Abstract
This work describes the performance of poly(methyl methacrylate) (PMMA) microfluidic DNA purification devices with embedded microfabricated posts, functionalized with chitosan. PMMA is attractive as a substrate for creating high surface area (SA) posts for DNA capture because X-ray lithography can be exploited for extremely reproducible fabrication of high SA structures. However, this advantage is offset by the delicate nature of the posts when attempting bonding to create a closed system, and by the challenge of functionalizing the PMMA surface with a group that invokes DNA binding. Methods are described for covalent functionalization of the post surfaces with chitosan that binds DNA in a pH-dependent manner, as well as for bonding methods that avoid damaging the underlying post structure. A number of geometric posts designs are explored, with the goal of identifying post structures that provide the requisite surface area without a concurrent rise in fluidic resistance that promotes device failure. Initial proof-of-principle is shown by recovery of prepurified human genomic DNA (hgDNA), with real-world utility illustrated by purifying hgDNA from whole blood and demonstrating it to be PCR-amplifiable.
Introduction
Polynucleic acid purification is the precursor to DNA analysis of any type. DNA must be separated from cellular components which can inhibit downstream analysis, including PCR amplification. While there are a number of different approaches to nucleic acid purification (e.g., phenol–chloroform extraction, density gradient centrifugation),1 solid phase extraction (SPE) using a silica solid phase has become the most common and easily adaptable to microdevices.2–11 Since microchip purification occurs in a closed environment, many of the sample transfer steps of traditional SPE are eliminated, decreasing the risk of contamination and sample degradation. In addition, reduced sample and reagent volumes associated with microchip SPE translate to reduced cost and analysis time.
A wide variety of silica phases have been developed for microfluidic DNA purification ranging from packed silica beads8 to fabricated silica posts.7 Most microfluidic DNA extraction methods, including those mentioned, are performed in glass microchips. In the case of fabricated silica posts, it is often difficult to consistently bond glass covers to the top of glass posts (with small areas for bonding) without damaging the posts, due to the requisite bonding pressures. These difficulties imply that glass devices are labor-intensive and thus, costly, discouraging their use as disposable devices for point-of-care applications. Moreover, silica-based extraction requires the use of reagents (e.g. guanidine or isopropyl alcohol) that can inhibit downstream processes such as PCR.
The inherent challenges associated with glass microfabrication have spurred efforts to develop devices with alternative materials, notably polymers (polyolefin, polycarbonate, epoxy, and PMMA12–16) that are amenable to low cost fabrication approaches. The integration of silica-based SPE into polymeric devices offers fabrication advantages and yields attractive extraction performance, but existing approaches still require chaotropes and/or organic reagents that interfere with downstream processes. Alternatives to silica-based extractions, such as carboxy-coated surfaces that use poly (ethylene glycol) (PEG) or tetra (ethylene glycol) (TEG) for DNA binding15 still utilize other organic reagents (i.e. ethanol) that lead to similar concerns regarding inhibition of downstream processing. Moreover, the use of alcohols with polymers creates additional challenges relating to the deleterious effects of alcohols on polymer surfaces and interface integrity.
While each of the polymer-based microdevices presented here represents valuable steps towards a single-use plastic microdevice for SPE, none have met the optimal criteria: a device that (i) demonstrates efficient extraction of DNA from complex biological samples, (ii) avoids the use of PCR-inhibitory reagents used in silica-based purifications, (iii) exhibits a highly reproducible solid phase, (iv) yields surfaces that are easily modified and robust, and (v) is amenable to scalable cost-effective fabrication (i.e., enabling disposable devices).
In this work, a polymeric device for DNA purification is presented which meets these requirements by utilizing an extraction method, previously described by Cao et al.,17 based on pH-induced DNA binding to a chitosan-based phase.17 Chitosan, the partially deacetylated form of chitin, was shown to bind DNA in a pH-dependent manner: charge–charge interactions bind the DNA to the protonated chitosan at pH 5; neutralization of the charge at pH 9 results in DNA release. Purification of nucleic acids using the chitosan-based phase has previously been adapted to the microchip for SPE of DNA from blood and RNA from buccal swabs and resulted in PCR and reverse transcriptase PCR (RT-PCR) amplifiable product.17,18 This phase has been shown to be reproducible and is completely aqueous (eliminates PCR-inhibitory reagents necessary for traditional silica-based purifications), making the method more compatible with PCR integration.8,9
The work presented here describes the combination of the chitosan-based purification chemistry with a PMMA microfluidic, fabricated using the LIGA process that exploits X-ray lithography to create the high surface area, complex 3-D structures (pillars). The high aspect ratio of the pillars necessitated development of a procedure for bonding PMMA to PMMA which (i) would not destroy the complex 3-D structures within the channels but (ii) would create a bond which can withstand the pressure of syringe-driven flow. The most advantageous device design was determined from six possibilities. Recovery of DNA using the aqueous, chitosan-based extraction method from both prepurified DNA and whole blood, a common clinical sample, is also demonstrated, proving the utility of the device for clinical applications. This work demonstrates the foundation/first steps for developing a fully integrated genetic analysis plastic microdevice.
Materials and methods
Reagents
2-(4-Morpholino)-ethanesulfonic acid (MES, enzyme grade), hydrochloric acid, sodium hydroxide, Taq DNA polymerase, 10× PCR buffer, dNTPs, and MgCl2 were purchased from Fisher (Fair Lawn, NJ). 2-Amino-2-(hydroxymethyl)-1,3-propanediol (Trizma Base, 99.9%), low molecular weight chitosan (chitosan oligosaccharide lactate), ethylenediaminetetraacetic acid (EDTA) disodium salt dihydrate, 1,2-dichloroethane anhydrous (99.8%), and bovine serum albumin were purchased from Sigma (St Louis, MO). Potassium chloride was purchased from Mallinckroft (Paris, KY). Triton X-100 for molecular biology was purchased from Fluka (St Louis, MO). Proteinase K was purchased from Qiagen® (Valencia, CA). Quant-iT™ PicoGreen® dsDNA reagent, an intercalating fluorescent dye, was purchased from Invitrogen™ (Carlsbad, CA). Ethanol was purchased from AAPER Alcohol and Chemical Company (Shelbyville, KY). DNA primers for amplification of a fragment of the gelsolin gene were purchased from MWG-Biotech, Inc. (High Point, NC). Purified human genomic DNA was obtained through in-house purification from whole blood. All solutions were prepared in Nanopure water (Barnstead/Thermolyne, Dubuque, IA).
