Abstract
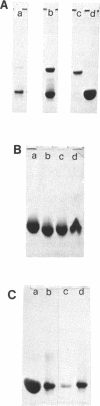
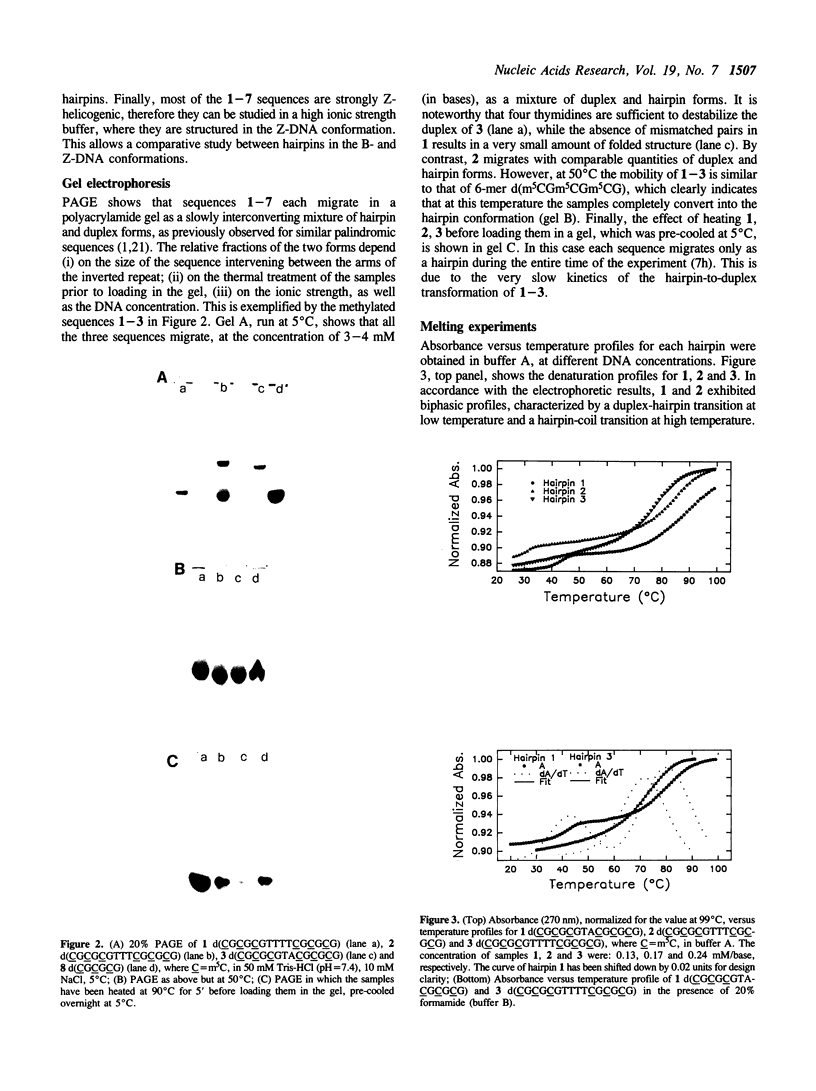
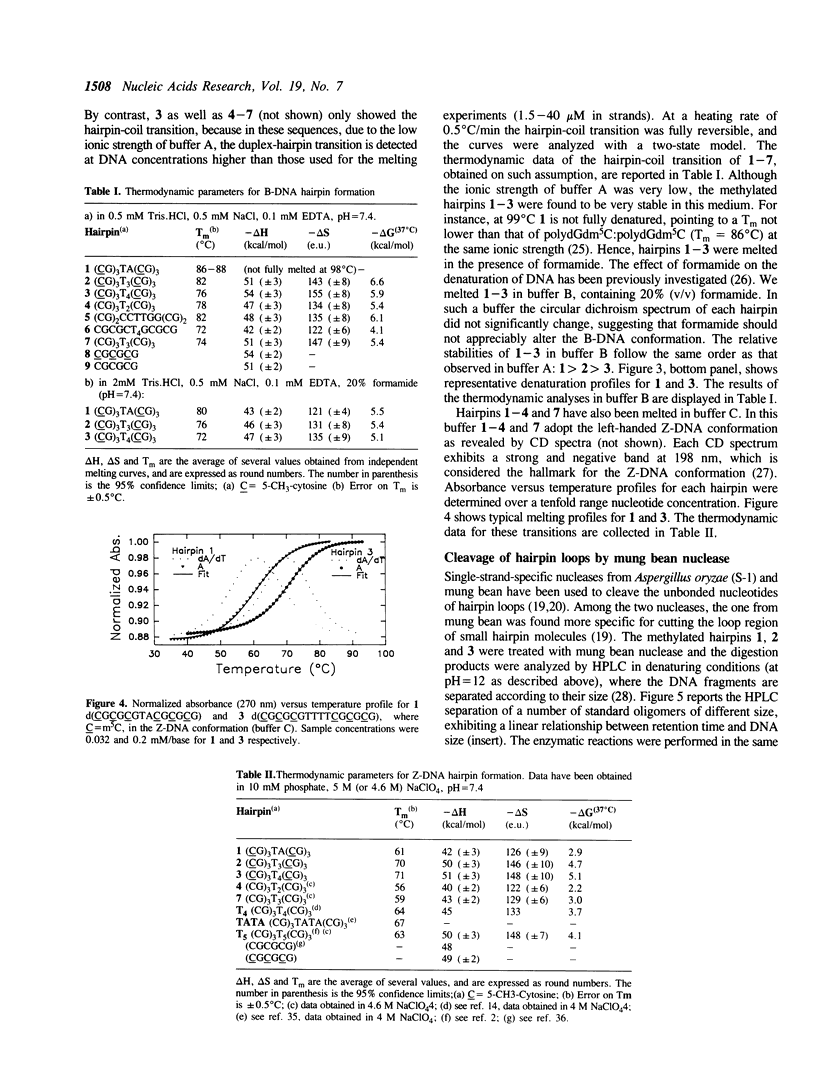
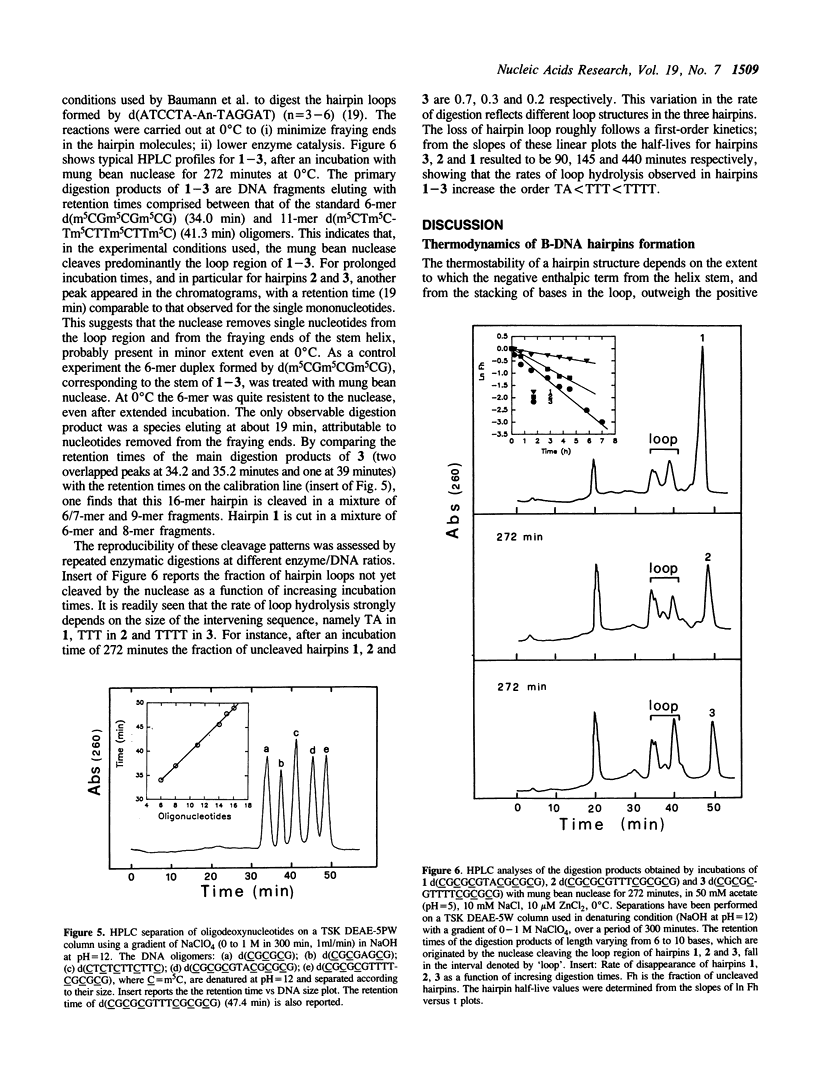
Hairpin structures formed by seven DNA inverted repeats have been studied by PAGE, UV(CD)-spectroscopy and nuclease cleavage. The hairpins consisted of (CG)3 stems and loops of 2, 3 and 4 residues. Thermal stabilities (Tm) have been determined in low and high ionic strength buffers, where the hairpins were structured in the B- and Z-DNA form respectively. The thermodynamic parameters of hairpin formation have been obtained by a two-state analysis of the hairpin-coil transitions. It is found that, on increasing the number of bases in the loop from 2 to 3 and 4, the Tms of the B-hairpins decrease, whereas the Tms of the same hairpins in the Z-form increase. This confirms previous evidence (1,2) that in a hairpin molecule the size and structure of the loop are modulated by the conformation of the helical stem. Moreover, B-hairpins with loops comprising 2, 3 and 4 bases have been digested with the single-strand-specific nuclease from mung bean. In our experimental conditions (0 degrees C) the nuclease preferentially cleaves the unbonded nucleotides of the loops. However, the rates of loop hydrolysis, which roughly follow a first-order kinetics, markedly depend on the size of the loop. At a ratio of 3 enzyme units/micrograms DNA, the half-lives of hairpins which are expected to form loops of 4, 3 and 2 residues are 90, 145 and 440 minutes respectively. Thermostability and enzymatic digestion data suggest that two-membered loops can be formed in B-hairpins but not in Z-hairpins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amaratunga M., Pancoska P., Paner T. M., Benight A. S. B to Z transitions of the short DNA hairpins formed from the oligomer sequences: d[(CG)3X4(CG)3] (X = A, T, G, C). Nucleic Acids Res. 1990 Feb 11;18(3):577–582. doi: 10.1093/nar/18.