Abstract
Purpose
To identify a disease-causing paired box 6 (PAX6) gene mutation in a Chinese family affected by autosomal dominant congenital aniridia.
Methods
All participants in the study, including the aniridia family and 100 unrelated senile cataract controls, received a comprehensive ophthalmic examination. Genomic DNA was extracted from their whole blood. Mutation screen in all exons and their adjacent splicing junctions of PAX6 was performed by direct sequencing of polymerase chain reaction (PCR) products. PCR products of heterozygous mutation were further cloned into T-vectors and confirmed by sequencing. Multiple alignments were performed using ClustalX to compare PAX6 protein sequences among vertebrates. MicroRNA binding sites were predicted by TargetScan.
Results
A novel heterozygous PAX6 deletion c.1251_1353del103 (p.Pro418Serfs*87) affecting exon 14 and the 3′-untranslated-region (3′-UTR) was identified in the congenital aniridia family. The mutation was exclusively observed in all affected family members but not in any unaffected family member or unrelated control. Bioinformatics analysis showed that the deletion led to remarkable changes of the PAX6 protein, including a frameshift, changes of protein sequence, and a COOH-terminal extension. Multiple alignments showed that the affected region of PAX6 shared high sequence identity (100%) among its vertebrate orthologs. The COOH-terminal extension might also affect microRNA binding sites in the 3′-UTR as predicted by TargetScan.
Conclusions
In the current study we reported a novel PAX6 deletion resulting in an abnormal PAX6 COOH-terminal extension in the Chinese family affected by aniridia. Our findings thus add to the mutation spectrum of PAX6.
Introduction
Congenital aniridia is a severe eye disease that occurs with a prevalence of 1 in 64,000–96,000 across different human populations [1]. Not only the iris but also the lens, optic nerve, cornea, anterior chamber, and retina can be affected by aniridia [2]. About two thirds of aniridia cases are familial, which are autosomal dominant, and the remaining one third are sporadic [3].
The majority of mutations in congenital aniridia have been found in the paired box 6 gene (PAX6) [4]. The gene is a member of the paired box gene family. Encoding a transcription factor, PAX6 spans more than 22 kb at chromosome 11p13, contains 14 exons and generates 3 isoforms of transcripts by alternative splicing [5]. The PAX6 protein consists of 3 domains as follows: a paired domain (PD) in the NH2-terminus, a proline-serine-threonine domain (PST) in the COOH-terminus and a homeodomain (HD) in-between [6]. At the present time, the total number of mutations reported is more than 700 [4]. Most of PAX6 mutations are found in exons 5–14, causing various severe phenotypes in the eye such as aniridia [3,5], cataract [7] and possibly myopia [8-10], all of which could eventually lead to blindness.
Although PAX6 mutations have been reported in Chinese [11-27], disease phenotypes vary among different PAX6 mutations. The phenotype-genotype correlation, which is important in the understanding of the disease mechanism, remains to be further elucidated. In the current study, we screened all exons and their adjacent splicing junctions in PAX6 in a Chinese family with severe disease phenotypes including congenital aniridia and myopia, and identified a novel 103 bp deletion, which caused a frameshift and an abnormal COOH-terminal extension at the protein level.
Methods
Subject recruitment and clinical examination
The family with congenital aniridia was recruited at the Joint Shantou International Eye Center, Shantou, China (Figure 1). One-hundred senile cataract controls without aniridia were recruited from surgical inpatients at the Hospital. Visual acuity, refraction error, intraocular pressure, and slit lamp examinations were performed and documented in all participants (clinical details of individual I-2 could not be obtained due to her old age). Spectral-domain optical coherence tomography (Cirrus HD-OCT; Carl Zeiss Meditec, Inc., Berlin, Germany) was conducted for individual III-2. Color Doppler ultrasound examination of the kidney for detection of Wilms’ tumor was performed at the Chaonan People Hospital for affected family members. Peripheral blood was collected from 6 members of the family (II-1, II-6, II-7, III-1, III-2, and III-3) and all senile cataract controls. Genomic DNA was extracted by using the QIAmp Blood kit (Qiagen, Hilden, Germany).
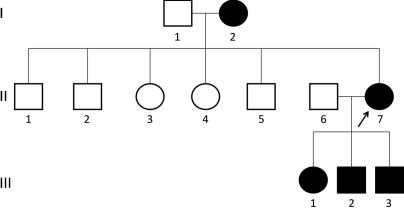
Figure 1.