Microdevice fabrication
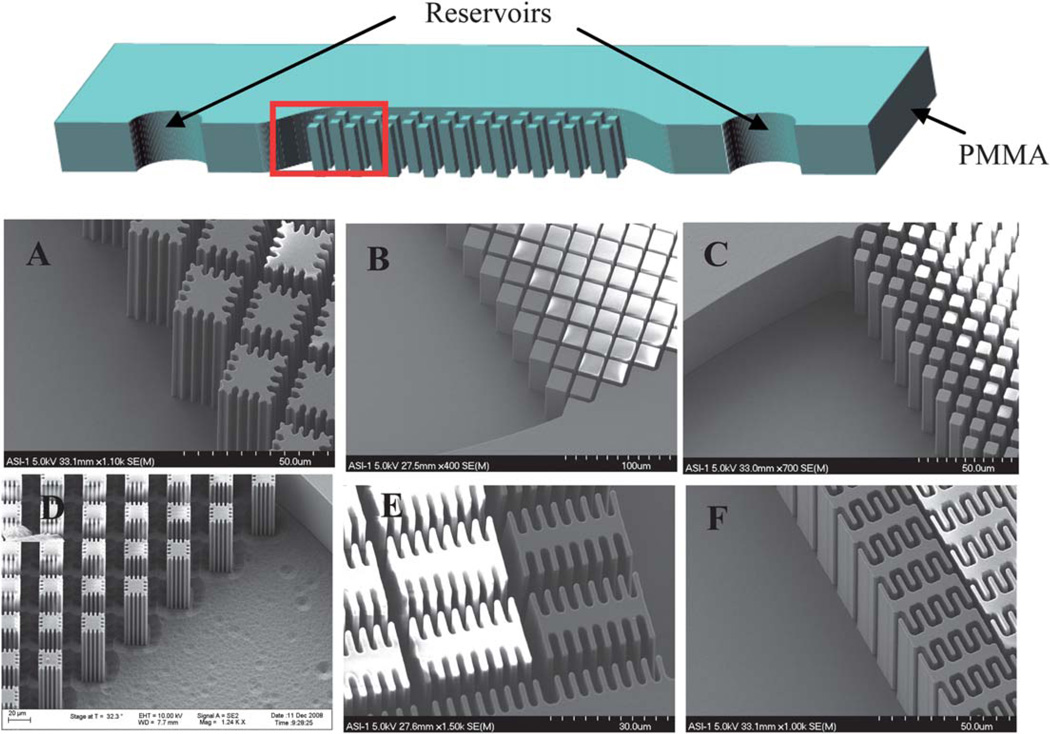
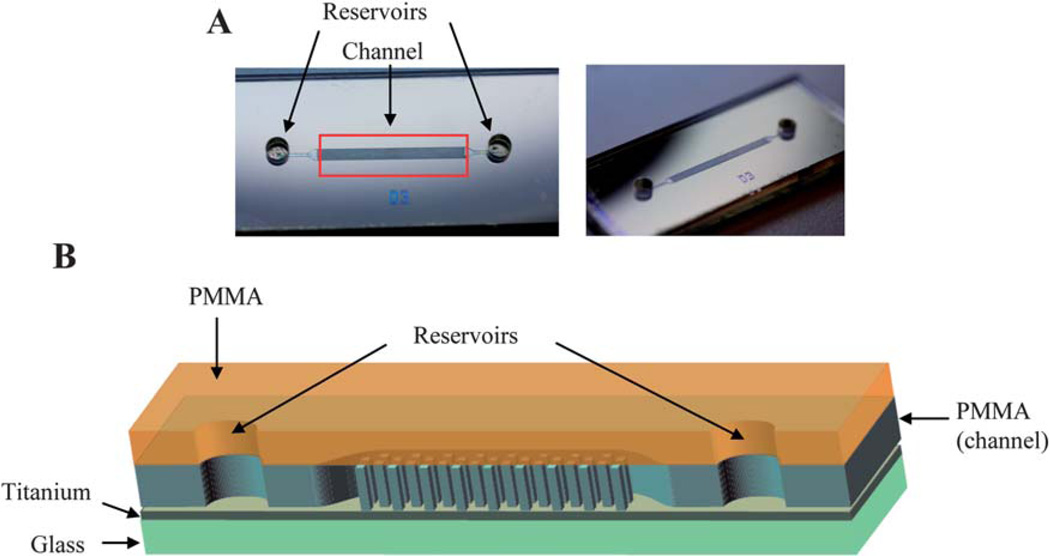
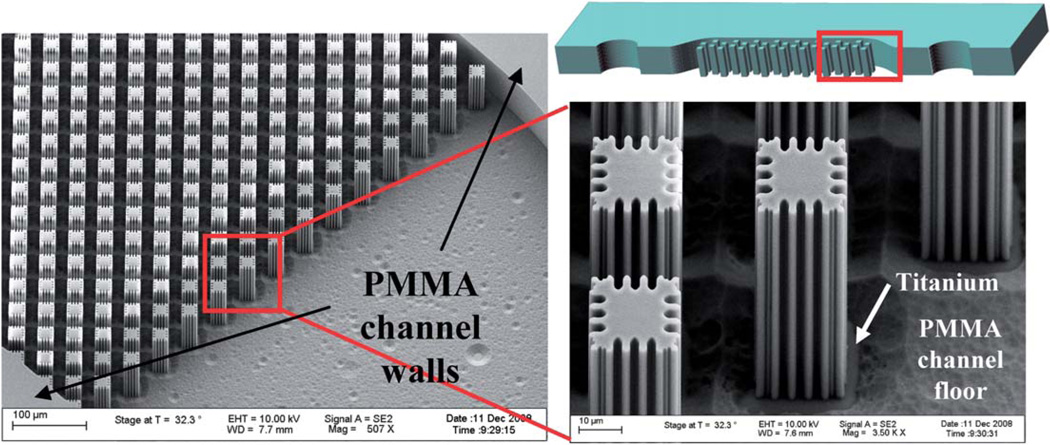
Microdevices were fabricated by HT Microanalytical, Inc., using the LIGA process that exploits X-ray lithography. Typically, fabrication using X-ray lithography uses a base substrate, such as glass, that a conductive material is deposited on. An X-ray photoresist material, such as PMMA, is then bonded to the conductive material. The desired pattern is translated to the photoresist through the use of a mask during X-ray exposure. The exposed photoresist is then developed/removed leaving the desired features of the device design.19 In this work glass was used as the base substrate onto which a layer of titanium was deposited. PMMA was then bonded to the titanium and the device exposed to X-rays, while a mask, previously generated, was used to define the features of the design. Following developing, a 1 cm long channel with a width of 800 µm and a depth of 50 µm (Fig. 1) resulted (Fig. 2A and B). PMMA cover pieces were fabricated with two 1 mm reservoirs drilled for the inlet and outlet.
Fig. 1.
