Abstract
The mission of the Pharmacogenomics Knowledge Base (PharmGKB; www.pharmgkb.org) is to collect, encode and disseminate knowledge about the impact of human genetic variations on drug responses. It is an important worldwide resource of clinical pharmacogenomic biomarkers available to all. The PharmGKB website has evolved to highlight our knowledge curation and aggregation over our previous emphasis on collecting primary data. This review summarizes the methods we use to drive this expanded scope of ‘Knowledge Acquisition to Clinical Applications’, the new features available on our website and our future goals.
Keywords: Clinical Annotations, Clinical Interpretations, genomic variation, pharmacogenetics, pharmacogenomics, Pharmacogenomics Knowledge Base, PharmGKB, Variant Annotations
The Pharmacogenomics Knowledge Base (PharmGKB) is a publicly available online worldwide resource [101]. The main objective of the PharmGKB is to aggregate and disseminate information and knowledge regarding pharmacogenomics in an online database website (alongside downloadable content), contributing to the drive towards personalized medicine for better therapeutics. The PharmGKB also plays an active role as an independent broker for international research consortia focused on pharmacogenomics. The Clinical Pharmacogenetics Implementation Consortium (CPIC), led by the PharmGKB and the NIH Pharmacogenomics Research Network (PGRN) was established to develop genetics-based dosing guidelines for specific drugs. The PharmGKB website has recently been redesigned to integrate these clinical dosing guidelines and Clinical Annotations with gene, drug and disease information in a user-friendly and accessible manner.
Pharmacogenomics research
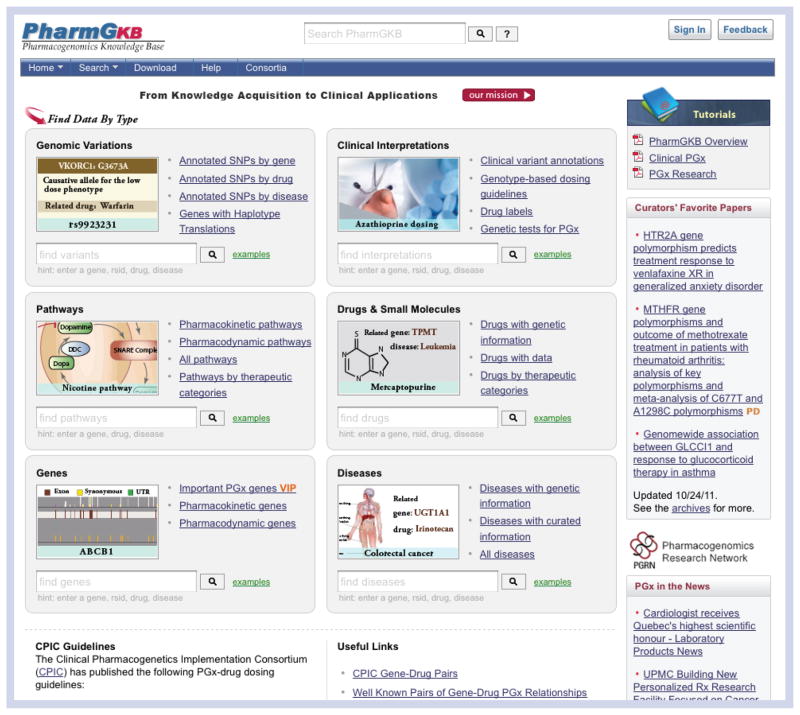
From the www.pharmgkb.org homepage, several search boxes can be utilized to find the gene, genetic variant or drug of interest (Figure 1). On each drug or gene page under the ‘Pharmacogenomics (PGx) Research’ tab, a table of variants associated with the drug or gene is presented (Figure 2). The first column of the table contains icons representing what kind of information is available for each variant (Table 1). This information can be found by clicking on the icon, variant, gene or drug links in these tables.
Figure 1. The new Pharmacogenomics Knowledge Base (PharmGKB) homepage.
The Pharmacogenomics Knowledge Base (PharmGKB) homepage is found at [101]. The ‘Clinical Interpretations’ box (top middle) contains links to genotype-based dosing guidelines, drug label information, Clinical Annotations and genetic test information. The search box can also be used to find clinically related information for a drug, gene or variant. The search boxes for ‘Genomic Variations’, ‘Pathways’, ‘Drugs & Small Molecules’, ‘Genes’ and ‘Diseases’ have been rearranged to increase ease of usability. Top center: more information regarding the mission of PharmGKB “From Knowledge Acquisition to Clinical Applications” can be found by clicking on ‘our mission’. Top right: users can learn more about how to utilize the PharmGKB website through our tutorials. Right side panel: the curator’s favorite papers and pharmacogenomics news are constantly updated to keep users up to date with the most recent pharmacogenomic research and policies.