Pedigree of a Chinese family with aniridia. Filled squares and circles denote affected males and females, respectively. Normal individuals are shown as empty symbols. The proband is indicated by an arrow.
This study was approved by the Ethics Committee of Joint Shantou International Eye Center and was conducted in accordance with the Declaration of Helsinki. Written consent was obtained from each participating subject after explanation of the nature of the study.
Mutation screening
Thirteen pairs of primers (Table 1) for amplicons targeting all exons and their adjacent splicing junctions in PAX6 (NCBI human genome build 37.2, NC_000011 for gDNA, NM_001604, NP_001595), were modified from a previously study [21]. The polymerase chain reaction (PCR) amplification was performed using the GeneAmp PCR System 9700 (ABI, Foster City, CA) in a 25-µl mixture containing 1.5 mM MgCl2, 0.2 mM of each dNTP (Sangon, Shanghai, China), 1 U Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.2 µM primers and 20 ng of genomic DNA. Bidirectional sequencing of PCR products was performed using the BigDye Terminator Cycle Sequencing v3.1 kit (ABI) and the 3130xl Genetic Analyzer (ABI). Haplotyping was used to confirm the mutation sequence. PCR products of heterozygous mutants were ligated to pMD®18-T vectors (Takara, Dalian, China), and subsequently sequenced by the 3130xl Genetic Analyzer. Mutation naming followed the nomenclature recommended by the Human Genomic Variation Society (HGVS).
Table 1. Primers used for PCR and sequencing of PAX6.
| Target | Exon ID | Primer sequence (5′-3′) | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|---|
| 1a* | E1aF | AGCTGTGCCCAACTCTAGCC | 57 | 399 |
| E1aR | TTCCATCTTTGTATGCCTCCTT | |||
| 1 | E1F | CGGAGCCGAAAACAAGTG | 57 | 388 |
| E1R | GAGTGTGGGTGAGGGAAGTG | |||
| 2 | E2F | CCACTTCCCTCACCCACAC | 60 | 422 |
| E2R | CTCCTGCGTGGAAACTTCTC | |||
| 3 | E3F | AAGTGGGATCCGAACTTGC | 57 | 349 |
| E3R | CAGCCACCACAGAACTTGC | |||
| 4 | E4F | CAAGCCCCAAAGGGTAGATT | 57 | 286 |
| E4R | CGAAGTCCCAGAAAGACCAG | |||
| 5 | E5F | GGCTGGTGGTCCTGTTGTCCTT | 58 | 441 |
| E5R | GAGGGCGTTGAGAGTGGAG | |||
| 6,7 | E6–7F | AAGCAAGGTCAGCACAAAAATAAATT | 64 | 648 |
| E6–7R | GGAGGAGGTAAAGAGGAGAGAGCATT | |||
| 8 | E8F | TAAGGTTGTGGGTGAGCTGAGATG | 66 | 315 |
| E8R | GGGAGAGTAGGGGACAGGCAAAGG | |||
| 9 | E9F | TTTGGTGAGGCTGTCGGGATATAAT | 58 | 415 |
| E9R | TGCCCAGAGAAATAAAAAGACAGAAA | |||
| 10 | E10F | TTGGTTGGAGGTAATGGGAGTGG | 61 | 334 |
| E10R | TGGCAGCAGAGCATTTAGCAGAC | |||
| 11,12 | E11–12F | GGGGCTGGGCTCGACGTAG | 62 | 438 |
| E11–12R | GCCACCACCAGCCGCACTTA | |||
| 13 | E13F | GGGGCTGTGGCTGTGTGATGT | 61 | 333 |
| E13R | CCCCAGGGACAAGGAAAGCAA | |||
| 14 | E14F | CCAAACATGCAAACAAACAGAGGA | 52 | 570 |
| E14R | TTCCAACTGATATCGTGCCTTCTG |
*: This extra exon is located at 5′ upstream of exon1, existing in another alternatively spliced transcript variant NM_001127612.1 but not in NM_001604.4.
Bioinformatics analysis
Sequences of PAX6 orthologs in other vertebrate species were retrieved from the NCBI Reference Sequence database. Multiple alignment of PAX6 orthologs from different vertebrate species was conducted using ClustalX version 2.0 [28]. MicroRNA targets in the PAX6 3′-UTR was predicted by TargetScan release 6.0 [29].
