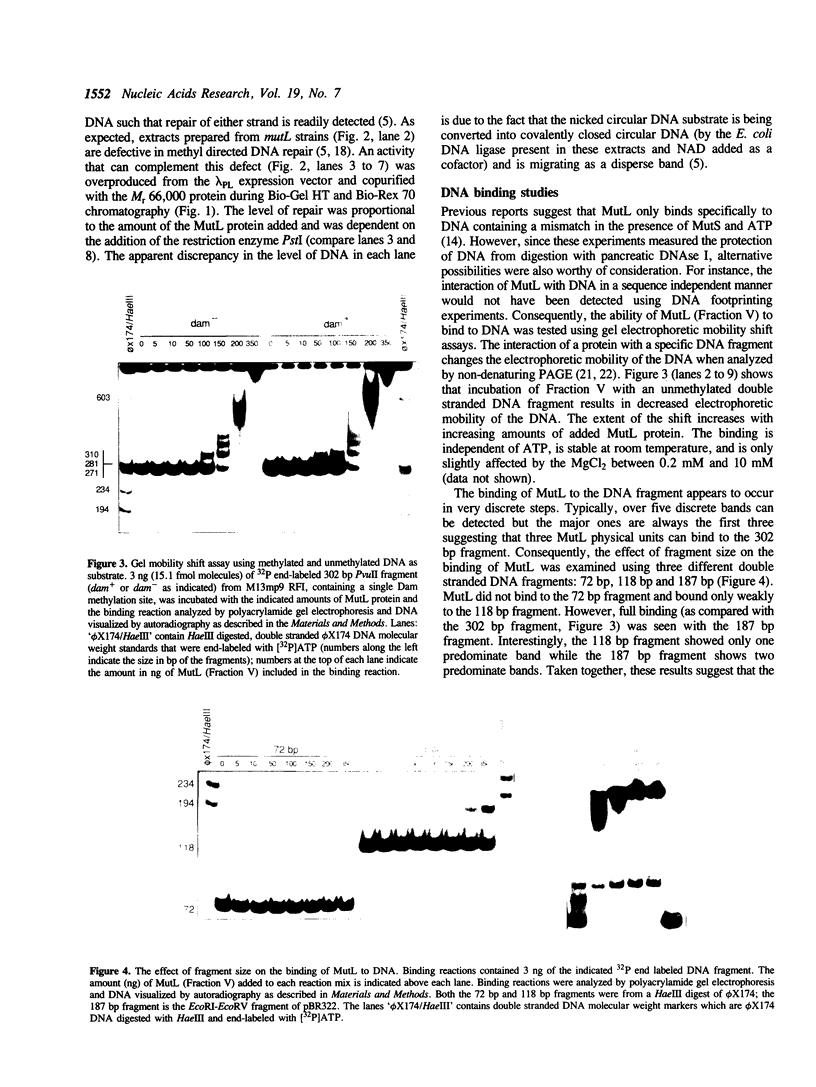
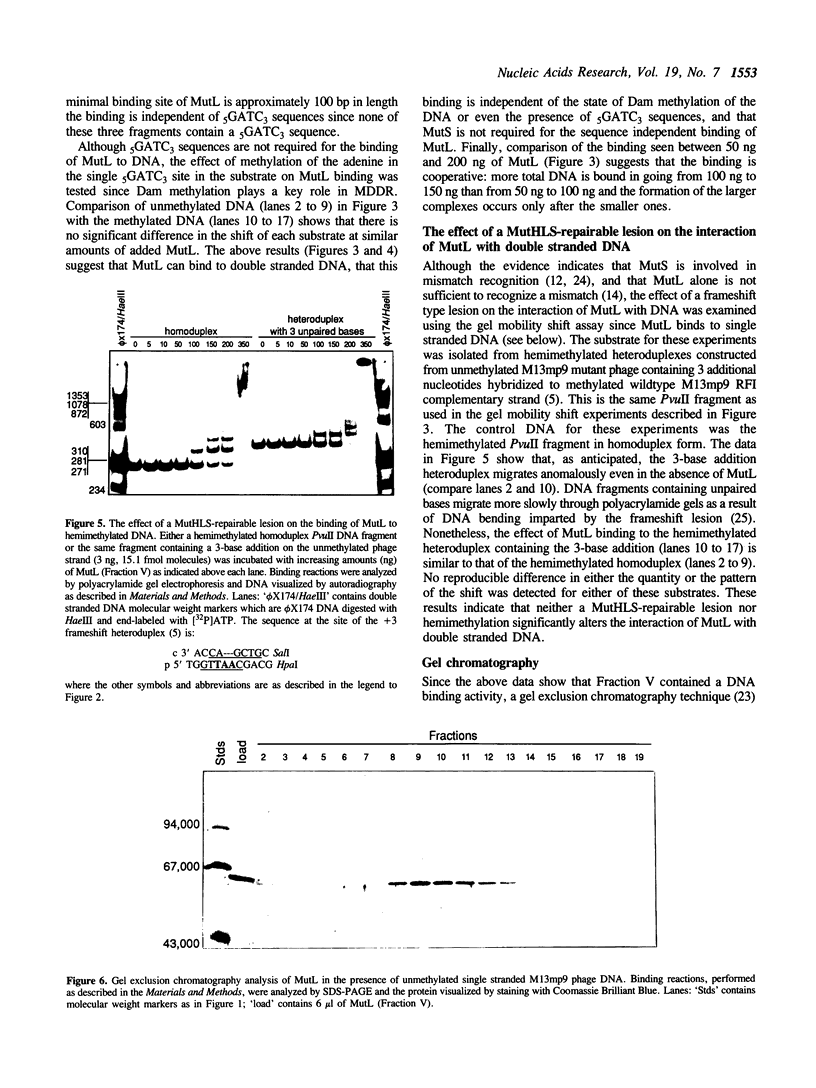
Abstract
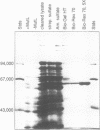
The mutL gene of Escherichia coli, which is involved in the repair of mispaired and unpaired nucleotides in DNA, has been independently cloned and the gene product purified. In addition to restoring methyl-directed DNA repair in extracts prepared from mutL strains, the purified MutL protein binds to both double and single stranded DNA. The affinity constant of MutL for unmethylated single stranded DNA was twice that of its affinity constant for methylated single stranded DNA and methylated or unmethylated double stranded DNA. The binding of MutL to double stranded DNA was not affected by the pattern of DNA methylation or the presence of a MutHLS-repairable lesion.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Ackers G. K., Smith H. O. Measurement of DNA-protein equilibria using gel chromatography: application to the HinfI restriction endonuclease. Biochemistry. 1985 Jun 4;24(12):3049–3054. doi: 10.1021/bi00333a037. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Hoess R. H. Cloning of mutH and identification of the gene product. Gene. 1983 May-Jun;22(2-3):245–253. doi: 10.1016/0378-1119(83)90109-9. [DOI] [PubMed] [Google Scholar]
- Grilley M., Holmes J., Yashar B., Modrich P. Mechanisms of DNA-mismatch correction. Mutat Res. 1990 Sep-Nov;236(2-3):253–267. doi: 10.1016/0921-8777(90)90009-t. [DOI] [PubMed] [Google Scholar]
- Grilley M., Welsh K. M., Su S. S., Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989 Jan 15;264(2):1000–1004. [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J., Su S. S., Wood S. G., Modrich P. Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 1988 Aug 25;16(16):7843–7853. doi: 10.1093/nar/16.16.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kumura K., Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984 Feb 10;259(3):1560–1565. [PubMed] [Google Scholar]
- Learn B. A., Grafstrom R. H. Methyl-directed repair of frameshift heteroduplexes in cell extracts from Escherichia coli. J Bacteriol. 1989 Dec;171(12):6473–6481. doi: 10.1128/jb.171.12.6473-6481.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Zimm B. H. Separations of open-circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987 Apr;6(4):1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Pang P. P., Lundberg A. S., Walker G. C. Identification and characterization of the mutL and mutS gene products of Salmonella typhimurium LT2. J Bacteriol. 1985 Sep;163(3):1007–1015. doi: 10.1128/jb.163.3.1007-1015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simatake H., Rosenberg M. Purified lambda regulatory protein cII positively activates promoters for lysogenic development. Nature. 1981 Jul 9;292(5819):128–132. doi: 10.1038/292128a0. [DOI] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]