Schematic of device design without a bonded PMMA cover piece (top). Dimensions: 1 cm long channel, 800 µm wide and 50 µm deep. Red box indicates location of SEM images A–F. (A) Device design A with dimensions: 14 µm square posts with ~3 µm extensions with 4 µm distance between posts. (B) Device design B with dimensions: 22 µm square posts with 4 µm distance between posts. (C) Device design C with dimensions: 8 µm square posts with 4 µm distance between posts. (D) Device design D with dimensions: post structure similar to design A but with 17 µm between each post. (E) Device design E with dimensions: 6µm × 30 µm posts with 8 µm extensions. (F) Device design F with dimensions: 6µm × 30 µm posts with 5 µm extensions.
Fig. 2.
(A) Bonded PMMA microdevice. (B) Schematic of bonded PMMA microdevice with each layer of device labeled.
SPE apparatus
Hamilton (250 µL) gas-tight syringes (Hamilton, Reno, NV) were connected to the microdevice through 0.25 mm i.d. PEEK™ tubing and mini-tight fittings (Upchurch, Oak Harbor, WA). The PEEK™ tubing was connected to the microdevice by pressfit into the inlet reservoir. Pressure driven solution flow was achieved with a SP100i syringe pump (WPI, Sarasota, FL). All experiments were performed at a flow rate of 1.6 µL min−1.
Device bonding
Bonding of the PMMA cover to the channel containing the PMMAposts was achieved using a method modified from that of Lin et al., which uses a low azeotropic solvent for bonding.20 The cover and channel were held together and 1–2 µL of 20% 1,2- dichloroethane (DCE), by weight, 80% ethanol solution (EtOH/ DCE solution) was pipetted along one contact edge and allowed to wick in. Pressure was applied for 5 min. The same volume of the EtOH/DCE solution was applied to the other contact edge of the substrates followed by 5 min of pressure. Glass slides along with C-clamps were tightened down to both sides of the substrates and allowed to sit for ≥2 h. The bonded device was removed from the clamps and water was flowed through the device for 15 min.
Chitosan functionalization
Functionalization of the PMMA with chitosan was based upon work by Witek et al.15 and achieved by first flushing water through the device for 15 min. Approximately 50 µL of 10 mM chitosan/10 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC)/100 mM K2HPO4, pH 7 coating solution prepared fresh for each coating, was flowed into the device manually with a syringe. The solution was allowed to incubate in the chip for 5 min. An additional 50 µL of coating solution was manually flowed onto the chip. The chip was submerged in 10 mL of the coating solution and vortexed at 1200 rpm for ≥11 h. The device was removed from the coating solution and water was flowed (by syringe and syringe pump) through the device for 30 min.
Sample preparation
Human genomic DNA (hgDNA) was purified in-house from whole blood obtained from the University of Virginia Medical School. To prepare lysed whole blood samples, a volume of blood containing 500 ng of DNA (calculated from the known white blood cell (WBC) count of the whole blood and the assumption of 6.25 pg DNA/WBC21) was added to 20 µL of 20 mg mL−1 proteinase K (Qiagen) and 50 mM MES pH 4.24 to bring the volume up to 500 µL. The solution was vortexed for 1 min and incubated in a water bath at 56 °C for 10 min.
Chitosan stripping
Following each extraction on a chitosan-coated PMMA device the chitosan phase was stripped by flowing 1 M NaOH through the device for 1 h. The device was then flushed with water for 1 h.
SPE procedure for DNA purification using an unfunctionalized PMMA device
A device that had never been coated with chitosan was used and first conditioned with 10 mM MES, pH 5 for 10 min. A load solution containing 40 ng hgDNA in 25 µL 10 mM MES, pH 5, was then flowed into the device and five 5 µL fractions were collected from the outlet. The channel was then washed with 10 µL of 10 mM MES, pH 5, while five 2 µL fractions were collected. The DNA was then eluted from the phase by flowing 10 mM Tris/50 mM KCl, pH 9, and ten 2 µL fractions were collected. An additional 100 µL of 10 mM Tris/50 mM KCl, pH 9, was flushed through the device as a rinse. All flow rates were 1.6 µL min−1. All fractions collected were stored for later fluorescence analysis.
SPE procedure for DNA purification using a chitosan coated and stripped PMMA device
Devices were conditioned with 10 mM MES, pH 5, for 10 min. A 25 µL load solution containing 10 ng hgDNA in 10 mM MES, pH 5, was loaded onto the device and five 5 µL fractions were collected. The phase was washed with 10 µL of 10 mM MES, pH 5, and five 2 µL fractions were collected. DNA was eluted by flowing elution buffer, 10 mM Tris/50 mM KCl, pH 9, through the device and collecting ten 2 µL fractions. All fractions were stored for fluorescence analysis.
SPE procedure for DNA purification from whole blood
A lysed whole blood sample was prepared as described above in Sample preparation. A chitosan-coated device was conditioned with 50 mM MES/1% Triton X-100, pH 4.24, for 10 min. Sample (25 µL) was then loaded onto the device and five 5 µL fractions were collected. The phase was washed with 10 µL of 10 mM MES, pH 5, and five 2 µL fractions were collected. The DNA was eluted from the phase with 10 mM Tris/50 mM KCl, pH 9, and ten 2 µL fractions were collected. All fractions were stored for later fluorescence or PCR analysis.
Fluorescence analysis
All fractions collected were prepared for analysis using a Pico-Green® intercalating dye, fluorescence assay.22 The samples were analyzed using a NanoDrop 3300 Fluorospectrometer (Nano-Drop, Wilmington, DE).
PCR analysis
Extracted DNA elution fractions were added to PCR master mix containing 3 mM MgCl2, 0.2 mM dNTPs, 0.8 µM forward and reverse primers (5′-AGTTCCTCAAGGCAGGGAAG-3′ and 5′- CTCAGCTGCACTGTCTTCAG-3′, MWG BioTech, High Point, NC)for a portion of the gelsolin gene, 1 × PCR buffer, 10 µg mL−1 BSA, 0.5 units µL−1 Taq DNA polymerase, and autoclaved water up to 25 µL. Samples were amplified using standard PCR protocols developed in the lab and a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA) for thermal cycling. The protocol used an initial denaturation step of 95 °C for 2 min, followed by 35 cycles of 95 °C, 30 s/64 °C, 30 s/72 °C, 30 s, and a final extension of 72 °C for 2 min. DNA 1000 Series II kit and a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA) were used for separation and analysis of the PCR products.