Results
Clinical data
As illustrated by Table 2 and Figure 2, four affected patients (II-7, III-1, III-2, and III-3) receiving ophthalmic examination showed similar clinical symptoms, including congenital aniridia, foveal hypoplasia, and nystagmus (Figure 2). Intraocular hypertension without evident glaucoma was also observed (except the youngest child, III-3). Other clinical features were characterized by normal corneas and transparent lens. The proband (II-7) had low visual acuity, and had high myopia with refractive errors equal to −18 diopters (OD) and −12 diopters (OS). Highly myopic status indicated by relative elongated eye axial length was found in the eldest child (III-1; OD: 22.82 mm; OS: 24.10 mm) with regards to his age of 6 years. Myopia was not observed in the other two children (III-2 and III-3) probably due to their younger ages. Color Doppler ultrasound examination showed that there was no Wilms’ tumor in the four affected patients.
Table 2. Demographic and clinical data of the aniridia family.
| Ophthalmic anomalies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject* | Age (years) | Gender | Complete aniridia | Foveal hypoplasia | Nystagmus | High myopia | Congenital cataract | Ocular Hypertension | Wilms’ tumor |
| I-2 | 71 | F | + | NA | NA | NA | - | NA | NA |
| II-7 | 31 | F | + | + | + | + | - | + | - |
| III-1 | 6 | F | + | + | + | + | - | + | NA |
| III-2 | 5 | M | + | + | + | - | - | + | - |
| III-3 | 2 | M | + | + | + | NA | - | - | - |
M: male; F: female; NA: not available; * All of the other families members have complete iris, without ocular hypertension or high myopia or other major eye diseases, and thus are not listed the table.
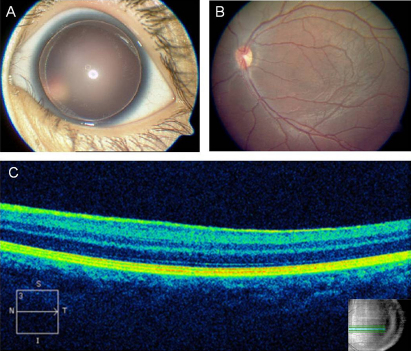
Figure 2.
Photos and images showing the clinical features of affected patients. A: Complete absence of iris in III-2 (OS). B: Foveal hypoplasia observed in III-2 (OS). C: A flat fovea in III-2 (OS) demonstrated by optical coherence tomography.
Mutation analysis
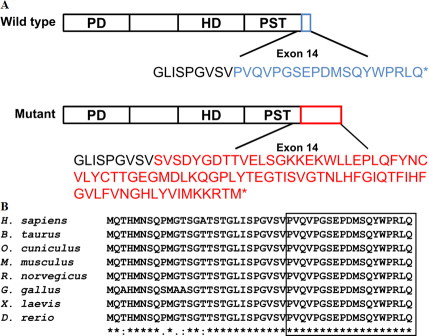
By the means of direct sequencing and haplotyping, a heterozygous 103 bp deletion (c.1251_1353del103, p.Pro418Serfs*87) was identified in all affected family members, which consisted of part of exon 14 and 3′-UTR (Figure 3 and Figure 4). The mutation was not found in any unaffected family member or senile cataract control. No other mutation was detected from any other PAX6 exon in the affected family members. The mutation caused a frameshift and was predicted to generate proteins with an abnormal COOH-terminal extension, in which 19 amino acid residues starting from codon 418 were replaced by a peptide of 86 amino acid residues (Figure 5A). Multiple alignments of PAX6 sequences from different vertebrate species revealed 100% identity of the deleted region at PAX6 COOH-terminus, which suggested that it was highly conserved during evolution (Figure 5B). TargetScan analysis predicted a target site of hsa-miR-365 and another target site of has-miR-375 at the wild type PAX6 3′-UTR, which was converted into coding region by the shifted open reading frame in the mutant (Figure 4).
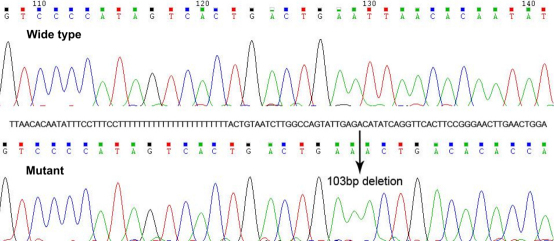
Figure 3.
Confirmation of the PAX6 deletion in the aniridia family by direct-sequencing. The sequence in the plus strand of chromosome 11 is shown. The upper panel is the chromatogram of the PAX6 wild type, and the lower panel is the chromatogram of the mutant. The arrow indicates the position of the deletion, and the deleted sequence is shown above the arrow.