Reprinted with permission from PharmGKB.
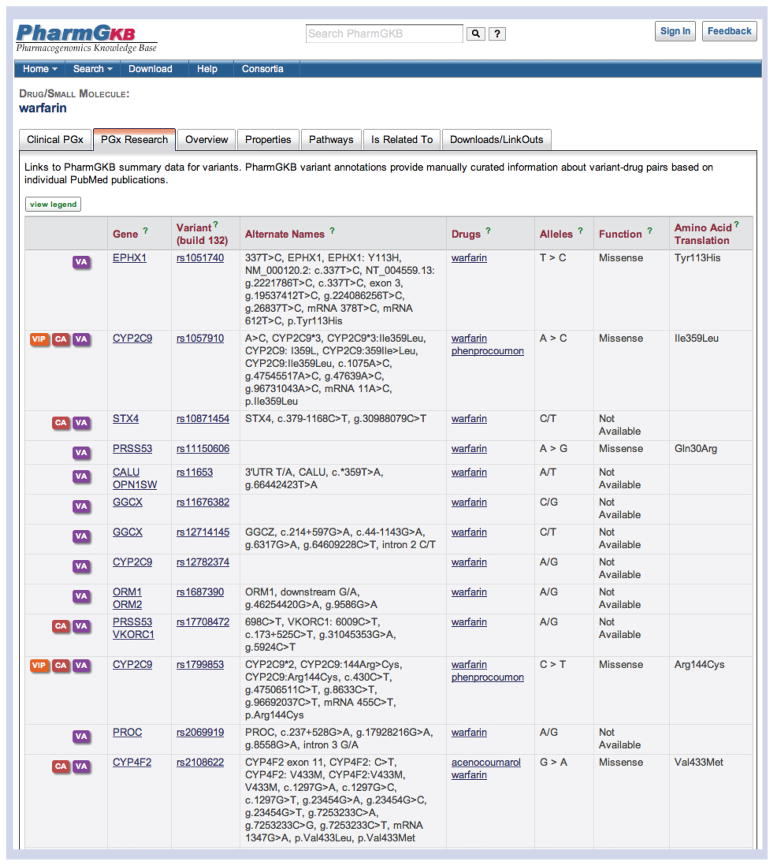
Figure 2. The ‘Pharmacogenomics (PGx) Research’ tab for the drug warfarin.
Under the ‘Pharmacogenomics (PGx) Research’ tab on a drug page, a list of variants with associations with the drug can be found. On gene pages, a similar format is found, with a list of variants within that gene and the drugs they have been associated with. If curated Variant Annotations and Clinical Annotations are available for the variant, this is represented by an icon in the first column of the table (Table 1). If the variant is described in a Very Important Pharmacogene (VIP) summary of the gene, this symbol is also provided (Table 1). As well as the reference single nucleotide polymorphisms identification (rsID), alternative names for the variant are provided. Useful information includes the variant alleles, the amino acid change (if applicable) and the features of the variant (e.g., a missense amino acid change). Alleles, Function and Amino Acid Translation are all sourced from single nucleotide polymorphisms database build 132 [103].
Reprinted with permission from PharmGKB.
Table 1.
Symbols used on the Pharmacogenomics Knowledge Base (PharmGKB) website.
| Symbol† | Definition |
|---|---|
 |
Pharmacokinetic relationship |
 |
Pharmacodynamic relationship |
 |
Very Important Pharmacogene summary information is available for this variant/gene |
 |
Variant Annotations are available for this variant/gene/drug |
 |
Clinical Annotations are available for this variant/gene/drug |
 |
Literature Annotations are available for this gene/drug |
 |
Genotype-based dosing guideline information is available for this gene/drug |
 |
Drug labeling information is available for this gene/drug |
 |
A dataset is available for this variant/gene/drug |
 |
When you move the mouse over a symbol, it provides information about what this |
Legends are also available on the Pharmacogenomics Knowledge Base (PharmGKB) website pages. The boxed icons can be clicked on to directly access the indicated information.
Reprinted with permission from PharmGKB.
One of the many tasks of the PharmGKB curators is to review the past and current literature and add any relevant pharmacogenetic or genomic articles to the PharmGKB database. Manual coverage of the huge volume of pharmacogenetic literature is not feasible given the size of our curation staff, and we expect ongoing efforts in natural language processing (NLP) research will aid this in the future (see ‘Future perspective’ section). The curator team routinely curates literature from several major pharmacogenetic journals and from publications by the PGRN. Additional pharmacogenomic literature is annotated in the course of creating pathways (see ‘Pathway summaries’ section), Very Important Pharmacogene (VIP) summaries (see ‘VIP summaries’ section) and further curation tasks.
