Summary
Crossovers (COs) between homologous chromosomes ensure their faithful segregation during meiosis. We identify C. elegans COSA-1 as a key component required to convert double-strand breaks (DSBs) into COs. COSA-1 localizes to foci during late meiotic prophase that correspond to the single CO site on each homolog pair. These foci represent sites of eventual concentration of other conserved CO proteins that initially exhibit broader distribution. Chromosomes gain and lose competence to load CO proteins at DSBs during meiotic progression, with competence to load COSA-1 requiring a prior licensing event. Our data further suggest a self-reinforcing mechanism maintaining CO designation. Modeling of a non-linear dose-response relationship between IR-induced DSBs and COSA-1 foci reveals efficient conversion of DSBs into COs when DSBs are limiting, and a robust capacity to limit the number of cytologically-differentiated CO sites when DSBs are in excess. COSA-1 foci serve as a biodosimeter for DSB levels and a unique live-cell readout of CO interference.
Introduction
Genetic recombination is an integral part of the sexual reproduction program by which most diploid organisms generate haploid gametes. In addition to promoting reassortment of genetic traits, crossover (CO) recombination between homologous chromosomes plays a key mechanical role in directing their segregation at the meiosis I division (Martinez-Perez and Colaiacovo, 2009). Crossing over is initiated by double-strand DNA breaks (DSBs), a subset of which is converted into COs by a specialized meiotic repair pathway that uses the homolog as a recombination partner. COs mature during the pachytene stage of meiotic prophase in the context of the synaptonemal complex (SC), a meiosis-specific structure that assembles at the interface between lengthwise-aligned homologs. During the subsequent diplotene and diakinesis stages, the SC disassembles and chromosomes condense and reorganize around each CO site to reveal a chiasma, a structure resulting from the CO in conjunction with sister chromatid cohesion flanking the CO site. Chiasmata maintain connections between homologs through metaphase of meiosis I, when they enable reliable biorientation of homologs toward opposite spindle poles.
Despite reliance on COs to ensure homolog segregation, most organisms make very few COs per homolog pair (on the order of 1-3 per chromosome arm), even though DSBs occur in substantial excess (Martinez-Perez and Colaiacovo, 2009). Moreover, the distribution of COs along chromosomes reflects a propensity of (nascent) COs to inhibit formation of other COs nearby on the same chromosome pair, a phenomenon known as CO interference (Muller, 1916). These properties imply that meiotic recombination must be governed by a robust CO control system that can guarantee the formation of sufficient COs while simultaneously limiting their numbers.
Although CO control remains poorly understood, many components of the machinery that promotes the CO outcome of meiotic DSB repair have been identified. One key player is a heterodimer of conserved meiosis-specific MutS family members MSH4 and MSH5. MSH4-MSH5 is implicated in formation and/or stabilization of CO intermediates in vivo (Baudat and de Massy, 2007; Lynn et al., 2007) and can load onto Holliday junction substrates in vitro (Pochart et al., 1997; Snowden et al., 2004). Further, localization of MSH4-MSH5 to DSB-dependent foci implies function at the sites of nascent recombination events. Notably, while the number of Msh4 foci in S. cerevisiae corresponds well with the number of COs generated by this pathway (Hollingsworth et al., 1995; Ross-Macdonald and Roeder, 1994), MSH4/MSH5 foci in several other species are initially detected in significant excess of COs (Higgins et al., 2004; Higgins et al., 2008; Kneitz et al., 2000; Lenzi et al., 2005; Santucci-Darmanin et al., 2000). Another key player, Zip3/ZHP-3, is a predicted SUMO or ubiquitin ligase (Lynn et al., 2007). S. cerevisiae Zip3 functions in regulating SC assembly, and localizes in foci that substantially overlap with Msh4, consistent with its role in CO formation (Agarwal and Roeder, 2000; Macqueen and Roeder, 2009). C. elegans ZHP-3 also functions to promote COs, but in contrast to its S. cerevisiae ortholog, it is dispensable for SC assembly and instead participates in organizing SC disassembly (Bhalla et al., 2008; Jantsch et al., 2004). Moreover, C. elegans ZHP-3 initially localizes along the lengths of the SCs, and gradually shrinks down to become localized at presumptive CO sites only very late in prophase. The fact that both MSH4/MSH5 and ZHP-3 show initial localization that differs substantially from the eventual distribution of COs raises a key question: how does CO-promoting activity become concentrated at and restricted to bona fide CO sites?
Here, we investigate the execution and regulation of CO formation during C. elegans meiosis, exploiting several advantageous attributes of this system. CO control is highly robust in C. elegans, with most chromosome pairs undergoing only a single CO (Hammarlund et al., 2005; Hillers and Villeneuve, 2003; Nabeshima et al., 2004). Further, while some organisms generate a significant fraction of their COs using alternative pathways, essentially all COs in C. elegans are formed using the canonical MSH-4-MSH-5-dependent, interference-sensitive pathway (Zalevsky et al., 1999). Moreover, the “production-line” organization of the germ line enables visualization of cytological correlates of CO progression simultaneously at all stages of meiotic prophase, both under normal conditions and after acute induction of DSBs.
This study is driven by our discovery of COSA-1, a CO promoting factor widely conserved in metazoa that functions in conjunction with MSH-4-MSH-5 and ZHP-3. GFP::COSA-1 localizes in foci specifically at presumptive CO sites, illuminating a key transition during CO maturation and providing evidence for a self-reinforcing mechanism that sequesters CO factors at designated CO sites. Further, combined experimental analysis and modeling of the response of COSA-1 foci to varying DSB levels reveals highly efficient conversion of DSBs into COs when DSBs are limiting, as well as a robust capacity to limit the number of cytologically-differentiated CO sites to one per homolog pair when DSBs are in excess. These and other findings indicate that COSA-1 foci represent a reliable surrogate for the events that are distributed by the CO control system. Thus our ability to visualize GFP::COSA-1 in live worms creates an unprecedented opportunity, making it possible to apply genetic screening strategies to investigate the elusive basis of CO interference.
Results
COSA-1 is required to convert meiotic DSBs into interhomolog COs
The cosa-1(me13) mutation was isolated based on frequent mis-segregation of sex chromosomes: cosa-1 hermaphrodites (XX) produce 38% XO male self progeny (compared to 0.2% for wild type (Hodgkin et al., 1979) ). cosa-1 hermaphrodites also produce a high frequency of inviable embryos (97%, n=2737), indicative of autosomal missegregation. These segregation defects reflect a lack of chiasmata connecting homologous chromosomes (Figure 1A). Whereas wild-type diakinesis oocytes contain 6 DAPI stained bodies (n=172), corresponding to 6 pairs of homologs held together by chiasmata, cosa-1 oocytes contain an average of 11 resolvable DAPI stained bodies (n=177), indicating absence of chiasmata.