Results and discussion
Development of device bonding
In order to produce devices that can withstand the pressure of the syringe-driven flow at a flow rate of 1.6 µL min−1, a bonding method is required to form a tight PMMA–PMMA bond (cover to top of posts) without damaging the intricate post structures that form the backbone of the solid phase. It is important to first note the basis for the selection of 1.6 µL min−1 as the flow rate required. Previous work had determined the optimal linear flow rate for DNA purification on a chitosan solid phase to be 0.67 mm s−1 (data not shown). Based upon this, the optimal volumetric flow rate to operate DNA purification in these devices was calculated to be 1.6 µL min−1. Initial bonding attempts employed epoxy (a thin layer applied to the PMMA cover followed by bonding to the channel) to bind the top PMMA cover to the channel that contained the PMMA fabricated post structures. This method resulted in only partial bonding at the top of the posts, as was evident when black dye was flowed through the device (Fig. 3A). Black printer dye, used for easy visualization of bonded regions, can be seen in Fig. 3A where the tops of the posts are not bonded to the top PMMA cover piece (i.e., the dye flows over the posts). Although the posts remained intact with this method, the bonding was not capable of withstanding the pressure generated by the syringe-driven flow, as evidenced by leaking at the contact edges of the bonded surfaces.
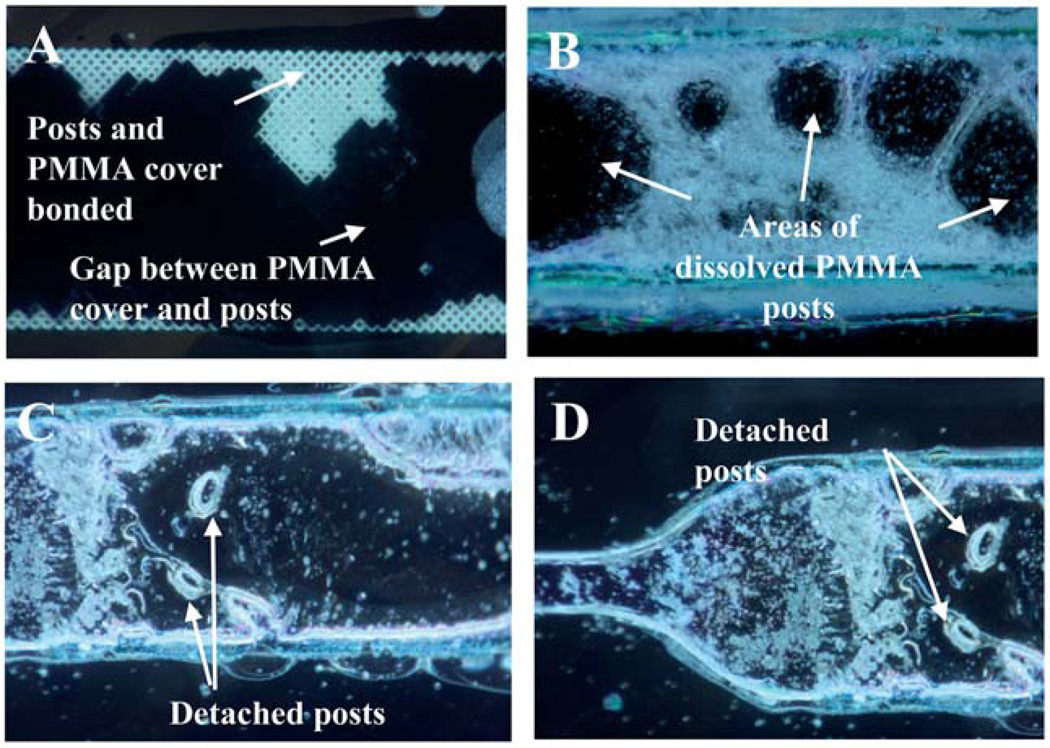
Fig. 3.
Images from devices bonded using epoxy (A) which resulted in insufficient bonding or solvent bonding (B–D) which resulted in damage to the post structures.
Bonding using a sacrificial layer (agarose and photoresist) as described previously,23 solvent vapor (hexanes and chloroform) with applied pressure,24 or solvent coating (isopropanol, acetone, and chloroform) with applied pressure were all tested. Both sacrificial layer and solvent vapor with pressure resulted in nonbonded (in all areas of the chip) devices (data not shown), while solvent coating with applied pressure resulted in damage to the 3-D microstructures (Fig. 3B–D). Damage to the posts is clear in Fig. 3B–D where posts have either been detached from the bottom of the channel or dissolved, leaving gaps within the channel that significantly reduce the separation efficiency of the device by dramatically lowering the surface area utilized during extraction. This clearly demonstrates that previous methods that successfully bond plastic devices are unsuitable for devices with post structures.
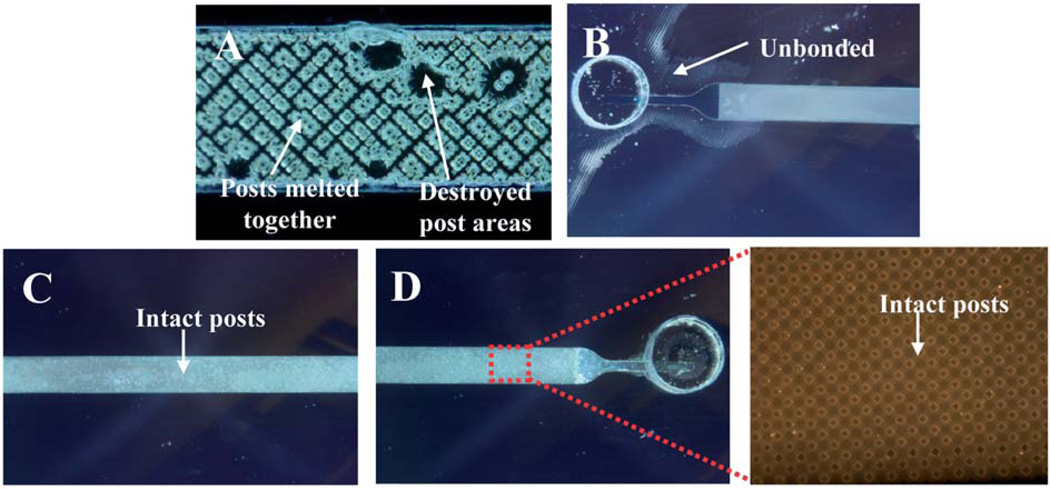
A device bonding method was then devised by modifying that described by Lin et al.20 where bonding was completed at room temperature using a low azeotropic solvent. In that work, the authors characterized the bonding method and showed it successful for bonding channels as wide as 3 mm without causing the channel to collapse. Additionally, the bonding method was shown to have a 17-fold greater bonding strength than conventional thermal bonding techniques which was tested by applying force directly to the bonded surfaces until bond failure occurred. For bonding of the devices in this work, the volume of bonding solution was adjusted to −20 µL of the DCE/EtOH mixture, from the 2–3 drops (equivalent to −50–75 µL) suggested by Lin et al. due to the delicate nature of the complex 3-D structures present in the channel. This volume resulted in some destruction of the post structures (Fig. 4A). In an attempt to account for this and to prevent excess solvent from contacting the posts in the devices used in this work, the volume of solvent was decreased to 1–2 µL and pressure was applied with C-clamps (to the point where the chip remained stationary without sliding in the clamp) as before. This method resulted in a portion of the device being unbonded (Fig. 4B). By applying five minutes of directed pressure (handheld) before switching to C-clamps, fully bonded devices were achieved (Fig. 4C and D).