Variant Annotations
A Variant Annotation is a summary created for a particular genetic variant associated with a drug described in a single publication. Essential information regarding the pharmacogenetic association is provided, such as the alleles or genotypes described, the phenotypic effect or effect on drug metabolism, study characteristics of the population, and statistics. Variant Annotations summarize the findings of a variety of study types, including clinical trials, clinical case studies, genome-wide association studies, and functional in vivo and in vitro studies. Curators also endeavor to cover pharmacogenetic studies mentioned within a review, introduction or discussion. Variant Annotations are represented by the ‘VA’ symbol throughout the PharmGKB website (Table 1 & Figure 2). Links to PubMed® IDs (PMIDs) [102] within pathway descriptions and VIP summaries also link to curated Variant Annotations.
An example of a Variant Annotation in our new standardized format is shown in Figure 3, for PMID 21383771 [1]. Below the article title and abstract, the gene, drug and diseases mentioned in the article are displayed. These link directly to the individual gene, drug and disease pages. A symbol is provided to tag whether a pharmacokinetic (PK) or pharmacodynamic (PD) relationship is discussed (Table 1). In this example, the journal discusses the genes CYP2C9, CYP4F2 and VKORC1 and response to the drug warfarin. Although the CYP2C9 and CYP4F2 genes are involved in the metabolism of warfarin, the annotated article studies responses to warfarin (PD) and not the PK of the drug, and therefore, it has been labeled with a PD tag by the curator.
Figure 3. A Pharmacogenomics Knowledge Base (PharmGKB) Literature Annotation, with Variant Annotations.
The title and reference of the journal are found at the top of the page, along with a link to the journal within PubMed® [102] (reference [1] is shown as an example). If available, the abstract is also provided. The Pharmacogenomics Knowledge Base (PharmGKB) curated sections are ‘Relationships’ and ‘Variant Annotations’. The genes, drugs and diseases discussed in the journal are provided in the ‘Relationships’ section, and link to each feature page in the database. Where possible, associations between a drug and a genetic variant reported in the journal are represented in the form of Variant Annotations in the table below. The variant reference single nucleotide polymorphism identification (rsID) is used as an identifier and whether the association was related to pharmacokinetics or pharmacodynamics is given in the ‘Paper discusses’ column (Table 1). Study parameters relevant to the association are also provided for each Variant Annotation in the sub-table.
Reprinted with permission from PharmGKB.
Individual Variant Annotations for any genetic variants investigated for a drug interaction are provided in the table (Figure 3). Variant Annotations include those for single nucleotide polymorphisms (SNPs), insertions or deletions (indels), copy number variants and haplotypes. Full haplotype spreadsheets can be downloaded from the ‘Haplotype’ tab of a gene page, if available. For PMID 21383771, three different Variant Annotations with a PD relationship with warfarin are given (Figure 3). The National Center for Biotechnology (NCBI) SNP database (dbSNP) [103] reference SNP identification (rsID) is used as a reference for each variant. A standardized sentence is provided, describing the association of the allele or genotype with a drug response. For the CYP2C9 variant rs1057910 “Genotype AA is associated with increased dose of warfarin in people with a stable therapeutic international normalized ratio between two and three as compared to genotypes CC + AC”. This individual association is labeled with a PD relationship, as response to warfarin is discussed in the article. Study parameter details relevant to the significance of the association are collected, including study size, race, allele frequencies and the p-value of the association. For example, the variant associations described in PMID 21383771 were found in a study of 248 Asian individuals, and the association between rs1057910 genotype AA and increased dose of warfarin was statistically significant at p = 0.000161. This information is then stored in our database, and is valuable to both users as well as to curators when compiling Clinical Annotations (see ‘Clinical Annotations’ section below).
Our previous Variant Annotations allowed free text and were subject to the curator’s own approach. The aim of this new feature is to standardize the annotations, provide the most essential information relevant to the pharmacogenetic association, and allow all the information to enter our database so it can easily be searched or downloaded by users. We have integrated internal microdictionaries from which standardized terms can be selected. Disease and phenotype ontologies are sourced from the National Library of Medicine’s Medical Subject Headings (MeSH browser) [104], and drugs, compound and substance ontology from DrugBank [2–4,105], the WHO’s Anatomical Therapeutic Chemical (ATC) classification system [106] or PubChem® [5,107]. Unfortunately, the rsID is often not used in the paper, and we urge authors to use this form of identification to standardize genetic variant nomenclature throughout the scientific community in their future published works, facilitating the creation of useful annotations by our curators. If not provided in the article, identifying the rsID for a gene variant can entail several resources including use of PharmGKB Variant Annotations, literature cross-referencing, HapMap [108], Online Mendelian Inheritance in Man® (OMIM®) [109] or mapping to the human reference genome sequence, to ensure the correct position and, thus, the correct variant identification is obtained. If a gene or locus is associated with a drug, but no specific variant is mentioned in the article, the publication is curated by adding the relationships into our database as a Literature Annotation, represented by the ‘LA’ symbol (Table 1). These can be accessed by a variety of means, by clicking on the LA icon, by searching directly for a publication using the PMID or searching for a drug, gene or disease, and found under the ‘Is Related To’ tab (Figure 2).