Figure 1. COSA-1 is required to convert meiotic DSBs into interhomolog COs.
(A) Full karyotypes of individual diakinesis-stage oocytes. 6 DAPI-stained bodies in the wild-type nucleus correspond to 6 pairs of homologs connected by chiasmata; 12 individual chromosomes (univalents) in the cosa-1 mutant nucleus reflect a lack of chiasmata. (B) cosa-1mutant pachytene nuclei in which pairing was assessed either by FISH at the 5S rDNA locus (chromosome V) or by immunostaining for X-chromosome pairing center (X-PC) binding protein HIM-8. A single FISH or HIM-8 signal in each nucleus indicates successful pairing. (C) cosa-1 mutant pachytene nuclei, showing colocalization of SC lateral element protein HIM-3 and SC central region protein SYP-1 between parallel tracks of DAPI-stained chromatin. (D) Immunolocalization of RAD-51 in mid to late pachytene nuclei in the cosa-1 mutant. RAD-51 foci indicative of DSB formation are abundant in mid-pachytene, and are greatly reduced or absent in most late pachytene nuclei (asterisk indicates an apoptotic nucleus). (E) Early diplotene nuclei. In WT, SYP-1 and HTP-1/2 are localized to reciprocal domains on each chromosome pair; in cosa-, this indicator of CO formation is not observed, as SYP-1 and HTP-1/2 remain extensively colocalized. Scale bar = 5 μm.
Several lines of evidence together indicate that lack of chiasmata in cosa-1 mutants reflects a defect in the process of meiotic recombination, specifically in the conversion of initiated recombination events into COs. First, cosa-1 mutants are proficient for homolog pairing, as assessed by fluorescence in situ hybridization (FISH) at the 5S rDNA locus on chromosome V and by HIM-8 immunofluorescence (IF) to evaluate pairing at a specific region of the X chromosome known as the pairing center (MacQueen et al., 2005; Phillips et al., 2005) (Figure 1B). Further, cosa-1 mutants are proficient for assembly of the SC between paired homologs, as immunostaining revealed colocalization of HIM-3 (a component of the lateral elements of the SC) and SYP-1 (a component of the SC central region) (MacQueen et al., 2002; Zetka et al., 1999) between parallel tracks of DAPI-stained DNA (Figure 1C).
Moreover, a combination of cytological and genetic data indicate that cosa-1 mutants initiate recombination but fail to repair DSBs as COs. DNA strand exchange protein RAD-51 (Colaiacovo et al., 2003) is detected in abundant foci beginning in late zygotene/early pachytene, indicating that DSBs form in the cosa-1 mutant (Figure 1D). These foci eventually disappear at late pachytene, suggesting that DSBs are repaired but are not converted into interhomolog COs. Further, whereas COs trigger relocalization of SYP-1 and chromosome axis protein HTP-1/2 to reciprocal domains during late pachytene in wild-type meiosis (Martinez-Perez et al., 2008), cosa-1 mutants lack this prominent cytological indicator of COs (Figure 1E). Finally, measurement of genetic recombination frequency showed that absence of COSA-1 reduces the incidence of COs to less than 1% of control levels (Table S1).
This phenotypic analysis indicates a role for COSA-1 in the formation of meiotic COs. Moreover, these phenotypes closely parallel those of mutants lacking HIM-14 (MSH4), MSH-5 and ZHP-3 (Jantsch et al., 2004; Kelly et al., 2000; Zalevsky et al., 1999), suggesting that COSA-1 functions in conjunction with these conserved CO-promoting proteins.
cosa-1 encodes a cyclin-related protein conserved in metazoa
SNP mapping and sequencing identified a G-to-A transition in the me13 mutant resulting in a premature stop at codon 148 of predicted gene Y71H2AM.7 (360 codons total). An independently generated deletion allele (tm3298) fails to complement me13, confirming the identity of cosa-1 as Y71H2AM.7 (Figure 2A).
Figure 2. COSA-1 is a distant member of the cyclin superfamily with orthologs in metazoa.
(A) Top, predicted gene structure of C. elegans cosa-l with mutant alleles indicated. Gray, UTR; magenta, coding exons. Bottom, construct used to express GFP::COSA-1. Green, GFP coding sequence; blue, extra tags and linker sequences. (B) Phylogenetic tree depicting a sampling of metazoan species. Green indicates lineages where COSA-1, MSH-4 and MSH-5 orthologs are all present; red indicates the absence of all three from Drosophilid species. (C) Predicted structure of residues 56-360 of C. elegans COSA-1 (yellow and magenta) aligned with crystal structure of residues 167-426 of human cyclin B1 (cyan). N-terminal residues of COSA-1 and cyclin B1 were removed to aid visualization of the two core cyclin fold motifs. In canonical cyclins, cyclin-fold motifs consist of 5 α-helices, with well-conserved interhelical angles in the N-terminal cyclin box motif. In the predicted COSA-1 structure, the N-terminal cyclin box is interrupted by an insertion of 33 amino acids, modeled here as an extension of α-helix 2 and an additional helix (magenta, α-2.5). Predicted α-helices 3-5 of COSA-1 align well with the corresponding helices of cyclin B1 and cyclin A, which contribute to the cyclin/CDK interface in Cyclin A/CDK2 (Jeffrey et al 1995).
Three notable observations arose from our search for COSA-1 homologs (Supplemental Analysis). First, COSA-1 orthologs are found throughout the metazoan lineage (Figure 2B), but are not detected in plants and fungi, suggesting either that COSA-1 arose in metazoa or that divergence in plants and fungi has rendered it unrecognizable. Second, while COSA-1 orthologs are present in other diptera, they are absent in the Drosophila genus (Figure 2B), which also lacks both MSH4 and MSH5 (Schurko et al., 2010). Together with our phenotypic data, this phylogenetic distribution suggests that COSA-1 and MSH-4-MSH-5 may act as a functional module. Third, expression data for the mouse and human orthologs, provisionally named Cntd1 (Cyclin n-terminal domain containing 1), are consistent with conservation of function in meiosis (Supplemental Analysis).