Fig. 4.
Images showing development of bonding method where 20 µL of DCE/EtOH solvent was used (A) damaging some of the post structures or where 1–2 µL was used but with too little pressure (B) and then the optimized bonding where the device is intact (C and D) along with posts (inset).
Following bonding, a 50 µm deep, 1 cm long, and 800 µm wide channel with microscale posts resulted (Fig. 2A and B). The strength of the bonding was then tested by flowing water through the device at 1.6 µL min−1. For design D, the device remained bonded with no observable loss of solution from the device (Table 2). In contrast, the other designs debonded and solution began to leak between the cover and PMMA channel layers upon flowing water through the device. This demonstrated that, using the developed bonding procedure, the devices can withstand a head pressure (Table 1) of 0.8 kPa (head pressure generated with design D) whereas at 27.2 kPa (head pressure generated with design C) the bond no longer holds. This led to the selection of design D for all subsequent studies—this architecture is described in depth in the next section. The adjustments made to the bonding procedure provided a method for bonding PMMA to PMMA for devices that contain 3-D microstructures that are susceptible to damage when exposed to excessive solvent, heat and/or pressure.
Table 2.
Surface and aperture dimensions of each PMMA device design in addition to 30 and 15 µm silica for comparison
| Sphere diameter/µm | Chamber SA/V ratio/cm2 cm−3 | Total chamber SA including chamber walls/cm2 |
Largest diameter inscribed circular aperture/µm |
|---|---|---|---|
| 30 | 1480 | 0.67 | 4.64 |
| 15 | 2960 | 1.26 | 2.32 |
| Micropost designs | |||
| Micropost design A | 3007 | 1.29 | 4 |
| Micropost design B | 3363 | 1.44 | 4 |
| Micropost design C | 2218 | 0.98 | 4 |
| Micropost design D | 1281 | 0.63 | 17.1 |
| Micropost design E | 3737 | 1.58 | 4 |
| Micropost design F | 4028 | 1.69 | 2 |
Table 1.
Head pressure of each device design and whether each was capable of withstanding the pressure generated during syringe-driven flow (denoted by yes and no)a
| Device design | Head pressure/kPa | Withstood pressure during flow |
|---|---|---|
| A | 41.6 | No |
| B | 62.4 | No |
| C | 27.2 | No |
| D | 0.8 | Yes |
| E | 32 | No |
| F | 360 | No |
No—device bond failed and solution leaked from the device.
Selection of device design
The effectiveness with which DNA can be extracted using the PMMA microdevices depends not only on the surface chemistry but also on the design of the device. The device must have ample surface area for functionalization with chitosan to ensure sufficient sites for DNA binding and a design that provides minimal back pressure during syringe-driven flow. Six post designs were fabricated in order to determine the optimal design with regard to both surface area and flow. The surface area and largest aperture were determined for each design as well as for 30 and 15 µm beads packed in a channel (1 cm × 200 µm × 200 µm), to allow for comparison to previously described methods for DNA extraction (Table 2).2,5,8,9,25,26 The device design chosen was based upon a number of factors including flow resistance through the device and surface area for binding. The chosen design (as previously mentioned) has a similar surface area (0.63 cm2) to that of 30 µm beads (0.67 cm2), which would allow for sufficient quantities of DNA to bind to the phase, resulting in adequate DNA for ensuing analyses. Another factor that led to the selection of design D was the aperture diameter of 17.1 µm, significantly larger than the other designs at ~2–4 µm. The larger aperture of design D leads to less flow resistance through the channel when using pressure-driven flow. Head pressure calculations were completed for each design, as previously mentioned, and show that less flow resistance occurs with design D during syringe-driven flow at 1.6 µL min−1 (Table 1). It was determined that design D resulted in the lowest head pressure, 0.8 kPa at a flow rate of 1.6 µL min−1, compared to 27.2 to 360 kPa for the other designs considered (Table 1). Flow of water through each device design was attempted at 1.6 µL min−1 and the effect on the device was observed. It was found that designs A, B, C, E and F resulted in solution leaking from the side of the device, an indication that the PMMA bond failed under the pressure conditions associated with that flow rate. Thus only design D, with the lowest calculated head pressure and largest aperture, was found to be amenable to separations using syringe-driven flow at the rate required for efficient separation (Table 1), 1.6 µL min−1. Although a slower flow rate could potentially be used, which would result in less bond failure of the other designs, an increase in the time required for purification would follow. Therefore, to minimize the time required for the extraction, design D was chosen as the most advantageous design for the remaining studies (Fig. 5).
Fig. 5.
SEM images of device design D.