Clinical Interpretations
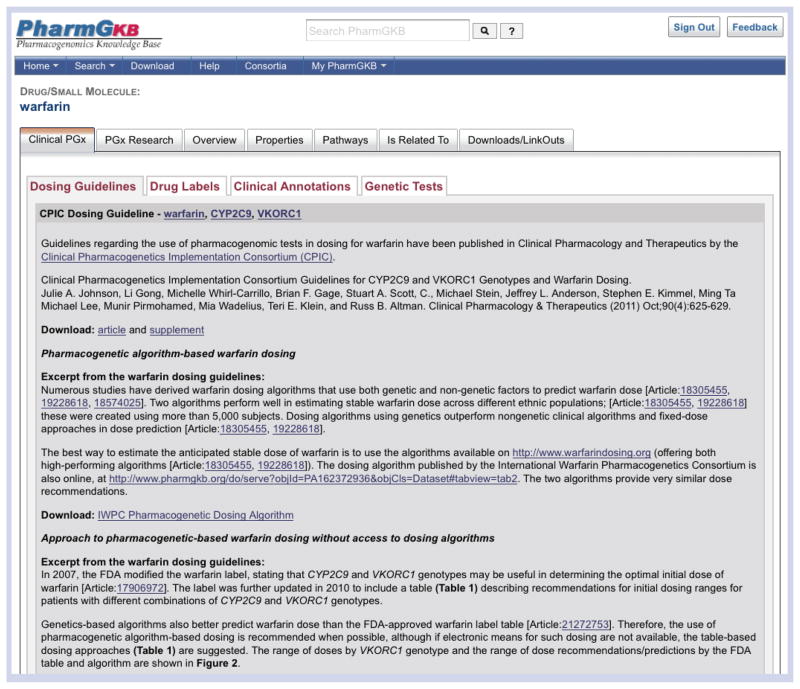
The PharmGKB Clinical Interpretations are the latest addition to the PharmGKB knowledge repertoire, a product of the direction that PharmGKB is taking towards its expanded mission “From knowledge acquisition to clinical applications”. The homepage of the PharmGKB website now features a new search box entitled ‘Clinical Interpretations’ (Figure 1). Within this box are four useful links to clinically related pharmacogenetic information on the PharmGKB website. The ‘Clinical Variant Annotations’ link takes users to a list of our curated Clinical Annotations for genetic variants with the highest level of evidence (for further information see ‘Clinical Annotations’ section below). The link to ‘Genotype-based dosing guidelines’ leads the user to a list of published genetically influenced dosing guidelines, including those by CPIC [6] and The Royal Dutch Association for the Advancement of Pharmacy Pharmacogenetics Working Group [7]. This list is updated as genotype-based dosing guidelines are released, and further information can be obtained by clicking on each gene–drug pair individually (Figure 4). A list of drug labels that have pharmacogenetic information highlighted by the US FDA [110] can be found using the ‘Drug labels’ link. We anticipate expanding the drugs label link to include international drug labels, such as pharmacogenetic information highlighted by the European Medicines Agency (EMA) and the Pharmaceuticals and Medical Devices Agency (PMDA), Japan. The ‘Genetic tests for PGx’ link takes the user to a noncomprehensive list of example pharmacogenetic diagnostic tests. Within the Clinical Interpretations box users can search for a gene, variant rsID, drug or disease in order to retrieve related clinical information. Clinically related information can also be found under the ‘Clinical PGx’ tab found on every drug and gene page on the PharmGKB website. Under this tab are four subtabs containing the relevant information for that drug or gene (Figure 4): ‘Dosing Guidelines’; ‘Drug Labels’; ‘Clinical Annotations’; and ‘Genetic Tests’.
Figure 4. The ‘Clinical PGx’ tab on the warfarin drug page.
