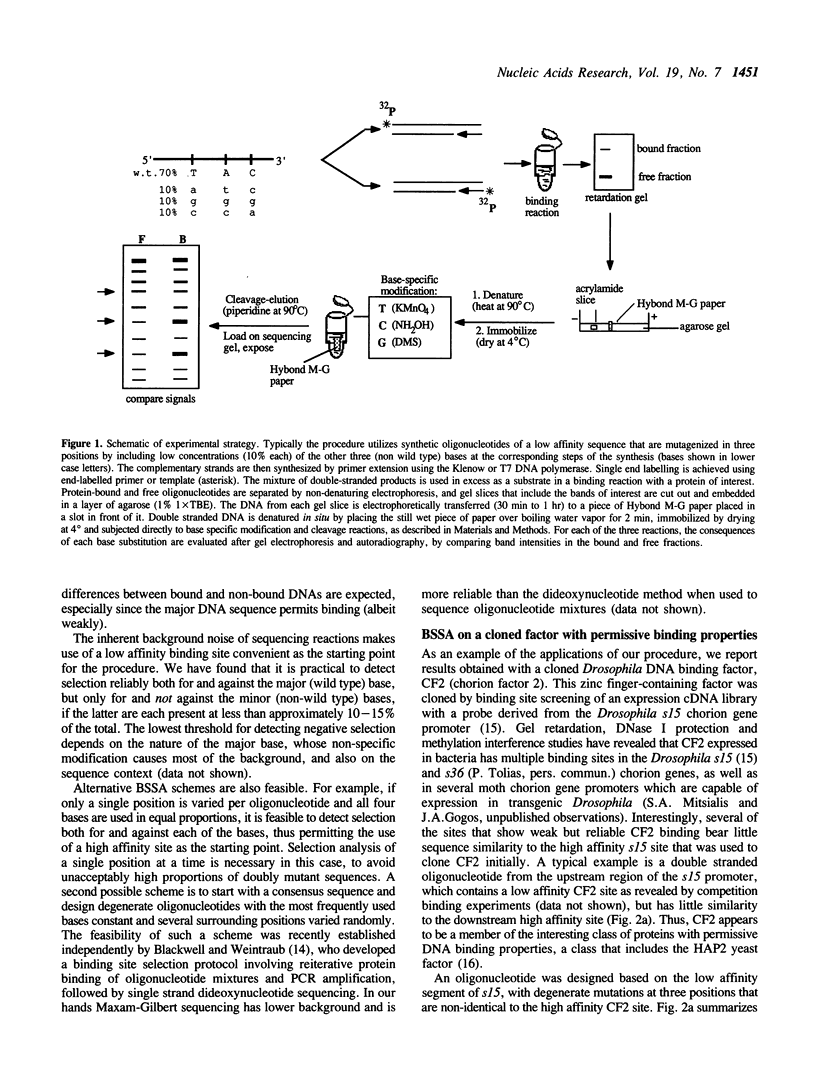
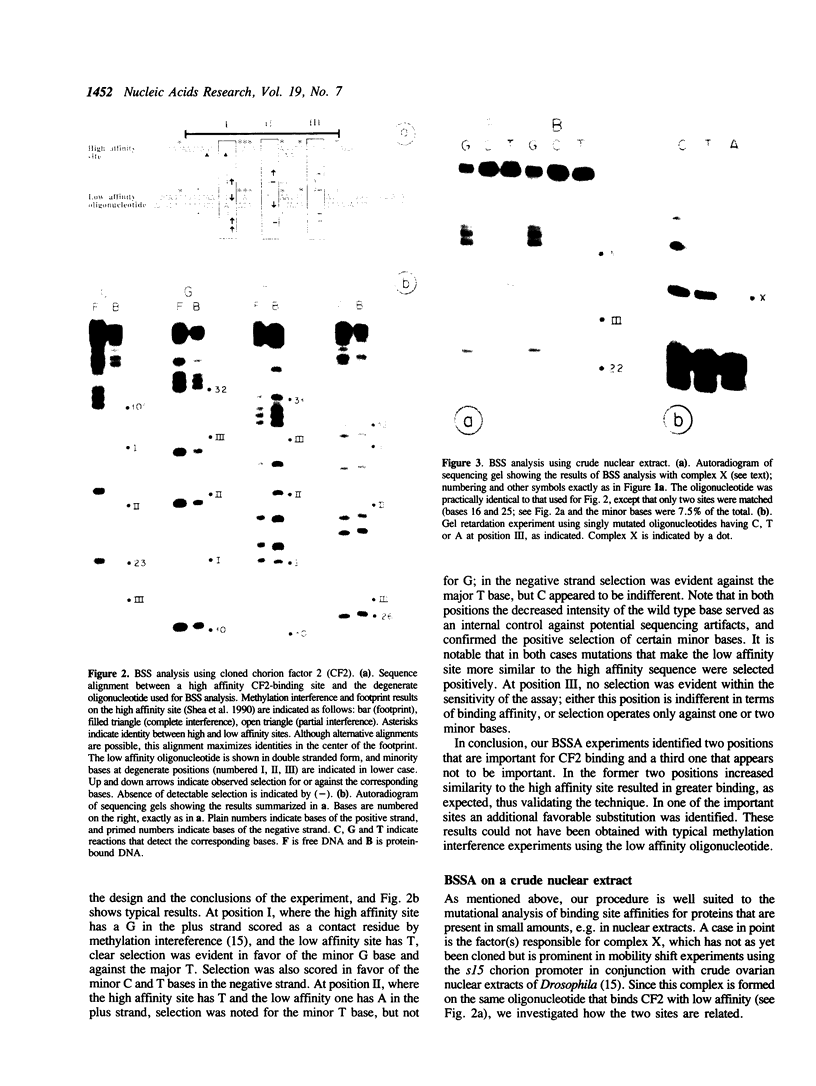
By combining the concept of degenerate oligonucleotide mutagenesis (1,2,3,4) and the convenience of solid phase chemical DNA sequencing (5), we have developed a rapid procedure for determining the specificity of DNA-binding proteins in vitro. Starting with a degenerate oligonucleotide mixture, the technique assays for alternative nucleotides in fractions that are bound or non-bound to the protein of interest. In contrast to previous approaches using degenerate oligonucleotides, it does not involve cloning but rather employs direct sequencing of the oligonucleotide mixtures after attachment to a solid support. Solid state processing obviates the need for both DNA extractions from polyacrylamide gels and time-consuming ethanol precipitations. Because of its convenience and sensitivity, this binding site selection analysis is well suited to determining rapidly the sequence preference of DNA-binding proteins that are available in small amounts, and complements well established approaches like methylation interference or missing contact assays. The solid phase reaction protocol we propose can also improve these latter approaches.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos J. A., Karayiorgou M., Aburatani H., Kafatos F. C. Detection of single base mismatches of thymine and cytosine residues by potassium permanganate and hydroxylamine in the presence of tetralkylammonium salts. Nucleic Acids Res. 1990 Dec 11;18(23):6807–6814. doi: 10.1093/nar/18.23.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Ellington A. D., Szostak J. W. In vitro genetic analysis of the Tetrahymena self-splicing intron. Nature. 