COSA-1 is predicted to have a cyclin-like structure based on analyses using PSI-BLAST and the PHYRE and I-TASSER structure prediction servers (see Supplemental Analysis). Alignment of the predicted COSA-1 structure with the crystal structure of human cyclin B1 shows a high degree of similarity (Figure 2C), including conservation of the region corresponding to the Cyclin/CDK interface. However, an insertion of 33 amino acids results in an extra predicted alpha helix in the highly conserved N-terminal cyclin-box domain (Petri et al., 2007). This feature is also present in human CNTD1 (Figure S1) and distinguishes the COSA-1/CNTD-1 family from conventional cyclins, indicating that it represents a distinct branch of the cyclin superfamily.
GFP::COSA-1 marks sites of presumptive COs
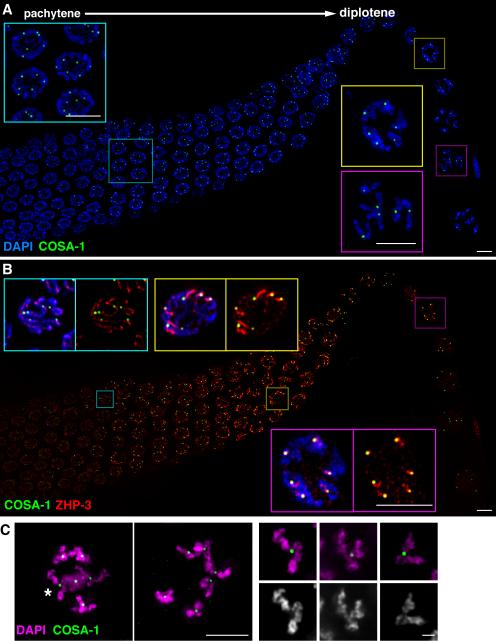
We generated a transgenic strain expressing a functional GFP::COSA-1 fusion to assess localization of COSA-1 during meiotic prophase progression (Figures 2A, 3A, S2). GFP::COSA-1 is detected as a diffuse nucleoplasmic signal beginning in early pachytene, then displays a striking localization pattern later in pachytene that is consistent with expectations for a protein that marks CO sites. Starting at the transition from mid to late pachytene and persisting through diplotene, GFP::COSA-1 localizes to 6.0 ± 0.2 bright foci per nucleus (n=76), corresponding in number to the 6 COs, 1 per chromosome pair, that form during wild-type meiosis (Figure 3A). At diplotene, GFP::COSA-1 localizes at the site of the single emerging chiasma on each homolog pair (Figure 3A and C). We conclude that COSA-1 localizes to CO sites, and have named the protein Crossover-site-associated-1.
Figure 3. GFP::COSA-1 localizes to foci corresponding to CO sites.
(A and B) IF images of a portion of a gonad extending from mid-pachytene through diplotene and early diakinesis. GFP::COSA-1 foci are detected from late pachytene through early diakinesis. (A) Left inset: late pachytene nuclei, each containing 6 bright foci. Right insets: diplotene nuclei, with one focus on each chromosome pair; bottom panel shows COSA-1 foci positioned at the site of the single emerging chiasma on each chromosome pair. (B) Relationship between localization of GFP::COSA-1 and ZHP-3. Top left inset: 6 COSA-1 foci in a mid-pachytene nucleus with ZHP-3 in long stretches along the chromosomes. Top right inset: COSA-1 localized at one end of each comet-like stretch of ZHP-3. Bottom inset: COSA-1 and ZHP-3 colocalization in a diplotene nucleus. (C) Representative images of GFP::COSA-1 localization in late diplotene/early diakinesis nuclei, highlighting the location of GFP::COSA-1 at the site of the single emerging chiasma on each chromosome pair. Large panels, full projections of entire nuclei showing all 6 bivalents; asterisk indicates a bivalent depicted in smaller panels, which shows partial projections of individual bivalents. Scale bars = 5 μm except in the single bivalent panels in (C), where scale bar = 1 μm.
COSA-1 foci and RAD-51 foci are detected in largely reciprocal domains within the pachytene region of the germ line: Prior to late pachytene, nuclei only have RAD-51 foci, while in late pachytene, most nuclei only have COSA-1 foci (Figure S3). Even in nuclei with both, RAD-51 and COSA-1 do not colocalize. This pattern is consistent with a major transition in recombination progression and indicates that COSA-1 is loaded after the majority of RAD-51 is removed.
GFP::COSA-1 foci are sites of eventual concentration of multiple CO-promoting factors
GFP::COSA-1 foci represent sites where two other conserved CO-promoting proteins, MSH-5 and ZHP-3, also eventually become concentrated. MSH-5 is first detected during mid-pachytene, appearing as faint foci that accumulate in excess of eventual COs (Figure 4A). Upon transition to late pachytene, MSH-5 foci decrease in number to 6 per nucleus, colocalizing with COSA-1 foci and exhibiting an increased intensity that presumably reflects increased local concentration of MSH-5 at these sites. ZHP-3 initially localizes in stretches along the full length of the SCs but begins to concentrate to one side of each presumptive CO site during late pachytene, initially forming comet-like structures stretching from the presumptive CO site to one end of the chromosome then progressively shrinking to foci by mid-diplotene (Bhalla et al., 2008; Jantsch et al., 2004). COSA-1 begins to appear as foci just as ZHP-3 begins to redistribute, then localizes at the head of each ZHP-3 comet during late pachytene progression and colocalizes with each ZHP-3 focus at diplotene (Figure 3B). Thus, while both MSH-5 and ZHP-3 initially exhibit broader localization patterns, both eventually become concentrated at COSA-1-marked sites.
Figure 4. MSH-5 colocalizes with and depends on COSA-1.
(A) IF images showing that MSH-5 foci are detected in mid-pachytene nuclei in excess of eventual COs (cyan inset), then decline by late pachytene, when they co-localize with GFP::COSA-1 foci (yellow inset). (B) Left, late pachytene nuclei from a wild-type germ line, showing comet-like localization of ZHP-3 with COSA- foci at the comet heads. Right, late pachytene nuclei from a cosa-1 mutant, showing persistence of ZHP-3 localization along the length of the chromosomes and a lack of MSH-5 foci. Scale bars = 5 μm; for insets, scale = 1 μm.
MSH-5, ZHP-3 and COSA-1 not only colocalize at presumptive CO sites, they also exhibit interdependence for this localization. First, COSA-1 foci were not observed in a msh-5 mutant (Figure S4A), and conversely, late pachytene MSH-5 foci were not detected in a cosa-1 mutant (Figure 4B). Mid-pachytene MSH-5 foci were also diminished or lost in the cosa-1 mutant, suggesting that COSA-1 may facilitate or stabilize the association of MSH-5 with nascent recombination events even before COSA-1 is detected on chromosomes. Second, ZHP-3 persisted along the lengths of SCs during late pachytene in the cosa-1 mutant (Figure 4B), and conversely, 6 COSA-1 foci were not detected in a zhp-3 mutant (Figure S4B).