Clinical interpretation information can be found on every drug or gene page of the Pharmacogenomics Knowledge Base (PharmGKB) website. This is exemplified here with the drug warfarin. A drug page can be found in a number of ways, for example using the search boxes on the homepage or by simply searching for ‘warfarin’ in the search box at the top of the PharmGKB website. More information about our searches can be found in our tutorials (Figure 1). Under the ‘Clinical PGx’ tab is information related to genotype-based warfarin dosing guidelines (as shown here [8]), warfarin drug labeling information, curated PharmGKB Clinical Annotations for genetic variants involved in warfarin response, and genetic tests available for warfarin-related genotyping.
Reprinted with permission from PharmGKB and Clinical Pharmacology & Therapeutics.
Clinical Annotations
The PharmGKB Clinical Annotations are created by the PharmGKB curators and aim to combine accumulated Variant Annotations to provide an evidence-rated genotype profile for a particular pharmacogenetic variant. The objective is to present a succinct clinical interpretation that is a summary of the literature evidence for an association between a genetic variant and a drug, but is also useable for clinicians, researchers and the general public. A growing number of people have purchased their genotype profile from a private company and are interested in learning more about their personalized pharmacogenetics. We hope to provide a source of information for anyone interested in pharmacogenetics. The PharmGKB curators use specific criteria to assess the collective Variant Annotations and determine the level of evidence (Table 2). Clinical Annotations are reviewed routinely, and therefore can move up or down the scale as further evidence is published, or as contradictory findings are released.
Table 2.
Level of evidence for Pharmacogenomics Knowledge Base (PharmGKB) Clinical Annotations
| Level | Definition |
|---|---|
| 1 | The highest level of evidence for a Clinical Annotation. Requires replication in populations of at least 1000 cases and 1000 controls of the same ethnicity with statistically significant p-values <0.05 after correction. Occasional exceptions are made by curators for associations that are widely accepted in the pharmacogenetics community and for which large studies are not feasible |
| 2 | A medium level of evidence for a Clinical Annotation. Associations that are found in at least one population of 100 with a statistically significant p-value of <0.05, and may or may not be replicated |
| 3 | Lower level of evidence for a Clinical Annotation. Associations that do not meet the above criteria for population size or p-value, or may be in vitro or pharmacokinetic studies only |
Reprinted with permission from PharmGKB.
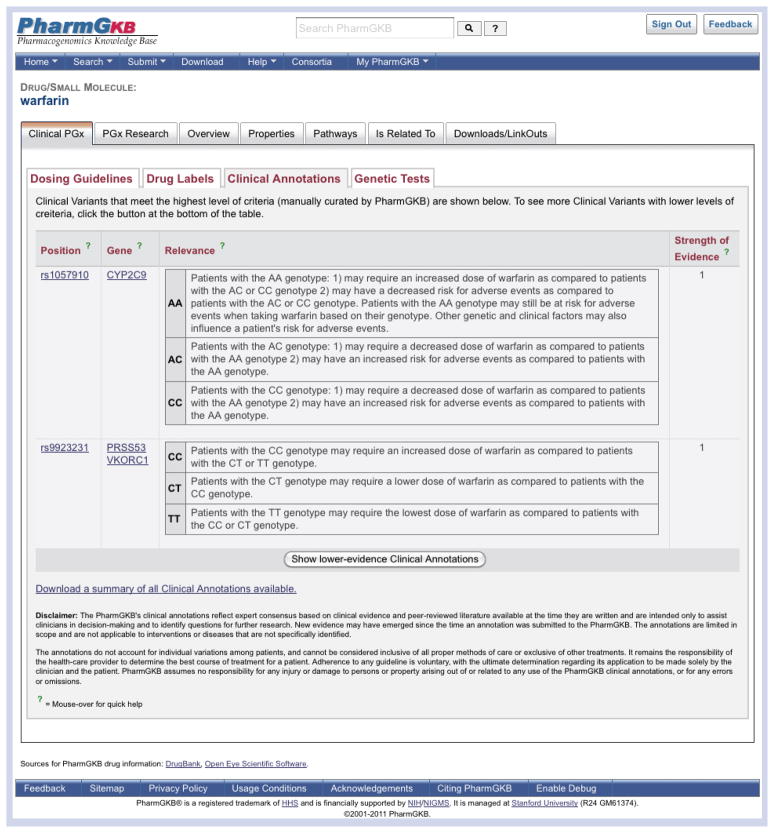
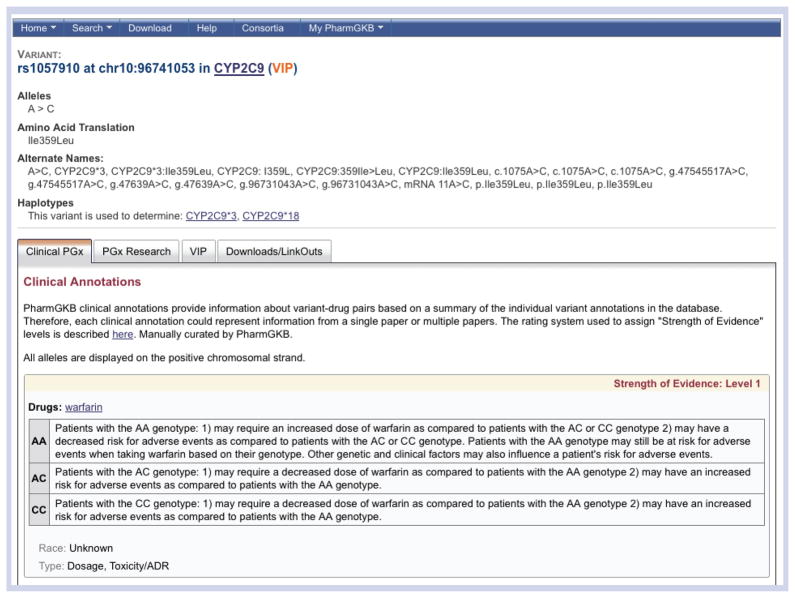
Clinical Annotations are found under the ‘Clinical PGx’ tab (Figure 5). A table of Level 1 Clinical Annotations for warfarin is shown, with the option to show the full list. A downloadable file with a summary list of all the PharmGKB Clinical Annotations is also available on this page. The table provides the variant rsID and gene, the ‘relevance’ of the association between warfarin and each genotype of the variant, and the level of evidence for this association. An example of a Level 1 Clinical Annotation for warfarin and the variant rs1057910 in the CYP2C9 gene is shown in Figure 5. A summary of information for each genotype is provided:
Figure 5. The ‘Clinical Annotations’ tab on the warfarin drug page.
Clinical Annotations are created by the Pharmacogenomics Knowledge Base (PharmGKB) curators, weighing levels of evidence to provide an association between the drug and each genotype (Table 2). They are found on drug and gene pages under the ‘Clinical Pharmacogenomics (PGx)’ tab, as shown here for the drug warfarin.