1990 Sep 27;347(6291):406–408. doi: 10.1038/347406a0. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Oliphant A. R., Struhl K. Mutagenesis with degenerate oligonucleotides: an efficient method for saturating a defined DNA region with base pair substitutions. Methods Enzymol. 1987;155:558–568. doi: 10.1016/0076-6879(87)55036-4. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. Promoters selected from random DNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7405–7409. doi: 10.1073/pnas.83.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. The use of random-sequence oligonucleotides for determining consensus sequences. Methods Enzymol. 1987;155:568–582. doi: 10.1016/0076-6879(87)55037-6. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Rosenthal A., Jung R., Hunger H. D. Solid-phase methods for sequencing of oligodeoxyribonucleotides and DNA. Methods Enzymol. 1987;155:301–331. doi: 10.1016/0076-6879(87)55022-4. [DOI] [PubMed] [Google Scholar]
- Shea M. J., King D. L., Conboy M. J., Mariani B. D., Kafatos F. C. Proteins that bind to Drosophila chorion cis-regulatory elements: a new C2H2 zinc finger protein and a C2C2 steroid receptor-like component. Genes Dev. 1990 Jul;4(7):1128–1140. doi: 10.1101/gad.4.7.1128. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer V. L., Wobbe C. R., Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990 Apr;4(4):636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Truss M., Chalepakis G., Beato M. Contacts between steroid hormone receptors and thymines in DNA: an interference method. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7180–7184. doi: 10.1073/pnas.87.18.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]