Kinetics of and stage-dependence of competence for loading CO proteins at IR-induced recombination sites
We used GFP::COSA-1 to investigate changes that occur as recombination intermediates form and mature into COs, conducting a time course analysis to assess the temporal kinetics and developmental constraints governing recruitment of recombination proteins to DSBs induced by ionizing radiation (IR). For these experiments, spo-11 mutant worms (which lack endogenous DSBs) (Dernburg et al., 1998) expressing GFP::COSA-1 were exposed to 1000 rads of γ-irradiation, then assessed at various time points for localization of recombination proteins. These experiments exploited the fact that C. elegans chromosomes undergo homologous pairing and synapsis in the absence of DSBs, so that pachytene nuclei in the spo-11 mutant are already poised to engage in interhomolog recombination once DSBs are introduced.
In untreated control spo-11 germ lines, most nuclei lack MSH-5 and COSA-1 foci, and ZHP-3 remains localized along the length of the SC throughout pachytene and into diplotene, as expected given the lack of DSBs and COs (Figure 5A) (Bhalla et al., 2008). However, a subset of late pachytene nuclei have 1 or 2 atypical COSA-1 aggregates in which MSH-5 can also be detected; these tend to correspond to spots of higher ZHP-3 concentration, and may reflect an inherent tendency of these proteins to colocalize.
Figure 5. Time course of localization of CO proteins at IR-induced recombination sites.
Immunolocalization of CO proteins (GFP::COSA-1, MSH-5 and/or ZHP-3) in pachytene nuclei from gfp::cosa-1; spo-11 worms, either in the absence of IR (A, left) or at the indicated times following exposure to 1 kRad IR. Scale bars = 5 μm. (A) Localization of COSA-1 and ZHP-3 or MSH-5 in late pachytene nuclei in the absence of IR (pre-IR) and 8 hr post-IR. In the unirradiated spo-11 control, ZHP-3 persists along the lengths of the chromosomes and the majority of nuclei lack COSA-1 and MSH-5 foci; a subset of nuclei have one or two COSA-1/MSH-5 aggregates (indicated by asterisks). 8 hr post-IR: 6 bright COSA-1 foci localize at the heads of comet-like ZHP-3 signals. (B) Mid to late pachytene region of a 1 hr post-IR germ line. Abundant IR-induced MSH-5 foci are detected specifically in mid-pachytene nuclei (left), while MSH-5 foci are not detected above baseline in late pachytene nuclei (right; 0, 1 or 2 MSH-5 signals colocalize with COSA-1, as in unirradiated controls). (C) GFP::COSA-1 localization in nuclei within the late pachytene region at 2.5 and 4 hrs post-IR; fields also include a few mid-pachytene nuclei (at the left) and a few early diplotene nuclei (at the right). Circles indicate nuclei in which 6 COSA-1 foci are detected. At 2.5 hrs post-IR, nuclei with 6 COSA-1 foci are limited to a narrow zone near the start of the late pachytene region. At 4hrs post-IR, the zone of nuclei with 6 COSA-1 foci has expanded, presumably reflecting movement into and progression through late pachytene of nuclei that had been exposed to IR during mid-pachytene. (D) Localization of MSH-5 and COSA-1 at 8 hrs post-IR in a region spanning the mid-to-late pachytene transition. Left inset: Mid-pachytene nuclei, showing MSH-5-only foci, in excess of eventual COs. Right inset: Late pachytene nuclei, showing 6 MSH-5 foci that co-localize with 6 COSA-1 foci.
By 1 hr post-IR, both MSH-5 and RAD-51 foci were detected in irradiated germ lines (Figure 5B, data not shown). Whereas RAD-51 foci were present throughout the germ line, MSH-5 foci were restricted to a limited region. Induced MSH-5 foci were abundant in the mid-pachytene region, where MSH-5 foci first appear during wild-type meiosis, but were not detected in late pachytene nuclei, which appeared similar to unirradiated spo-11 controls. This suggests that DSBs per se are not sufficient to recruit MSH-5, but must occur during the appropriate stage of meiotic prophase to be competent to load MSH-5 de novo. Notably, at 1 hr post-IR there was little change in COSA-1 localization, indicating that the conditions for COSA-1 loading were not yet met.
At 2.5 hrs post-IR, a few nuclei with 6 COSA-1 foci were visible near the mid-to-late pachytene border (Figure 5C). These likely correspond to nuclei that had been in mid-pachytene at the time of irradiation, had loaded MSH-5, and then subsequently progressed into late pachytene and became competent to load COSA-1.
At 4 hrs post-IR, the region containing nuclei with 6 COSA-1 foci had expanded further into the late pachytene region and a subset of nuclei had begun to display reorganization of ZHP-3 (Figures 5C and S5B). Notably, a distinct boundary is visible within the late pachytene region between a zone containing nuclei with 6 COSA-1 foci and a zone without such nuclei, creating the impression of a wave of nuclei with 6 COSA-1-marked CO-sites progressing through late pachytene.
By 8 hrs post-IR, the wave front had reached early diplotene and ZHP-3 signals had substantially retracted toward the COSA-1 foci (Figure 5A). 8 hr post-IR gonads stained for MSH-5 and COSA-1 display a striking cytological transition at the mid-to-late pachytene boundary (Figure 5D). Mid-pachytene nuclei exhibit abundant MSH-5 foci but lack COSA-1 foci, while late pachytene nuclei have only 6 MSH-5 foci, each colocalizing with COSA-1.
In summary, 1) only nuclei in mid-pachytene are competent for rapid de novo loading of MSH-5 at IR-induced DSBs. 2) DSBs induced in germ cells that are already in late pachytene are not competent to load either MSH-5 or COSA-1. 3) Germ cells can acquire competence to load COSA-1 upon transit into late pachytene if they had been in mid-pachytene at the time of DSB induction (and presumably had loaded MSH-5 at that time). 4) Loading of COSA-1 appears to stabilize MSH-5, as MSH-5 foci not colocalized with COSA-1 are lost following transit to late pachytene.