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann U., Frank R., Blöcker H. Conformational analysis of hairpin oligodeoxyribonucleotides by a single-strand-specific nuclease. Eur J Biochem. 1986 Dec 1;161(2):409–413. doi: 10.1111/j.1432-1033.1986.tb10460.x. [DOI] [PubMed] [Google Scholar]
- Benight A. S., Wang Y. S., Amaratunga M., Chattopadhyaya R., Henderson J., Hanlon S., Ikuta S. Conformation and dynamics of a left-handed Z-DNA hairpin: studies of d(CGCGCGTTTTCGCGCG) in solution. Biochemistry. 1989 Apr 18;28(8):3323–3332. doi: 10.1021/bi00434a030. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R., Ikuta S., Grzeskowiak K., Dickerson R. E. X-ray structure of a DNA hairpin molecule. Nature. 1988 Jul 14;334(6178):175–179. doi: 10.1038/334175a0. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988 Dec 23;16(24):11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Thermal stability of RNA hairpins containing a four-membered loop and a bulge nucleotide. Biochemistry. 1989 Jan 24;28(2):742–747. doi: 10.1021/bi00428a049. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Van Kimmenade J. M., van Knippenberg P. H., Hinz H. J. Calorimetric measurements of the destabilisation of a ribosomal RNA hairpin by dimethylation of two adjacent adenosines. Nucleic Acids Res. 1983 Jan 11;11(1):203–210. doi: 10.1093/nar/11.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta S., Chattopadhyaya R., Ito H., Dickerson R. E., Kearns D. R. NMR study of a synthetic DNA hairpin. Biochemistry. 1986 Aug 26;25(17):4840–4849. doi: 10.1021/bi00365a018. [DOI] [PubMed] [Google Scholar]