Reprinted with permission from PharmGKB.
“AA: Patients with the AA genotype: 1) may require an increased dose of warfarin as compared to patients with the AC or CC genotype 2) may have a decreased risk for adverse events as compared to patients with the AC or CC genotype. Patients with the AA genotype may still be at risk for adverse events when taking warfarin based on their genotype. Other genetic and clinical factors may also influence a patient’s risk for adverse events.
AC: Patients with the AC genotype: 1) may require a decreased dose of warfarin as compared to patients with the AA genotype 2) may have an increased risk for adverse events as compared to patients with the AA genotype.
CC: Patients with the CC genotype: 1) may require a decreased dose of warfarin as compared to patients with the AA genotype 2) may have an increased risk for adverse events as compared to patients with the AA genotype.”
Each genetic variant has a page on the PharmGKB website. By clicking on the variant link from the Clinical Annotation page, more information regarding the rs1057910 variant can be seen (Figure 6). This includes curated Clinical and Variant Annotations involving the variant, information from the VIP summary (if available) and information about haplotypes containing this variant (if available).
Figure 6. The variant page for rs1057910 in the CYP2C9 gene.
Each variant annotated on the Pharmacogenomics Knowledge Base (PharmGKB) has a page that includes information regarding the alleles, amino acid change (if relevant), alternative names that have been used to describe the variant and links to haplotypes that include the variant (if available). The ‘Clinical Pharmacogenomics (PGx)’ tab includes Clinical Annotations involving the variant and the ‘PGx Research’ tab lists Variant Annotations involving the variant. The ‘VIP’ tab is shown if Very Important Pharmacogene (VIP) summary information is available for the variant, and the ‘Downloads/Linkouts’ tab includes links to resources of further information.
Reprinted with permission from PharmGKB.
VIP summaries
The objective of a VIP summary is to provide a succinct pharmacogenetic-based overview of the literature of an important gene involved in drug response. These cover background information including gene structure, physiological role of the encoded protein and disease associations, as well as providing a collective view on PK and PD relationships with variants in the gene. The VIP summary also includes more in-depth information of particularly important variants and haplotypes involved in drug response. They provide an excellent starting point for those new to the field wanting to learn more. VIPs are chosen through a variety of means including genes referred to within FDA drug guidelines or dosing guidelines, important pharmacogenetic developments in recent literature, historically known pharmacogenes and those that have a large list of Variant Annotations on the PharmGKB website. The curator begins with a thorough search of the literature and combines Variant and Literature Annotations already available on the PharmGKB website. Each pharmacogene is assigned to a curator, and the summaries are reviewed by at least one other curator before posting on the PharmGKB website. Many of the VIP summaries are also reviewed or contributed to by experts in the field for their input and approval. We currently have 44 VIP summaries available online, many of which have been published in the Pharmacogenetics and Genomics journal. See “PharmGKB summary: very important pharmacogene information for PTGS2” for a recent example [9]. A direct link to a full list of our VIP annotations can be found on the homepage in the ‘Genes’ search box named ‘Important PGx genes’ (Figure 1). The orange ‘VIP’ symbol used throughout the PharmGKB website indicates a link to a gene or variant that has a VIP summary (Table 1).