Dose-response analysis reveals efficient DSB utilization and a robust ability to limit COSA-1-marked sites to one per homolog pair
The consistent localization of COSA-1 to 6 foci both in wild-type meiosis and in our IR time course experiments prompted us to conduct a dose-response analysis to investigate the relationship between DSB number and COSA-1-marked sites. For these experiments, we exposed spo-11 mutant worms expressing GFP::COSA-1 to different IR doses, then assessed COSA-1 foci in late pachytene nuclei fixed at 8 hrs post-IR (Figure 6A and 6B).
Figure 6. Dose-response analysis reveals a highly non-linear relationship between IR-induced DSBs and COSA-1 foci.
(A) Paired IF images showing GFP::COSA-1 foci in late pachytene nuclei from gfp::cosa-1; spo-11 germ lines exposed to the indicated IR doses, fixed 8 hrs post-IR, with numbers of foci in each nucleus indicated. Scale bar = 5 μm. (B) Stacked bar graph showing percentages of nuclei with the indicated numbers of COSA-1 foci at different IR doses. (C) Graph showing the highly non-linear relationship between IR dose and the mean number of COSA-1 foci per nucleus. Experimental data points are plotted in red, with error bars indicating standard deviation. Our mathematical model (μ = 6(1-e-cr) is plotted in blue; see Results and Supplemental Analysis. (D) Graph depicting linear relationship between IR dose and inferred mean number of DSBs per chromosome pair, calculated from our empirical data based on the postulates of our model. Empirical data points are in red, with linear regression in blue.
This analysis revealed a striking non-linear relationship between IR dose and COSA-1 foci (Figure 6C). From 100-1,000 rads, the average number of foci per nucleus increased with increasing IR dose, suggesting that DSB number limits the number of COSA-1 foci at doses below 1 kRad. At 1,000 rads, 90% of nuclei scored had exactly 6 foci, indicating that this dose is sufficient for 99% of chromosome pairs to receive at least 1 DSB. The number of foci then plateaued, so that even when the dose was increased 10 fold, exactly 6 foci were still detected in 90% of nuclei, indicating that most excess DSBs do not yield COSA-1 marked sites.
The observed dose-response relationship closely matches that predicted by a model in which: 1) irradiation results in a random distribution of DSBs, 2) a chromosome pair with 0 DSBs will have 0 COSA-1 foci, and 3) the presence of ≥1 DSB on a chromosome pair yields exactly 1 COSA-1 focus. We used these postulates and assumptions to derive a function (see Supplemental Analysis) to model the relationship between IR dose and the mean number of COSA-1 foci per nucleus:
where μ = mean number of COSA-1 foci per nucleus, r = IR dose, and c = constant describing the relationship between DSB number and IR dose. The best-fit dose-response curve generated by this function (using a value of 0.0039134 for c) is shown in blue in Figure 6C, together with the observed data in red. Given the relationship:
where λ = mean number of DSBs per chromosome pair, this fitted model predicts that an average of 3.9 DSBs per chromosome pair would be generated at the 1000 rad IR dose. This translates to a yield of 1 DSB/ 17 Mb / krad, which is identical to the DSB yield previously estimated for mammalian cells based on physical detection assays (Thompson and Limoli, 2000). The correspondence of these estimates for efficacy of IR in DSB induction further supports the validity of the relationship between DSBs and COSA-1-marked CO sites deduced from our experimental/modeling analysis. Moreover, it implies that most or all DSBs induced by IR in our system are competent to enter the meiotic recombination pathway.
We also took a complementary approach, directly calculating empirical values for λ (here, the inferred mean number of DSBs per chromosome pair) using the data for the lower IR doses and the Poisson equation P(0)=e--λ, where P(0) = the fraction of chromosome pairs inferred to have 0 COSA-1 foci and thus 0 DSBs based on the postulates stated above (see Supplemental Analysis). A linear regression analysis describing the relationship between IR dose and λ (Figure 6D) provided strong validation for our initial assumptions (R2=0.997). Moreover, extrapolation of the line (slope = 0.00390) yields an average of 3.9 DSBs per chromosome pair at the 1 kRad dose, matching the value generated above.
In summary, our dose response data provide strong support for a model in which chromosome pairs lacking DSBs will lack COSA-1 foci, while chromosome pairs with one or more DSBs will receive a single COSA-1 focus regardless of DSB number. This indicates the operation of a robust CO control system that is both: 1) highly efficient at converting a single DSB into a CO when DSBs are limiting, and 2) highly effective at limiting the number of cytologically-differentiated CO sites to one per chromosome pair when DSBs are in excess.
COSA-1 foci exhibit interference under conditions that alter CO number
To further characterize the relationship between COSA-1 foci and COs, we assessed COSA-1 foci under several circumstances previously shown to alter CO number:
mnT12 fusion chromosome homozygotes
Worms homozygous for mnT12, an X;IV fusion chromosome, have 5 rather than 6 chromosome pairs. Previous work showed that the fusion chromosome pair undergoes only one CO in the majority of meioses, indicating that interference operates on this extended chromosome to limit CO number (Hillers and Villeneuve, 2003). In line with the prior CO analysis, 5 COSA-1 foci were observed in the majority of late pachytene nuclei in mnT12 homozygotes (Figure 7A), consistent with COSA-1 foci being responsive to CO interference operating over distances that exceed the length of a normal chromosome.
Figure 7. Relationship of COSA-1 to COs in conditions that alter CO number.
(A) Graph showing percentages of nuclei with the indicated numbers of COSA-1 foci in strains with altered numbers of COs. Numbers of foci in the rtel-1(tm1866) and dpy-28(s939) mutants did not differ significantly from the control (Mann-Whitney test). Worms homozygous for the mnT12(X;IV) fusion chromosome have only 5 chromosome pairs, and mnT12 undergoes only one CO in the majority of meioses; an average of 5.3 COSA-1 foci per nucleus were observed in mnT12 homozygotes. Numbers of COSA-1 foci in rtel-1; mnT12 worms did not differ significantly from mnT12 controls. *WT control contains the gfp::cosa-1 transgene in an otherwise wild-type background. (B) The rtel-1 mutation does not suppress the lack of chiasmata caused by loss of cosa-1 function. Graph shows percent of diakinesis nuclei with a given number of DAPI-stained bodies. As in cosa-1(tm3298) single mutants, 11-12 DAPI bodies were detected in rtel-1; cosa-1 double mutants, reflecting a lack of chiasmata. Numbers of oocyte nuclei scored: wild type (n = 164), rtel-1 (n = 125), cosa-1 (n = 116), rtel-1; cosa-1 (n = 114). (C) Bar graph indicating genetic map distances (cM ± 95% C.I.) for the unc-60 dpy-11 interval measured for worms of the indicated genotypes (see Supplemental Experimental Procedures). ** indicates p < 0.001; * indicates p = 0.01. The CO frequency in the rtel-1 mutant (19.6 cM) was significantly elevated over wild type (13.4 cM, p = 0.0002; Fisher exact) and cosa-1/ + (13.6 cM, p = 0.0006) controls, which did not differ from each other. rtel-1 also differed significantly from rtel-1; cosa-1/+ (15.6 cM, p = 0.01), indicating that elevation in CO frequency was suppressed in rtel-1; cosa-1/+ worms. (D) Paired 3D volume renderings of a representative nucleus used to quantify X chromosome associated COSA-1 foci in the dpy-28 mutant. Staining for chromosome axis protein HTP-3 reveals the paths of synapsed chromosome pairs; arrow indicates the X chromosome, marked by X-PC associated protein HIM-8. Scale bar = 2 μm. Expected incidence of X chromosomes with 2 or more COSA-1 foci was estimated to be ≥ 36% based on frequencies of 2-CO and 3-CO products detected by genetic assay (Tsai et al., 2008).