Extraction of hgDNA on chitosan-coated PMMA device
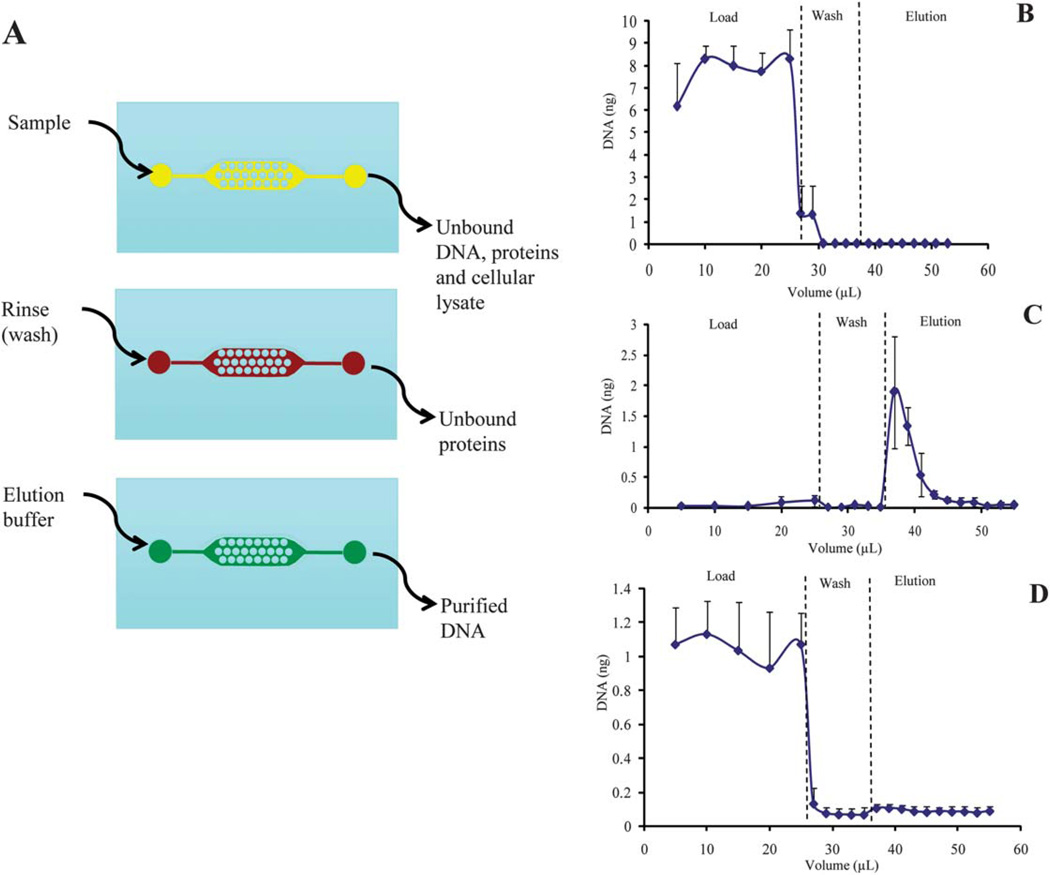
Prior to functionalizing the PMMA surface to yield a surface chemistry that would allow for DNA binding (chitosan), the interaction of DNA with the PMMA microstructures of the channel was examined during the three steps that comprise an extraction—the load, wash, and elution (Fig. 6A). Typically, during the load step, the DNA in the biological sample binds to the solid phase while unbound cellular and extracellular material is removed during the wash step. The purified DNA is then eluted from the solid phase during the elution step. A solution containing 40 ng human genomic DNA (the mass of DNA yielded from ~5700 cells) (hgDNA) in 25 µL 10 mM MES, pH 5, was loaded onto the device (after the device had been thoroughly rinsed for ~1 h with deionized water at 1.6 µL min−1) and the extraction performed as described in SPE procedure for DNA purification using an unfunctionalized PMMA device in Materials and methods. DNA, detected by fluorescence using a commercial intercalating reagent (PicoGreen®), flowed unretained through the device containing an underivatized surface during the load step (Fig. 6B), demonstrating that little to no DNA was binding to the PMMA micropost structures. Residual DNA from the load was recovered during the wash step with 10mMMES, pH 5. No DNA was recovered (as can be detected with PicoGreen® analysis) during the elution step where 10 mM Tris/50 mM KCl, pH 9, was infused through the device. This clearly demonstrates that the PMMA microdevice requires functionalization with a DNA binding phase, such as chitosan.
Fig. 6.
(A) Schematic of the DNA purification procedure where the lysed biological sample is loaded, the phase rinsed with buffer (wash step) to remove any unbound material, and finally, elution buffer flowed through the device to elute the purified DNA. (B) The extraction profile from the purification of hgDNA (40 ng) on a PMMA device not coated with chitosan (n = 3). (C) The extraction profile from the purification of hgDNA (10 ng) on a PMMA device coated with chitosan which resulted in an extraction efficiency of 47.8 (±9.3)% EE (n = 5). (D) The extraction profile from the purification of hgDNA (10 ng) on a chitosan-coated device that had been stripped with 1 M NaOH post-extraction to remove the chitosan phase (n = 4).
After determining that functionalization was necessary in order to reversibly bind and purify DNA using the PMMA device, a method developed for coating PMMA was tested. A device that had been coated with chitosan, as described in Chitosan functionalization in Materials and methods and based upon previous work by Witek et al.,15 was used to purify hgDNA (10 ng). After loading the sample, the device was washed with 10 mM MES, pH 5, and the DNA eluted with 10 mM Tris/50 mM KCl, pH 9. Fractions collected were analyzed using a fluorescence assay as described in Materials and methods. The resulting extraction profile shows that DNA was efficiently bound to the chitosan phase during the load and recovered during the elution, resulting in an extraction efficiency of 47.8 (±9.3) % (n = 5) (Fig. 6C). These data demonstrate that a PMMA device can be coated with chitosan and provide a reproducible, efficient extraction of hgDNA. These results were obtained with multiple devices, demonstrating chip-to-chip reproducibility of the method. These extractions represent the first use of a PMMA microdevice with chitosan functionalization for the extraction of DNA. Functionalization of glass microdevices using chitosan for the extraction of DNA has previously been shown by Cao et al.,17 but plastic (PMMA) microdevices provide the inherent advantage of being cost effective when in large scale production, thus moving toward a completely disposable integrated device for clinical or on-site analysis.
Chitosan stripping and extraction of stripped PMMA device
Because these devices are not yet mass produced in quantities to allow for single-use disposability, the chitosan coating needed to be stripped from the PMMA posts following the extraction of DNA to ensure that no DNA was carried over between multiple extractions on the same device. The multiple use of each device will be eliminated and changed to single-use when the devices become commercialized, but for research purposes the devices need to be used for multiple extractions. The removal of the chitosan phase post-extraction (chitosan stripping) was performed by flowing 1 M NaOH through the device for 1 h, followed by flowing water for 1 h. Sodium hydroxide was chosen for stripping due to the fact that it can hydrolyze the amide bonds that are formed between the PMMA and chitosan during the coating procedure.27 A concentration of 1 M was chosen to balance sufficient hydrolysis of the amide bond with preserving the fragile post structures, which are susceptible to highly concentrated solutions.
The efficiency of the base hydrolysis of the amide bond was determined to ensure that all chitosan had been removed, thus reestablishing the binding sites for recoating with chitosan for the next extraction. The efficiency of hydrolysis was determined by loading 10 ng of hgDNA onto the stripped PMMA device in 10 mM MES, pH 5, washing with 10 mM MES, pH 5, and eluting the DNA in 10 mM Tris/50 mM KCl, pH 9. Fig. 6D shows the extraction profile that resulted, demonstrating that all DNA passed through the device, unbound, during the load. This profile is like that of the underivatized device (Fig. 6B) and shows the absence of DNA binding. As further confirmation that little to no DNA bound during the load, little DNA is recovered during the elution step. These results demonstrate that 1 M NaOH is an effective method for removal of chitosan from the PMMA surface, returning the device to its original state where no DNA binding is seen.
While the ultimate goal of this work is to produce disposable SPE microdevices, only a limited number of devices were available during development, requiring devices to be used more than once. Therefore, experiments were performed to determine whether a device that had been stripped of the chitosan coating could then be recoated with chitosan and successfully purify DNA. Extractions of hgDNA were performed on devices that had undergone chitosan stripping and then recoated. This resulted in a reproducible elution profile with an extraction efficiency of 47.8 (±9.3)% (Fig. 6C), demonstrating that the chitosan stripping and coating procedure provides a reproducible solid phase generation method that results in the reproducible extraction of DNA. It also shows that the number of times a device is recoated has little effect on the performance as the extraction profile shown in Fig. 6B represents extractions on a device which had undergone multiple cycles of stripping and recoating.