- Ikuta S., Wang Y. S. Conformation and dynamics of Z-DNA oligomer duplex of d[(CG)3TATA(CG)3] in solution. Nucleic Acids Res. 1989 Jun 12;17(11):4131–4144. doi: 10.1093/nar/17.11.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump H. H., Löffler R. Reversible helix/coil transitions of left-handed Z-DNA structures. Comparison of the thermodynamic properties of poly(dG).poly(dC), poly[d(G-C)].poly[d(G-C)], and poly(dG-m5dC).poly(dG-m5dC). Biol Chem Hoppe Seyler. 1985 Apr;366(4):345–353. doi: 10.1515/bchm3.1985.366.1.345. [DOI] [PubMed] [Google Scholar]
- Manzini G., Xodo L. E., Quadrifoglio F., van Boom J. H., van der Marel G. A. dC-dG alternating oligonucleotides: thermodynamic and kinetic aspects of the B-Z transformation. J Biomol Struct Dyn. 1987 Feb;4(4):651–662. doi: 10.1080/07391102.1987.10507666. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res. 1986 May 27;14(10):4187–4196. doi: 10.1093/nar/14.10.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paner T. M., Amaratunga M., Doktycz M. J., Benight A. S. Analysis of melting transitions of the DNA hairpins formed from the oligomer sequences d[GGATAC(X)4GTATCC] (X = A, T, G, C). Biopolymers. 1990 Dec;29(14):1715–1734. doi: 10.1002/bip.360291405. [DOI] [PubMed] [Google Scholar]
- Riazance J. H., Baase W. A., Johnson W. C., Jr, Hall K., Cruz P., Tinoco I., Jr Evidence for Z-form RNA by vacuum UV circular dichroism. Nucleic Acids Res. 1985 Jul 11;13(13):4983–4989. doi: 10.1093/nar/13.13.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokita S. E., Romero-Fredes L. Facile interconversion of duplex structures formed by copolymers of d(CG). Biochemistry. 