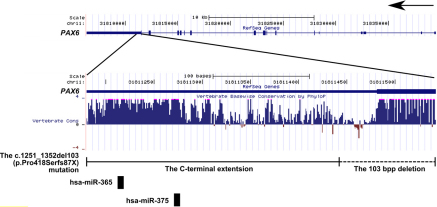
Figure 4.
Diagram of the human PAX6 gene and the deletion with the consequent COOH-terminal extension in the current aniridia family. MicroRNA targets are predicted by TargetScan.
Figure 5.
The deleted region in the PAX6 protein is highly conserved among vertebrates. A: Diagrams of the wild type and mutant PAX6 proteins. Upper panel represents a human wild type PAX6 protein. The Lower panel represents a mutant. Due to the frameshift and COOH-terminal extension generated by the DNA deletion, the peptide in blue encoded by exon 14 of the wild type is replaced by the peptide in red in the mutant. B: Multiple alignment of PAX6 COOH-terminal sequences from different species of vertebrates. The box indicated that the deleted peptide shares 100% identity among vertebrates.
Discussion
In the current study with a family affected by aniridia, we identified a novel 103 bp deletion (c.1251_1353del103, p.Pro418Serfs*87), which generated a frameshift and a COOH-terminal extension. The deletion was correlated with aniridia disease phenotypes in an autosomal dominant pattern, but did not affect the cornea or lens. These findings suggested that COOH-terminal extension mutations of PAX6 could also cause severe aniridia phenotype in the Chinese population, which was comparable to those reported in Caucasians [10].
In Caucasians it was reported that the most common PAX6 mutations were premature termination mutations, amino acid substitutions and COOH-terminal extensions [10]. There were only limited data of PAX6 COOH-terminal extension mutations reported in Chinese [25]. Therefore, COOH-terminal extensions of PAX6 in Chinese remained to be further investigated. The PAX6 deletion found in our study (c.1251_1353del103) led to a frameshift, and caused a COOH-terminal extension into the 3′-UTR (p.Pro418Serfs*87). It also changed the COOH-terminal amino acid sequence by replacing the last 19 amino acid residues with a peptide of 86 amino acid residues, and possibly affected the PST domain of PAX6 (Figure 5). Sequence analysis showed that the deleted residues in the PST domain were highly conserved among different vertebrate species from Zebrafish to humans (Figure 5), implicating its substantial role in ocular and neurologic development. The PST domain is a transactivation domain and participates in modulating the DNA binding function of PAX6 [30-33]. Brain-expressed proteins interact with PAX6 through the COOH-terminus and with the entire PST domain [34]. The COOH-terminus extension in our Chinese family possibly disrupted the function of PST domain, and could cause severe ocular anomalies including aniridia in an autosomal dominant pattern.
Phenotypic consequence in different types of PAX6 mutations was reported in Chinese aniridia patients [11-27], but the understanding of that in COOH-terminal extension mutations remained to be further studied. Previously Zhang et al. [25] reported a COOH-terminal extension mutation c.1268A>T (X423LeuextX*15) in aniridia patients, but the mutation was also found in a patient with cataract and intact iris, probably due to incomplete penetrance. In Caucasians the severity of anomalies in COOH-terminal extension mutations were comparable to that in premature termination codon mutations, including iris anomalies, foveal hypoplasia, keratopathy, cataracts and myopia [9,10]. Intriguingly, in our Chinese aniridia family the mutation had part of the last exon and the 3′-UTR in PAX6 deleted, and resulted in a COOH-terminal extension, probably without triggering nonsense-mediated mRNA decay [35]. Among the patients bearing the mutation, the anomalies included aniridia, intraocular hypertension and high myopia, although myopic status remained to be confirmed in the two younger children due to their young ages. If untreated, glaucomatous damage might also develop in these affected patients. Taken together, these findings possibly were in line with a suggestive role played by the PAX6 3′-UTR in myopia etiology. Single nucleotide polymorphisms in the PAX6 3′-UTR were associated with myopia in Chinese [36,37]. The PAX6 3′-UTR might be involved in myopia development by regulating the expression of PAX6 mRNAs, which was similar to short repeats in PAX6 promoter [8]. Analysis by TargetScan predicted targets of microRNAs hsa-miR-365 and has-miR-375 encompassed by the COOH-terminal extension (Figure 4). To date, there has been no report directly studying the role of microRNA targeting PAX6 in aniridia. Nevertheless, microRNAs targeting other genes in the PAX6 pathway have been reported to be involved in eye development. For example, Meis homeobox 2 (MEIS2) modulates PAX6 transcription activity, and in madaka fish morpholino-mediated ablation of miR-204 targeting Meis2 results in an eye phenotype characterized by microphthalmia, abnormal lens formation, and altered dorsoventral patterning of the retina [38]. Therefore, the conversion from microRNA binding sites at the wild type PAX6 3′-UTR to coding region in the mutant, may abolish the regulation capacity of has-miR-365 and has-miR-375, and further elicit increased PAX6 mRNA expression, and consequently aniridia and high myopia. In addition, our data showed that the mutation did not cause either anomalies in the cornea or lens, or systemic symptoms in the kidney such as Wilms’ tumor. Our genotype-phenotype analysis thus provided useful information for understanding functional consequences of COOH-terminal extension mutations in PAX6.