Extraction of hgDNA from whole blood
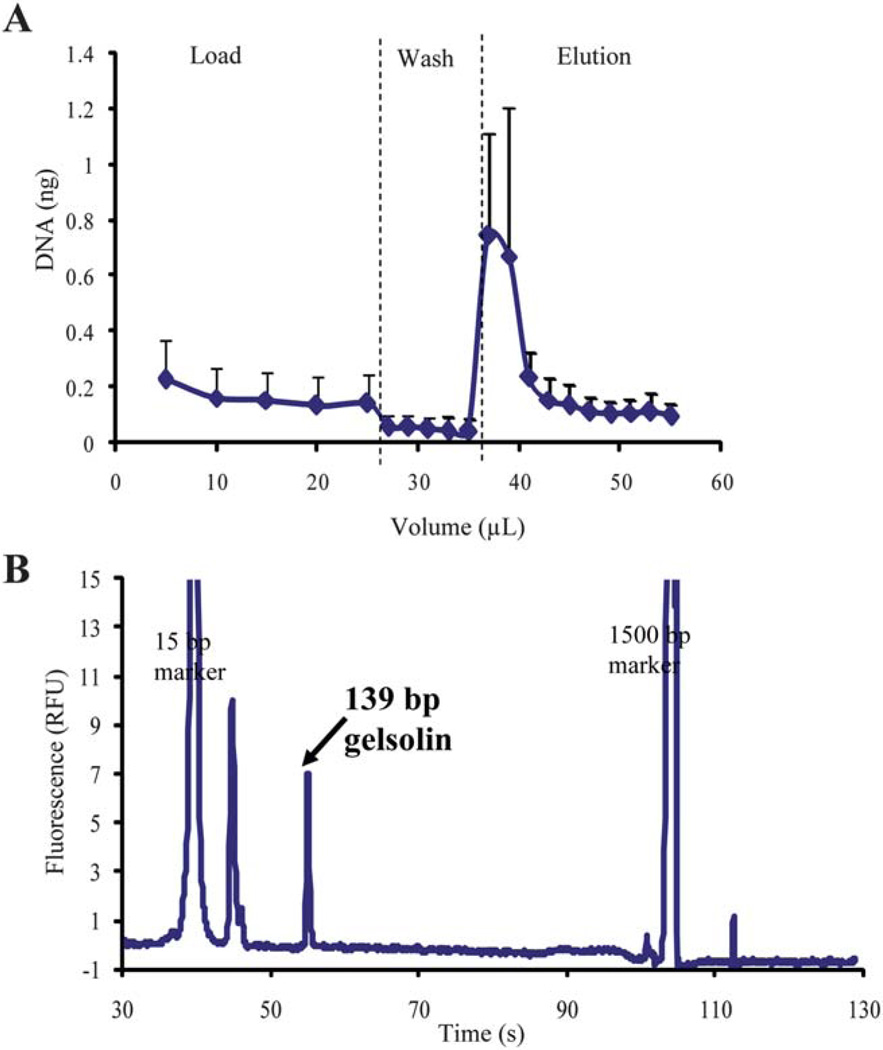
Although successful and reproducible extraction of prepurified hgDNA on the PMMA devices has been shown, the application of the device to commonly encountered clinical samples, such as whole blood, provides a more stringent measure of the method. A lysed whole blood sample was loaded (25 µL of a 1 ng µL solution; ~4000 WBCs) onto a chitosan-coated device. The device was washed with 10 mM MES, pH 5, and DNA eluted in 10 mM Tris/50 mM KCl, pH 9. Fractions collected were analyzed using a fluorescence assay and an extraction profile was generated (Fig. 7A, n = 5 with EE of 9.9 (±5.0)%). The extraction profile shows reproducibility from sample-to-sample and chip-to-chip despite the fact that multiple devices and blood donors were used. Although a decreased EE is seen, mainly due to overloading the DNA capacity (data not shown) of the phase which can later be increased with further coating optimization, sufficient quantities of DNA were recovered in a concentrated fraction during the elution for subsequent PCR analysis. The concentration of DNA recovered makes this DNA extraction method amenable to integration with on-chip downstream PCR. An additional advantage of the chitosan-coated PMMA device is the low protein binding nature of chitosan,17 which allows for the method to be applicable to complex biological samples, such as the whole blood demonstrated here, that contain variable and often large quantities of protein.
Fig. 7.
(A) The extraction profile from the purification of hgDNA from whole blood using a chitosan-coated PMMA device (n = 5). (B) Electropherogram from PCR amplification of a representative elution fraction of DNA purified from whole blood using a chitosan-coated PMMA device (n = 3). Successful purification is indicative by 139 bp amplicon for a portion of the gelsolin gene.
Amplification of extracted hgDNA from whole blood
Although sufficient quantities of hgDNA were eluted from the chitosan-coated PMMA device for PCR analysis, the suitability of this DNA for PCR amplification was unknown. To ensure that the extracted DNA was indeed PCR-amplifiable, an extraction of hgDNA was performed by loading 25 µL of lysed whole blood (sample prepared as described in Sample preparation in Materials and methods) onto the device. The phase was washed with 10 mM MES, pH 5, and DNA eluted with 10 mM Tris/50 mM KCl, pH 9. The elution fractions collected were then amplified for a portion of the gelsolin gene, whose gene product is known to play an important role in regulating the length of filaments involved in cell structure, apoptosis, and cancer,28,29 as described in PCR analysis in Materials and methods. PCR products were separated and analyzed on an Agilent 2100 Bioanalyzer. An electropherogram of a representative elution fraction is shown in Fig. 7B, where a 139 bp amplicon, indicative of the gelsolin gene fragment, is present. This confirms that not only can sufficient quantities of hgDNA for subsequent analysis be extracted using the device, but also the extracted DNA is of high quality allowing for successful PCR analysis to be completed. This method provides a clear advantage as the extraction is completely aqueous, eliminating the use PCR-inhibitory reagents (guanidine and isopropanol) which are required in silica-based extraction methods. Additionally, the chitosan phase used has been shown to be low protein binding which essentially increases the number of binding sites available for DNA on the phase and increases the quantity of DNA recovered. This advantage is not seen with silica-based purification where both the DNA and protein present in the sample bind the phase during the load, inherently decreasing the sites available for DNA.25
Conclusions and future work
The development of a plastic microdevice capable of extraction of hgDNA from whole blood was demonstrated, making the device viable for numerous clinical applications, as whole blood is a common clinical sample. By utilizing chitosan-based purification technology for reversible DNA binding, PCR inhibiting reagents, including guanidine hydrochloride and isopropyl alcohol, were eliminated. The DNA was also eluted in PCR buffer, making the extracted DNA PCR-ready and amendable to integration with PCR in efforts to develop a micro total analysis system (µTAS).30 The results shown in this work provide the next step towards a point-of-care, single-use device for use in clinical settings, such as physician offices. The desirable advantages of reduced analysis time and reagent consumption are achieved with this microdevice and technique. Decreased wait time for analysis could be vital for patients for whom delayed treatment can be disastrous. The results in this work also demonstrated sample-to-sample and chip-to-chip reproducibility owing to the fabrication process and functionalization method. This is a significant advantage over solid phase filled microdevices where variable packing can affect results.