Pathway summaries
The PharmGKB curated pathways aim to demonstrate the physiological interaction between genes and drugs in particular cells, and the relationships between different molecules with regard to their role in drug response. The pathways are documented through an extensive literature review, to provide an evidence-based summary of the PK and PD of a drug. They are drawn out using the PathVisio® software, and are available in Biopax format [10,111] to allow for this information to be more easily computationally searched, downloaded, shared across different databases and interchanged between different data platforms. A more illustrative representation of the pathway is also created. A pathway summary is included to explain the pathway and how the components interact, with links to Literature Annotations as evidence. A table of the individual components of the pathway is also provided, linking to individual gene and drug information for a more in-depth view. Many of the PharmGKB pathway summaries have also been published; see “PharmGKB summary: methotrexate pathway” for a recent example [11]. A full list of our pathways can be accessed via the homepage from the ‘Pathways’ search box, by selecting ‘All pathways’, or narrowing down the search query by selecting for PD and PK pathways (Figure 1). We currently have 79 curated pathways available online, many of which have been published in the Pharmacogenetics and Genomics journal. Through our feedback correspondence we know that the pathway diagrams are often used by researchers, students and pharmacologists for presentations and reports, citing Klein et al. [12], and provide an important resource of pharmacogenetic information.
Conclusion
A major challenge for the PharmGKB is to present the breadth and depth of material associated with human genetic variations on drug response. Our new interface is an attempt to integrate the Clinical Interpretations with our gene, drug and disease information in a straightforward layout for any user regardless of background. We encourage feedback from users of PharmGKB, and provide a ‘Feedback’ button at the top right-hand corner of each PharmGKB webpage.
Future perspective
The current challenge for curators is the huge exponential volume of pharmacogenetic literature being generated [13], keeping track of new drug labeling information and genetic test guidelines, updating previous VIP annotations as well as creating new variant and Clinical Annotations. A goal of the PharmGKB is to integrate NLP technology in an automated pipeline to identify relevant scientific publications and extract key pharmacogenomic information [14,15]. The information would be prioritized and displayed to the curators for their approval prior to entry into the knowledge base, thereby maintaining our high level of curated information while significantly increasing the speed and throughput of our current curation pipeline. This would enable broader coverage of the literature rather than the limited set of specialized journals covered today. As NLP summarization techniques improve, we expect to create automated Literature Annotations or pathway diagrams with supporting evidence automatically generated per pathway edge. NLP techniques will also be used to analyze our coverage of the pharmacogenomic space and to flag important variants and genes involved in drug response. The current role of NLP in the curation pipeline is minimal, but development is actively underway to enable more sophisticated methods of automatic knowledge aggregation and extraction. PharmGKB is currently creating an annotated corpus in which text providing the supporting evidence for Variant Annotations is highlighted in the full text articles in a semantically meaningful format. This annotated corpus will become a training set enabling collaborating text mining research groups to develop methods to automatically extract pharmacogenomic information from the text.
The heterogeneous and often confusing nomenclature for single variants, alleles and haplotypes for many gene families (e.g., CYP450 and UGT), makes curation harder but at the same time more valuable. The in-depth VIP and Clinical Annotations, which gather heterogeneous information and create a consensus, are valuable tools for researchers, students and clinicians alike, and we will continue with these endeavors. We also aim to further distribute PharmGKB-generated tools and information, aid in education of pharmacists, pharmacology and medical students, molecular scientists, drug regulators, bioinformaticians, and in schools, to help broaden our scope of users, contribute to scientific communication with the public, as well as contributing to driving personalized medicine forward.
Executive summary.
Background
The Pharmacogenomics Knowledge Base (PharmGKB) website has been redesigned to integrate Clinical Interpretations.
Pharmacogenomics research
The Pharmacogenomics (PGx) tab on each drug page provides an overview of the information available for genetic variants associated with that drug. On gene pages, the PGx tab provides an overview of the information available for genetic variants within the gene that have been associated with pharmacodynamics or pharmacokinetics.
Variant Annotations
Variant Annotations by the PharmGKB curators use a standardized sentence to give the bottom-line association between a drug and variant reported in a published article, and are integrated into our database.