In the present study, we found a novel PAX6 deletion in a Chines family affected by congenital aniridia. Our findings thus expanded the mutation spectrum of PAX6 gene in Chinese.
Acknowledgments
We would like to thank all patients and family members for their participation. This study was supported in part by the research grants from National Natural Science Foundation of China (No. 81000397), Natural Science Foundation of Guangdong Province, China (No. 8151503102000019), Science and Technology Planning Project of Guangdong Province, China (No. 2010B031600130) and Joint Shantou International Eye Center, Shantou University/The Chinese University of Hong Kong (No. 08-003, 10–020, 10–021 and 10–022). We also thank Dr. Gary Hin-Fai Yam for useful discussions.
References
- 1.Shaw MW, Falls HF, Neel JV. Congenital Aniridia. Am J Hum Genet. 1960;12:389–415. [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson LB, Spaeth GL, Nowinski TS, Margo CE, Jackson L. Aniridia. A review. Surv Ophthalmol. 1984;28:621–42. doi: 10.1016/0039-6257(84)90184-x. [DOI] [PubMed] [Google Scholar]
- 3.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1:328–32. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 4.Brown A, McKie M, van Heyningen V, Prosser J. The Human PAX6 Mutation Database. Nucleic Acids Res. 1998;26:259–64. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–9. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 6.Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong LC, Saunders GF. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–74. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 7.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 8.Ng TK, Lam CY, Lam DS, Chiang SW, Tam PO, Wang DY, Fan BJ, Yam GH, Fan DS, Pang CP. AC and AG dinucleotide repeats in the PAX6 P1 promoter are associated with high myopia. Mol Vis. 2009;15:2239–48. [PMC free article] [PubMed] [Google Scholar]
- 9.Hewitt AW, Kearns LS, Jamieson RV, Williamson KA, van Heyningen V, Mackey DA. PAX6 mutations may be associated with high myopia. Ophthalmic Genet. 2007;28:179–82. doi: 10.1080/13816810701356676. [DOI] [PubMed] [Google Scholar]
- 10.Hingorani M, Williamson KA, Moore AT, van Heyningen V. Detailed ophthalmologic evaluation of 43 individuals with PAX6 mutations. Invest Ophthalmol Vis Sci. 2009;50:2581–90. doi: 10.1167/iovs.08-2827. [DOI] [PubMed] [Google Scholar]
- 11.Song SJ, Liu YZ, Cong RC, Jin Y, Hou ZQ, Ma ZZ, Ren GC, Li LS. Mutation analysis of PAX6 gene in a large Chinese family with aniridia. Chin Med J (Engl) 2005;118:302–6. [PubMed] [Google Scholar]
- 12.Song S, Liu Y, Guo S, Zhang L, Zhang X, Wang S, Lu A, Li L. A novel PAX6 gene mutation in a Chinese family with aniridia. Mol Vis. 2005;11:335–7. [PubMed] [Google Scholar]
- 13.Wang P, Guo X, Jia X, Li S, Xiao X, Zhang Q. Novel mutations of the PAX6 gene identified in Chinese patients with aniridia. Mol Vis. 2006;12:644–8. [PubMed] [Google Scholar]
- 14.Zhu HY, Wu LQ, Pan Q, Liang DS, Long ZG, Dai HP, Xia K, Xia JH. Analysis of PAX6 gene in a Chinese aniridia family. Chin Med J (Engl) 2006;119:1400–2. [PubMed] [Google Scholar]
- 15.Yuan H, Kang Y, Shao Z, Li Y, Yang G, Xu N. Two novel PAX6 mutations identified in northeastern Chinese patients with aniridia. Mol Vis. 2007;13:1555–61. [PubMed] [Google Scholar]
- 16.Kang Y, Yuan HP, Li YY. A novel mutation of the PAX6 gene identified in a northeastern Chinese family with congenital aniridia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008;25:172–5. [PubMed] [Google Scholar]
- 17.Sun DG, Yang JH, Tong Y, Zhao GJ, Ma X. A novel PAX6 mutation (c.1286delC) in the patients with hereditary congenital aniridia. Yi Chuan. 2008;30:1301–6. doi: 10.3724/sp.j.1005.2008.01301. [DOI] [PubMed] [Google Scholar]