rtel-1 mutant
RTEL-1 is a DNA helicase that can disassemble D-loop recombination intermediates in vitro, and the rtel-1 mutant was reported to have a roughly 2-fold increase in COs (Barber et al., 2008; Youds et al., 2010). Despite this increase in COs detected by genetic assays, the numbers of COSA-1 foci in the rtel-1 mutant (6.0 ± 0.2) were indistinguishable from wild-type controls (6.0 ± 0.2; Figure 7A), indicating that COSA-1 does not concentrate in foci at the sites of excess COs in the rtel-1 mutant. Nevertheless, several lines of evidence suggest that the extra COs in rtel-1 mutants still require the canonical meiotic CO pathway for their formation. First, analysis of rtel-1; cosa-1 double mutants showed that the rtel-1 mutation does not rescue the lack of chiasmata or COs caused by loss of COSA-1 (Figure 7B and Table S1). Moreover, the elevation in CO frequency caused by loss of rtel-1 function was partially suppressed in rtel-1/rtel-1; cosa-1/+worms, while cosa-1 heterozygosity had no effect in the wild-type background (Figure 7C). Together these observations suggest that the excess COs in rtel-1 mutants are formed using the canonical COSA-1/MSH-5-dependent CO pathway (rather than an alternative pathway), despite the fact that cytologically detectable COSA-1 foci do not form at the extra CO sites. These findings are consistent with a mutually antagonistic relationship between RTEL-1 and COSA-1/MSH-5 at prospective recombination sites. COSA-1/MSH-5 may protect CO intermediates from RTEL-1-mediated disassembly at CO-designated sites, while RTEL-1 may be needed to efficiently disengage excess COSA-1/MSH-5-dependent intermediates at sites that were not designated to become COs. Thus, absence of RTEL-1 could allow such intermediates to progress to CO products without accumulating CO proteins at high concentration.
dpy-28 mutant
DPY-28 is a subunit of the condensin I complex, which associates with meiotic prophase chromosomes and influences both the length of the chromosomes and the number and positioning of DSBs (Mets and Meyer, 2009; Tsai et al., 2008). Previous genetic analysis revealed a high incidence of meiotic products with 2 or more COs on the X chromosomes in the dpy-28(s939) mutant. However, quantitation of COSA-1 foci specifically on the X chromosomes in the dpy-28 mutant (Figure 7D) did not reveal a commensurate incidence of X chromosome pairs with multiple COSA-1 foci. Whereas ≥36% of chromosome pairs are expected to have 2 or more COs based on the genetic data of (Tsai et al., 2008), 39 of 39 X chromosome pairs analyzed had only a single COSA-1 focus (p < 0.0001). Thus, despite the processing of extra intermediates into COs, interference is nevertheless still operating in the dpy-28 mutant to limit cytologically-differentiated CO sites to 1 per homolog pair in most meioses.
Discussion
Our identification of COSA-1 as an integral component of a conserved CO-promoting functional module highlights the utility of C. elegans for discovering previously unknown components of the meiotic machinery. Moreover, our ability to visualize COSA-1 as an in vivo marker of CO-designated sites provides an opportunity to investigate how prospective CO sites progressively differentiate, how these events are coupled to meiotic progression, and how events are regulated both to ensure the formation of COs and to limit their numbers.
Analysis of pro-CO proteins suggests a two-step process for CO specification
The localization dynamics of pro-CO factors during wild-type meiosis and in response to IR-induced DSBs support a model in which COs are specified via a two-step process. Together the data imply the existence of an initial “CO licensing” step, presumably involving de novo loading of MSH-5 to prospective recombination sites in excess of eventual COs, that must occur during mid-pachytene as a prerequisite for later concentration of COSA-1 at a single presumptive CO site per chromosome pair. Eligibility for de novo MSH-5 loading shuts down at the mid-to-late pachytene transition, coincident with a MAP-kinase-dependent transition in the mode of DSB repair (Hayashi et al., 2007). Further, subsequent concentration of COSA-1, MSH-5 and ZHP-3 at 6 sites following the mid-to-late pachytene transition implies that licensing is followed by a second “CO designation” step that limits the number of licensed sites that can ultimately mature into COs. While our data do not distinguish whether CO designation occurs at or just prior to the mid-to-late pachytene transition, it is clear that progression into late pachytene is required for its manifestation as cytologically-differentiated COSA-1 marked sites.
Integral to this two-step model for CO specification is that CO designation occurs after SC assembly, in agreement with recent findings of (Henzel et al., 2011) and (Rosu et al., 2011). This is illustrated both by the progressive differentiation of CO sites during wild-type meiosis and by the fact that DSBs induced in the context of assembled SCs are successful in maturing into COSA-1-marked sites in a regulated manner. While CO designation in S. cerevisiae may be complete prior to SC assembly, we speculate that CO designation after SC assembly may be a widespread feature of metazoan meiosis, as MSH4 foci in mice and humans occur in substantial excess of COs, then diminish to a number approaching the number of COs only after completion of synapsis (Baudat and de Massy, 2007).