1989 Dec 12;28(25):9674–9679. doi: 10.1021/bi00451a020. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by dAT oligomers. I. Hairpin and straight-chain helices. J Mol Biol. 1968 Sep 28;36(3):291–304. doi: 10.1016/0022-2836(68)90156-3. [DOI] [PubMed] [Google Scholar]
- Senior M. M., Jones R. A., Breslauer K. J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Cruciform transitions in DNA. J Biol Chem. 1984 May 25;259(10):6593–6600. [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer D. E., Chou S. H., Hare D. R., Reid B. R. Duplex-hairpin transitions in DNA: NMR studies on CGCGTATACGCG. Nucleic Acids Res. 1985 May 24;13(10):3755–3772. doi: 10.1093/nar/13.10.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk S. K., Hardin C. C., Germann M. W., van de Sande J. H., Tinoco I., Jr Comparison of the B- and Z-form hairpin loop structures formed by d(CG)5T4(CG)5. Biochemistry. 1988 Sep 6;27(18):6960–6967. doi: 10.1021/bi00418a043. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. Oligodeoxynucleotide folding in solution: loop size and stability of B-hairpins. Biochemistry. 1988 Aug 23;27(17):6321–6326. doi: 10.1021/bi00417a018. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. The B-Z conformational transition in folded oligodeoxynucleotides: loop size and stability of Z-hairpins. Biochemistry. 1988 Aug 23;27(17):6327–6331. doi: 10.1021/bi00417a019. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G., van Boom J. H. Hairpin structures in synthetic oligodeoxynucleotides: sequence effects on the duplex-to-hairpin transition. Biochimie. 1989 Jul;71(7):793–803. doi: 10.1016/0300-9084(89)90042-4. [DOI] [PubMed] [Google Scholar]