- 18.Li PC, Yao Q, Ren X, Zhang MC, Li H, Liu JY, Sheng SY, Wang Q, Liu MG. Analysis of PAX6 gene in a Chinese family with congenital aniridia. Zhonghua Yan Ke Za Zhi. 2009;45:931–4. [PubMed] [Google Scholar]
- 19.Lin Y, Li J, Yang Y, Yang JY, Zhang B, Tang X, Liu XQ, Lu F, Yang ZL. Mutation analysis of the PAX6 gene in a family with congenital aniridia and cataract. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:542–5. doi: 10.3760/cma.j.issn.1003-9406.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang LM, Ying M, Wang X, Wang YC, Hao P, Li ND. R240X mutation of the PAX6 gene in a Chinese family with congenital aniridia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:546–9. doi: 10.3760/cma.j.issn.1003-9406.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Cai F, Zhu J, Chen W, Ke T, Wang F, Tu X, Zhang Y, Jin R, Wu X. A novel PAX6 mutation in a large Chinese family with aniridia and congenital cataract. Mol Vis. 2010;16:1141–5. [PMC free article] [PubMed] [Google Scholar]
- 22.Kang Y, Yuan HP, Li X, Li QJ, Wu Q, Hu Q. A novel mutation of the PAX6 gene in a Chinese family with aniridia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:376–80. doi: 10.3760/cma.j.issn.1003-9406.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Cheng F, Song W, Kang Y, Yu S, Yuan H. A 556 kb deletion in the downstream region of the PAX6 gene causes familial aniridia and other eye anomalies in a Chinese family. Mol Vis. 2011;17:448–55. [PMC free article] [PubMed] [Google Scholar]
- 24.Luo F, Zhou L, Ma X, He Y, Zou L, Jie Y, Liu J, Pan Z. Mutation Analysis of PAX6 in a Chinese Family and a Patient with a Presumed Sporadic Case of Congenital Aniridia. Ophthalmic Res. 2012;47:27–31. doi: 10.1159/000327593. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Wang P, Li S, Xiao X, Guo X, Zhang Q. Mutation spectrum of PAX6 in Chinese patients with aniridia. Mol Vis. 2011;17:2139–47. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zhang Q, Tong Y, Dai H, Zhao X, Bai F, Xu L, Li Y. Large novel deletions detected in Chinese families with aniridia: correlation between genotype and phenotype. Mol Vis. 2011;17:548–57. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Tong Y, Xu W, Dong B, Yang H, Xu L, Li Y. Two novel mutations of the PAX6 gene causing different phenotype in a cohort of Chinese patients. Eye (Lond) 2011 doi: 10.1038/eye.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–21. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 31.Singh S, Tang HK, Lee JY, Saunders GF. Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J Biol Chem. 1998;273:21531–41. doi: 10.1074/jbc.273.34.21531. [DOI] [PubMed] [Google Scholar]
- 32.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–22. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 33.Singh S, Chao LY, Mishra R, Davies J, Saunders GF. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum Mol Genet. 2001;10:911–8. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- 34.Cooper ST, Hanson IM. A screen for proteins that interact with PAX6: C-terminal mutations disrupt interaction with HOMER3, DNCL1 and TRIM11. BMC Genet. 2005;6:43. doi: 10.1186/1471-2156-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–94. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang CL, Hsi E, Chen KC, Pan YR, Wang YS, Juo SH. A functional polymorphism at 3′UTR of the PAX6 gene may confer risk for extreme myopia in the Chinese. Invest Ophthalmol Vis Sci. 2011;52:3500–5. doi: 10.1167/iovs.10-5859. [DOI] [PubMed] [Google Scholar]
- 37.Jiang B, Yap MK, Leung KH, Ng PW, Fung WY, Lam WW, Gu YS, Yip SP. PAX6 haplotypes are associated with high myopia in Han chinese. PLoS ONE. 2011;6:e19587. doi: 10.1371/journal.pone.0019587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107:15491–6. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]