COSA-1 structure suggests a mechanism for reinforcement of CO designation
Several lines of evidence suggest that CO designation is maintained by a self-reinforcing mechanism. First, while MSH-5 foci decrease in number upon COSA-1 installation, the increased brightness of the remaining foci suggests an increase in the local concentration of MSH-5 at COSA-1-marked sites. Second, COSA-1 and MSH-5 have a propensity to colocalize in aggregates even in the absence of appropriate substrates (i.e. nascent recombination events). These observations suggest a self-reinforcing property that could simultaneously result both in enrichment of pro-CO factors at CO-designated sites and their depletion from sites elsewhere on the chromosome. Third, the identity of COSA-1 as cyclin-related protein suggests a specific model for self-reinforcement. COSA-1 may partner with a cyclin-dependent kinase (CDK) family member to form a dedicated COSA-1/CDK complex that promotes CO progression. Moreover, C. elegans MSH-5 represents a likely target substrate for this proposed CDK activity, as it has 15 potential CDK phosphorylation sites ([S/T]P) and can be phosphorylated by a Cyclin/CDK complex in vitro (Figure S6). We propose that upon transition to late pachytene, MSH-5 localized to CO-eligible sites could recruit COSA-1/CDK, which in turn could initiate a positive feedback loop that results in increased local concentration of both components.
The possibility that a CDK might play a direct role in CO progression had been suggested by the observation that CDK2 colocalizes with MLH1 foci in late pachytene mouse meiocytes (Ashley et al 2001). However, the functional significance of this localization with respect to CO formation was unknown, as apoptosis of Cdk2-/- spermatocytes prior to mid-pachytene precluded assessment of potential later roles (Ortega et al., 2003; Viera et al., 2009). (Ward et al., 2007) speculated that destruction of a B-type cyclin mediated by E3 ubiquitin ligase HEI10 could free CDK2 to localize at CO sites, presumably in partnership with a distinct cyclin. We suggest that mammalian CDK2 may indeed act locally at nascent CO sites, possibly partnered with COSA1/CNTD1.
COSA-1 foci illuminate robust CO control during C. elegans meiosis
Previous work analyzing the meiotic behavior of fusion chromosomes in C. elegans demonstrated the operation of a highly effective CO interference system that normally limits COs to 1 per homolog pair . This prior analysis probed the system by doubling the size of a chromosome, showing that the unusually large chromosome pair was nevertheless limited to a single CO in the majority of meioses (Hillers and Villeneuve, 2003) This finding showed that the frequency of COs per Mb was malleable, but did not address whether this reflected a change in DSB number or a change in the fraction of DSBs converted into COs. Here, we altered the substrate for the CO control machinery in a complementary manner, by varying levels of IR-induced DSBs in a background that lacks endogenous DSBs, and used COSA-1 foci as a readout for the response.
The highly non-linear response of COSA-1 foci to IR dose highlights the robustness of the C. elegans CO control system. First, our data and modeling provide insight into the relationship between DSBs and CO assurance, demonstrating that the C. elegans CO control system is highly efficient at converting DSB into COSA-1 marked sites when DSBs are present in limiting quantities. This finding dovetails with the recent finding of (Rosu et al., 2011) that CO formation is the preferred outcome of meiotic DSB repair when DSBs are limiting. High efficiency conversion of a single DSB into a COSA-1-marked CO implies that an average of 4 randomly distributed DSBs per chromosome pair would suffice to ensure a CO for >98% of chromosome pairs, raising the possibility that CO assurance during normal C. elegans meiosis could potentially be achieved largely through a random distribution of a modest number of DSBs.
Another striking feature of the dose-response analysis is that COSA-1 foci plateau at 6 per nucleus, even under conditions where DSBs are 5- or 10-fold more abundant than the number needed to ensure a chiasma for each homolog pair in nearly all nuclei. This implies that the interference mechanism limiting the number of cytologically-differentiated CO sites per bivalent is nearly impervious to a substantial excess of potential initiating events. Recently, the term “CO homeostasis” was coined to describe the ability to maintain a fixed number of CO events in the face of significant reduction in DSB levels (Martini et al., 2006). Our finding that C. elegans germ cells maintain a fixed number of COSA-1 foci in the face of a substantial excess of DSBs demonstrates homeostasis operating in the opposite direction, supporting the notion that interference and homeostasis are inextricably linked manifestations of the same underlying CO control mechanism. Moreover, the fact that we were able to recapitulate the entire spectrum of the dose-response by a highly constrained mathematical model assuming complete CO assurance and complete CO interference indicates that these two properties can fully explain CO homeostasis in this system.
Concordance between the DSB yield inferred from our dose-response analysis and the established estimate for mammalian cells further indicates that our system can be exploited as a “biological dosimeter” for DSB formation: We can use COSA-1 foci as a quantitative readout of DSB yield not only to estimate DSB number in meiotic mutants, but also to evaluate efficacy of treatments that either provoke or inhibit DSB formation.
GFP::COSA-1 as a tool to investigate CO interference
How CO designation at one site is communicated along a chromosome pair to decommission other prospective CO sites remains a major mystery. Although the phenomenon of CO interference has fascinated geneticists since its discovery a century ago, its mechanism has remained remarkably recalcitrant to experimental elucidation.
A major confounding factor in studying interference is that genetic assessment of COs can prove to be an ironically unreliable readout for evaluating the status of CO interference, especially in mutant situations. There are (at least) two pathways by which COs can be generated during meiosis (de los Santos et al., 2003), commonly referred to as Class I COs, which are subject to and confer interference, and Class II COs, which are interference-insensitive. Thus, lack of interference among residual COs in a mutant may simply reflect loss of events that are subject to interference. Conversely, an increase in CO number might reflect loss of a downstream effector that functions to facilitate non-CO repair after CO interference has been established, as is likely the case for the C. elegans rtel-1 mutant (Youds et al., 2010); this work). Cytological markers of presumptive CO sites (e.g. chiasmata, recombination nodules, and MLH1 foci) represent useful alternatives for evaluating CO distribution and interference (Anderson et al., 1999; Carpenter, 1975; Jones, 1984; Zickler et al., 1992). However, methods for detecting these features are not readily adapted to serve as the basis of genetic screens.
Our analyses suggest that GFP::COSA-1 foci may represent a crucial new experimental foothold for analyzing interference. Numbers of foci are robustly maintained even in the context of DSB levels that likely exceed normal levels by 10 fold, a level at which previously-reported SNP analysis would predict the formation of some 2-CO meiotic products (Mets and Meyer, 2009). Further, the ability to limit GFP:COSA-1-marked sites to one per chromosome pair is retained in some mutants where excess COs were detected by genetic assays. Thus, GFP::COSA-1 foci appear to represent a more reliable surrogate than COs per se for the chromosomal events that are actually being sensed and/or distributed by the interference mechanism. Moreover, GFP::COSA-1 foci are readily visualized in live worms. This creates an unprecedented opportunity to identify factors that contribute to the CO interference mechanism, making it possible to screen directly for impaired interference by screening for altered numbers of COSA-1 foci.