Future work will include optimization of the device coating with chitosan to determine whether greater quantities of DNA can be extracted from biological samples. Further optimization of the fabrication process will also be completed to allow for construction of a mold, for inexpensive fabrication of the delicate microstrucutres. Additionally, developments towards a µTAS30 will be investigated by integration with PCR on the plastic microfluidic device.
References
- 1.Price CW, Leslie DC, Landers JP. Lab Chip. 2009;9:2484–2494. doi: 10.1039/b907652m. [DOI] [PubMed] [Google Scholar]
- 2.Bienvenue JM, Duncalf N, Marchiarullo D, Ferrance JP, Landers JP. J. Forensic Sci. 2006;51:266–273. doi: 10.1111/j.1556-4029.2006.00054.x. [DOI] [PubMed] [Google Scholar]
- 3.Bienvenue JM, Legendre LA, Landers JP. Forensic Sci. Int.: Genet. 2010;4:178–186. doi: 10.1016/j.fsigen.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Boom R, Sol CJA, Saliman MMM, Jansen CL, Wertheim-Van Dillen PME, Van Der Noordaa J. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breadmore MC, Wolfe KA, Arcibal IG, Leung WK, Dickson D, Giordano BC, Power ME, Ferrance JP, Feldman SH, Norris PM, Landers JP. Anal. Chem. 2003;75:1880–1886. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- 6.Cady NC, Stelick S, Batt CA. Biosens. Bioelectron. 2003;19:59–66. doi: 10.1016/s0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 7.Christel LA, Petersen K, McMillan W, Northrup MA. J. Biomech. Eng. 1999;121:22–27. doi: 10.1115/1.2798037. [DOI] [PubMed] [Google Scholar]
- 8.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legendre LA, Bienvenue JM, Roper MG, Ferrance JP, Landers JP. Anal. Chem. 2006;78:1444–1451. doi: 10.1021/ac0516988. [DOI] [PubMed] [Google Scholar]
- 10.Reedy CR, Bienvenue JM, Coletta L, Strachan BC, Bhatri N, Greenspoon S, Landers JP. Forensic Sci. Int.: Genet. 2010;4:206–212. doi: 10.1016/j.fsigen.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Reedy CR, Hagan KA, Strachan BC, Higginson JJ, Bienvenue JM, Greenspoon SA, Ferrance JP, Landers JP. Anal. Chem. 2010;82:5669–5678. doi: 10.1021/ac100649b. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya A, Klapperich CM. Anal. Chem. 2006;78:788–792. doi: 10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Quijada GA, Peytavi R, Nantel A, Roy E, Bergeron MG, Dumoulin MM, Veres T. Lab Chip. 2007;7:856–862. doi: 10.1039/b700322f. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Cady NC, Batt CA. Biomed. Microdevices. 2007;9:769–776. doi: 10.1007/s10544-007-9088-9. [DOI] [PubMed] [Google Scholar]
- 15.Witek MA, Wei S, Vaidya B, Adams AA, Zhu L, Stryjewski W, McCarley RL, Soper SA. Lab Chip. 2004;4:464–472. doi: 10.1039/b317093d. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Vaidya B, Patel AB, Ford SM, McCarley RL, Soper SA. Anal. Chem. 2003;75:2975–2984. doi: 10.1021/ac030031n. [DOI] [PubMed] [Google Scholar]
- 17.Cao W, Easley CJ, Ferrance JP, Landers JP. Anal. Chem. 2006;78:7222–7228. doi: 10.1021/ac060391l. [DOI] [PubMed] [Google Scholar]
- 18.Hagan KA, Meier W, Ferrance JP, Landers JP. Anal. Chem. 2009;81:5249–5256. doi: 10.1021/ac900820z. [DOI] [PubMed] [Google Scholar]
- 19.Ford SM. Fabricating microfluidic devices in polymers for bioanalytical applications, Dissertation. Louisiana State University; 2002. [Google Scholar]
- 20.Lin C, Chao C, Lan C. Sens. Actuators, B. 2007;121:698–705. [Google Scholar]
- 21.Butler JM. Forensic DNA Typing: Biology, Technology, and Genetics of STR Markers. San Deigo: Elsevier Academic Press; 2005. [Google Scholar]
- 22.Ahn SJ, Costa J, Emanuel JR. Nucleic Acids Res. 1996;24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly RT, Pan T, Woolley AT. Anal. Chem. 2005;77:3536–3541. doi: 10.1021/ac0501083. [DOI] [PubMed] [Google Scholar]
- 24.Mair DA, Rolandi M, Snauko M, Noroski R, Svec F, Frechet JMJ. Anal. Chem. 2007;79:5097–5102. doi: 10.1021/ac070220w. [DOI] [PubMed] [Google Scholar]
- 25.Tian H, Huhmer AFR, Landers JP. Anal. Biochem. 2000;283:175–191. doi: 10.1006/abio.2000.4577. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe KA, Breadmore MC, Ferrance JP, Power ME, Conroy JF, Norris PM, Landers JP. Electrophoresis. 2002;23:727–733. doi: 10.1002/1522-2683(200203)23:5<727::AID-ELPS727>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Bakowies D, Kollman PA. J. Am. Chem. Soc. 1999;121:5712–5726. [Google Scholar]
- 28.Mielnicki LM, Ying AM, Head KL, Asch HL, Asch BB. Exp. Cell Res. 1999;249:161–176. doi: 10.1006/excr.1999.4461. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsu M, Sakai N, Fujita H, Kashiwagi M, Gasa S, Shimizu S, eguchi Y, Tsujimoto Y, Sakiyama Y, Kobayashi K, Kuzumaki N. EMBO J. 1997;16:4650–4656. doi: 10.1093/emboj/16.15.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz A, Harrison DJ, Verpoorte EMJ, Fettinger JC, Paulus A, Ldi H, Widmer HM. J. Chromatogr. 1992;593:253–258. [Google Scholar]