Clinical Interpretations
The ‘Clinical PGx’ tab found on each gene or drug page contains information about dosing guidelines, drug labeling, genetic tests and curated Clinical Annotations, all available directly from the homepage under the ‘Clinical Interpretations’ box.
Clinical Annotations
Clinical Annotations created by the PharmGKB curators provide an evidence-based summary of the association between a genotype and a drug.
Very Important Pharmacogene summaries
We continue to expand our collection of Very Important Pharmacogene (VIP) summaries.
Pathway summaries
Our PharmGKB pathways are now integrated into Biopax™.
Future perspective
Future directions include the integration of natural language processing technology for knowledge aggregation and information retrieval.
Acknowledgments
The authors would like to thank the PharmGKB team.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work is supported by the NIH National Institute of General Medical Sciences (R24 GM61374) and the National Library of Medicine (contract HHSN-276201000025C). RB Altman serves as a founder and consultant for Personalis and a consultant to 23andMe. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪ ▪ of considerable interest
- 1.Chan SL, Suo C, Lee SC, Goh BC, Chia KS, Teo YY. Translational aspects of genetic factors in the prediction of drug response variability: a case study of warfarin pharmacogenomics in a multiethnic cohort from Asia. Pharmacogenomics J. 2011 doi: 10.1038/tpj.2011.7. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Knox C, Law V, Jewison T, et al. DrugBank 3.0.: A comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolton E, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. In: Bolton E, Wang Y, Thiessen PA, Bryant SH, editors. Annual Reports in Computational Chemistry. American Chemical Society; Washington, DC, USA: 2008. [Google Scholar]
- 6▪▪.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. Provides an overview of the Clinical Pharmacogenetics Implementation Consortium (CPIC), the rationale behind its formation and the rating scheme for strength and evidence for each recommendation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7▪▪.Swen JJ, Nijenhuis M, De Boer A, et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi: 10.1038/clpt.2011.34. Provides pharmacogenetic-based therapeutic recommendations for 53 drugs. [DOI] [PubMed] [Google Scholar]
- 8▪.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. Provides an indepth evidence-based review of therapeutic dosing guidelines for warfarin integrating pharmacogenetic information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Thorn CF, Grosser T, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for PTGS2. Pharmacogenet Genomics. 2010;21(9):607–613. doi: 10.1097/FPC.0b013e3283415515. Example of a Pharmacogenomics Knowledge Base Very Important Pharmacogene (VIP) summary. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demir E, Cary MP, Paley S, et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28(9):935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪.Mikkelsen TS, Thorn CF, Yang JJ, et al. PharmGKB summary: methotrexate pathway. Pharmacogenet Genomics. 2011;21(10):679–686. doi: 10.1097/FPC.0b013e328343dd93. Example of a Pharmacogenomics Knowledge Base pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪▪.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenomics J. 2001;1(3):167–170. doi: 10.1038/sj.tpj.6500035. Explains the formation of the Pharmacogenomics Knowledge Base. [DOI] [PubMed] [Google Scholar]
- 13▪.Garten Y, Coulet A, Altman RB. Recent progress in automatically extracting information from the pharmacogenomic literature. Pharmacogenomics. 2010;11(10):1467–1489. doi: 10.2217/pgs.10.136. Provides a comprehensive review of recent developments in natural language processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. J Biomed Inform. 2010;43(6):1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulet A, Garten Y, Dumontier M, Altman RB, Musen MA, Shah NH. Integration and publication of heterogeneous text-mined relationships on the semantic web. J Biomed Semantics. 2011;2(Suppl 2):S10. doi: 10.1186/2041-1480-2-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.PharmGKB. www.pharmgkb.org.
- 102.PubMed®. National Center for Biotechnology Information, US National Library of Medicine; www.ncbi.nlm.nih.gov/ [Google Scholar]
- 103.dbSNP. National Center for Biotechnology Information, US National Library of Medicine; www.ncbi.nlm.nih.gov/snp/ [Google Scholar]
- 104.National Library of Medicine’s Medical Subject Headings (MeSH browser) www.nlm.nih.gov/mesh/2011/mesh_browser/MBrowser.html.
- 105.DrugBank. www.drugbank.ca.
- 106.WHO Anatomical Therapeutic Chemical (ATC) classification system. www.whocc.no/atc/structure_and_principles/
- 107.PubChem®. http://pubchem.ncbi.nlm.nih.gov/
- 108.International HapMap Project. http://hapmap.ncbi.nlm.nih.gov/
- 109.OMIM®, Online Mendelian Inheritance in Man®. www.ncbi.nlm.nih.gov/omim.
- 110.US FDA. Table of pharmacogenomic biomarkers in drug labels. www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- 111.Biopax. www.biopax.org.