Experimental Procedures
IR time course and dose-response experiments were conducted at 25°C; all other experiments were performed at 20°C. Experiments using GFP::COSA-1 also included the wild-type cosa-1 gene unless otherwise noted. Meiotic mutant homozygotes were derived from balanced heterozygotes by selecting progeny lacking a dominant marker associated with the balancer. FISH was performed as in (Dernburg et al 1998). Immunostaining was performed as in (Martinez-Perez and Villeneuve, 2005) with modifications. Dissections were performed on 24hrs post-L4 adults unless otherwise indicated. Except where noted, images are projections through 3D data stacks encompassing whole nuclei Lists of strains and antibodies and details of image acquisition are provided in the Supplement.
Quantitation of GFP::COSA-1 foci
Foci were quantified from deconvolved 3D data stacks; only nuclei completely contained within the stack were scored. Nuclei with features indicative of apoptosis (compact and DAPI-bright) were excluded. In unirradiated gonads (Figure 3 and 7), foci were counted in the last 5 rows of pachytene nuclei. In irradiated gfp::cosa-1; spo-11gonads, foci were counted within a 3-5 row zone of late pachytene nuclei bounded by 1 row of nuclei with consistent GFP::COSA-1 staining on either side. Numbers of nuclei scored for each strain are: gfp::cosa-1 (76), mnT12 fusion chromosome (150), rtel-1 (138), rtel-1; mnT12 (113), dpy-28 (79).
In the dpy-28 mutant, foci were also quantified specifically on the X chromosome pair. A HIM-8 antibody was used to mark the X-PCs, and axis marker HTP-3 was used to trace the chromosome paths. 3D volume renderings were generated using Volocity 5.5.1 to allow rotation of images. X chromosome-associated COSA-1 foci were quantified only in nuclei where X chromosome paths could be traced unambiguouosly, i.e., they had a HIM-8 focus near one end and exhibited a continuously traceable HTP-3 path that did not intersect with any other HTP-3 tracks. 49% of late pachytene nuclei examined met these criteria.
Irradiation experiments
gfp::cosa-1; spo-11 worms were cultured at 25°C to increase expression of GFP::COSA-1. γ-irradiation was performed at 18 hr post-L4 using a Cs-137 source. Unirradiated controls were dissected with the first experimental time point. For time course experiments, worms were exposed to 1 kRad of γ-irradiation, then dissected at various time points. For dose response experiments, worms were dissected 8 hrs after exposure to various IR doses. Numbers of nuclei scored for each dose: 0 rad (63), 100 rad (68), 250 rad (93), 500 rad (64), 1kRad (41), 5 kRad (83), 10 kRad (63).
Supplementary Material
Highlights.
-- COSA-1 is required to convert DSBs into COs and concentrates CO proteins at CO sites
-- Competence to install COSA-1 at designated CO sites requires a prior licensing event
-- Robust homeostasis and interference yield one marked CO site per chromosome pair
-- COSA-1 foci act as a DSB dosimeter and a live-cell reporter of CO interference
Acknowledgments
We thank S. Mitani (National Institute of Genetics, Japan) and S. Rosu (Villeneuve lab) for cosa-1(tm3298) and zhp-3(me95), N. Bhalla, A. Dernburg, A. la Volpe, B. Meyer, S. Boulton, M. Zetka and the Caenorhabditis Genetics Center for strains and antibodies, and J. Bessler for technical support. We thank A.D. Kaiser, M. AlQuaraishi, C. Janda, C. Thomas, D. Riordan, S. Clarke, and members of the Villeneuve lab for discussions and D. Libuda and A. Fire for comments on the manuscript. This work was supported by Stanford Genome Training Program NIH HG00044 Training Grant for RY, Leukemia and Lymphoma Society Postdoctoral Fellowship 5381-12 to KAZ, NIH grant R01GM67268 to AMV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AT. Electron microscopy of meiosis in Drosophila melanogaster females: II. The recombination nodule--a recombination-associated structure at pachytene? Proc Natl Acad Sci U S A. 1975;72:3186–3189. doi: 10.1073/pnas.72.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Davis MW, Nguyen H, Dayton D, Jorgensen EM. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics. 2005;171:1047–1056. doi: 10.1534/genetics.105.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzel JV, Nabeshima K, Schvarzstein M, Turner BE, Villeneuve AM, Hillers KJ. An asymmetric chromosome pair undergoes synaptic adjustment and crossover redistribution during Caenorhabditis elegans meiosis: implications for sex chromosome evolution. Genetics. 2011;187:685–699. doi: 10.1534/genetics.110.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FC, Jones GH. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 2008;55:28–39. doi: 10.1111/j.1365-313X.2008.03470.x. [DOI] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Jr., Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am J Hum Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen AJ, Roeder GS. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr Biol. 2009;19:1519–1526. doi: 10.1016/j.cub.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Colaiacovo MP. Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, Villeneuve AM. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The mechanism of crossing over. Am Nat. 1916;50:193–221. 284–305, 350–366, 421–434. [Google Scholar]
- Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Petri ET, Errico A, Escobedo L, Hunt T, Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–1349. doi: 10.4161/cc.6.11.4297. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart P, Woltering D, Hollingsworth NM. Conserved properties between functionally distinct MutS homologs in yeast. J Biol Chem. 1997;272:30345–30349. doi: 10.1074/jbc.272.48.30345. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–1289. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 2000;14:1539–1547. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- Schurko AM, Mazur DJ, Logsdon JM., Jr. Inventory and phylogenomic distribution of meiotic genes in Nasonia vitripennis and among diverse arthropods. Insect Mol Biol. 2010;19(Suppl 1):165–180. doi: 10.1111/j.1365-2583.2009.00948.x. [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Limoli CL. Origin, Recognition, Signaling and Repair of DNA Double-Strand Breaks in Mammalian Cells. Madame Curie Bioscience Database. 2000 [Google Scholar]
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera A, Rufas JS, Martinez I, Barbero JL, Ortega S, Suja JA. CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J Cell Sci. 2009;122:2149–2159. doi: 10.1242/jcs.046706. [DOI] [PubMed] [Google Scholar]
- Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, Langlais KK, Backus VL, Schimenti KJ, O'Brien MJ, et al. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 2007;3:e139. doi: 10.1371/journal.pgen.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Moreau PJ, Huynh AD, Slezec AM. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.