Molecular assessment has become a routinely-utilized and essential tool in modern medicine. Genetic analysis provides crucial, guiding information regarding inflammatory, infectious, inherited, and neoplastic diseases. In the dermatopathology laboratory, a molecular revolution has accelerated in past decade. In situ hybridization has enjoyed limited applicability in many laboratories for years, but recently fluorescence in situ hybridization (FISH) has enjoyed broad application to the interpretation of challenging melanocytic tumors.1 [Editors note: a detailed discussion of FISH is found elsewhere in this volume.2] Comparative genomic hybridization (CGH) has also emerged as a powerful instrument for detection of chromosomal copy number changes.3,4
When deploying CGH analysis, the detection of gains and losses of DNA in key chromosomal loci can be utilized to supplement conventional microscopy in the classification of ambiguous melanocytic tumors.
As a rule, conventional melanocytic nevi lack DNA copy number changes. In contrast, the majority of melanomas have chromosomal gains and losses in more than one locus. In the years following its development, CGH was first applied as a whole chromosomal assessment (chromosomal CGH or cCGH ). The emergence of CGH completed utilizing genetic arrays (array-based CGH or aCGH ) promises broader applicability and greater timeliness.
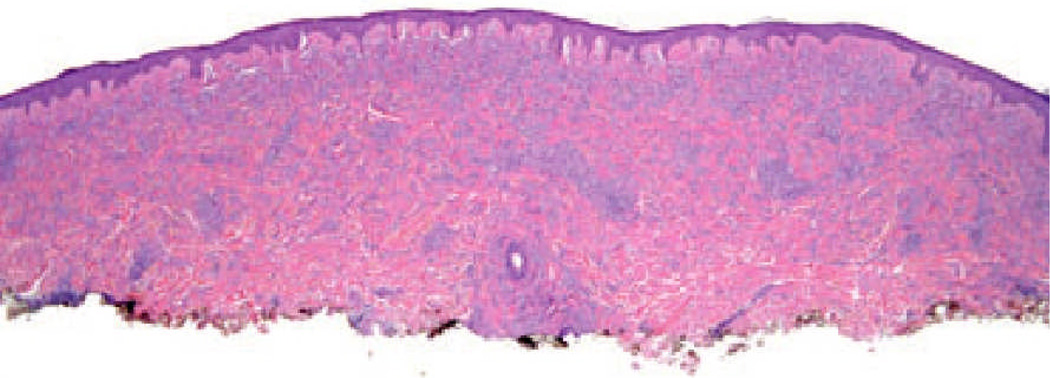
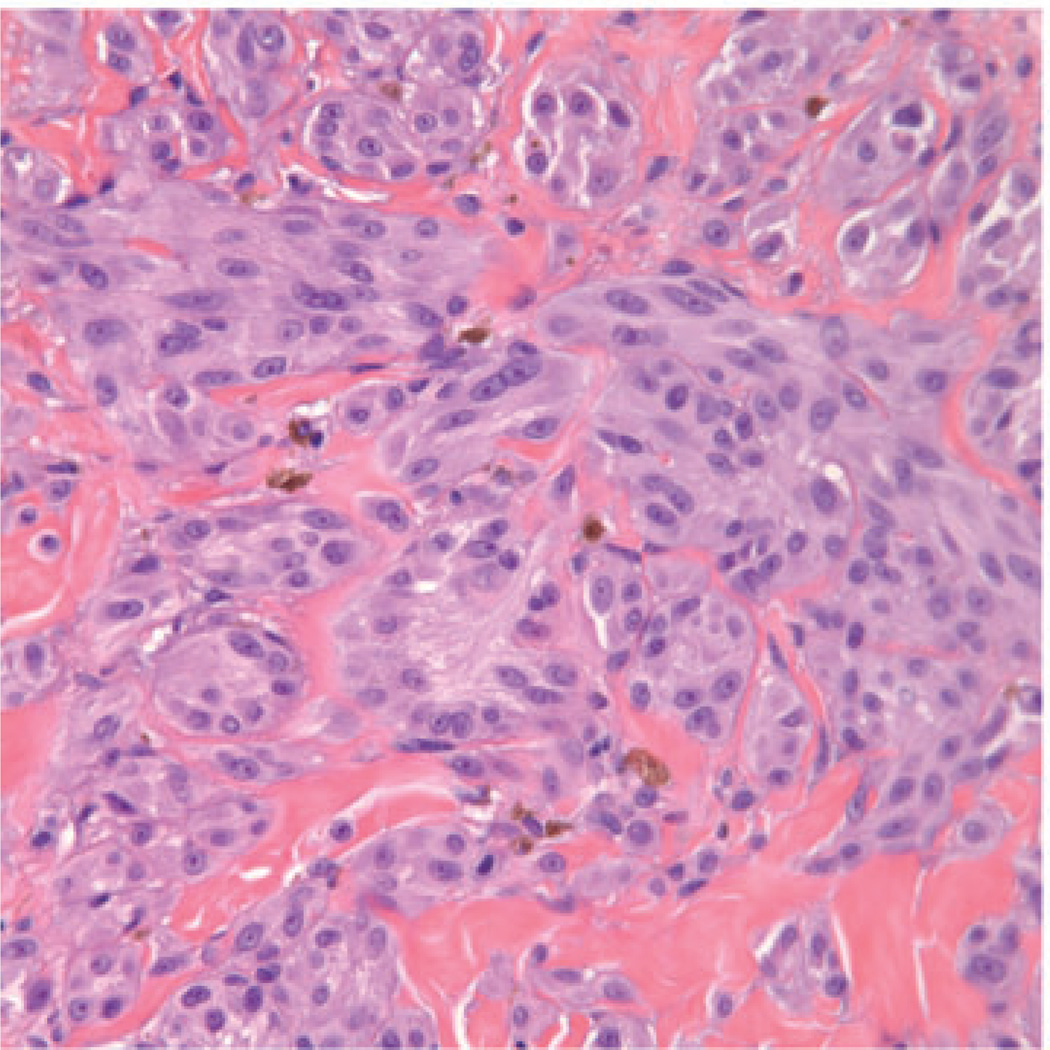
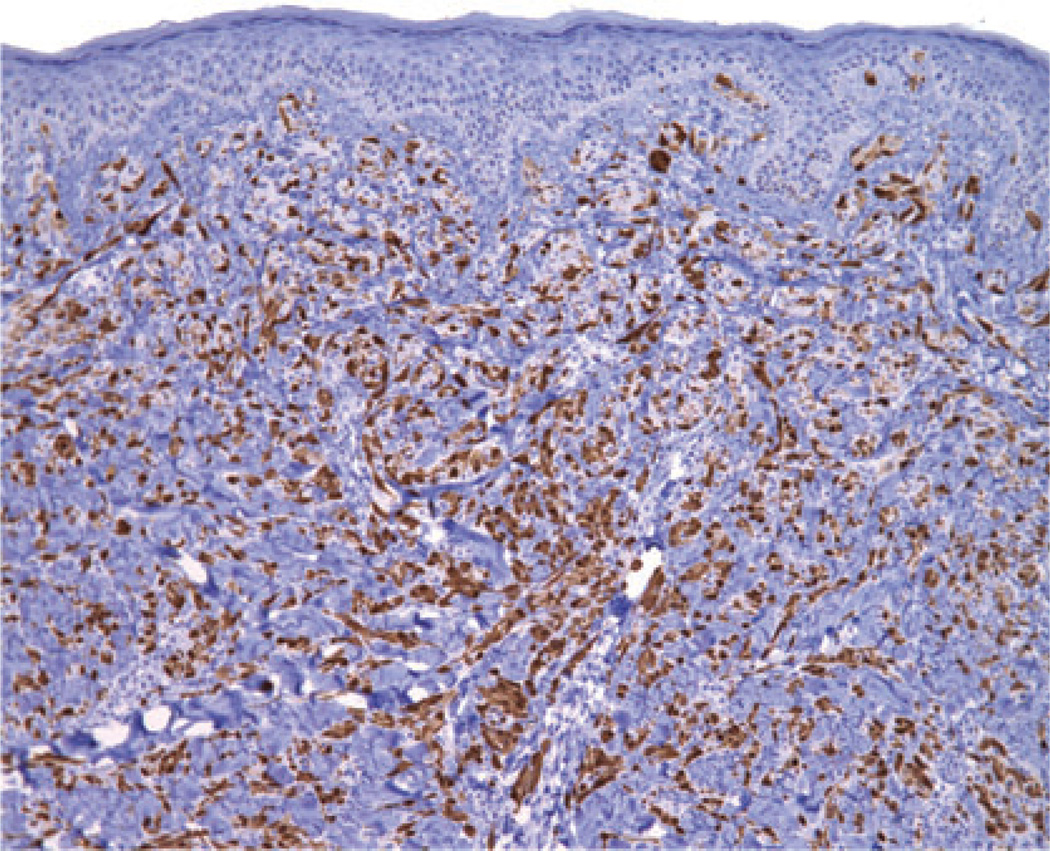
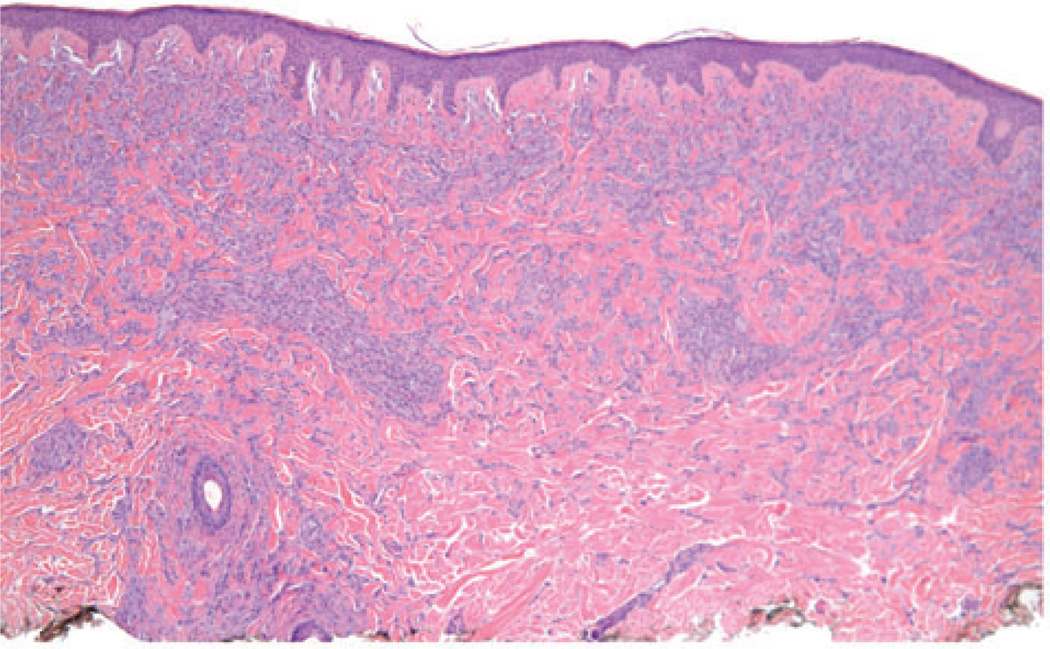
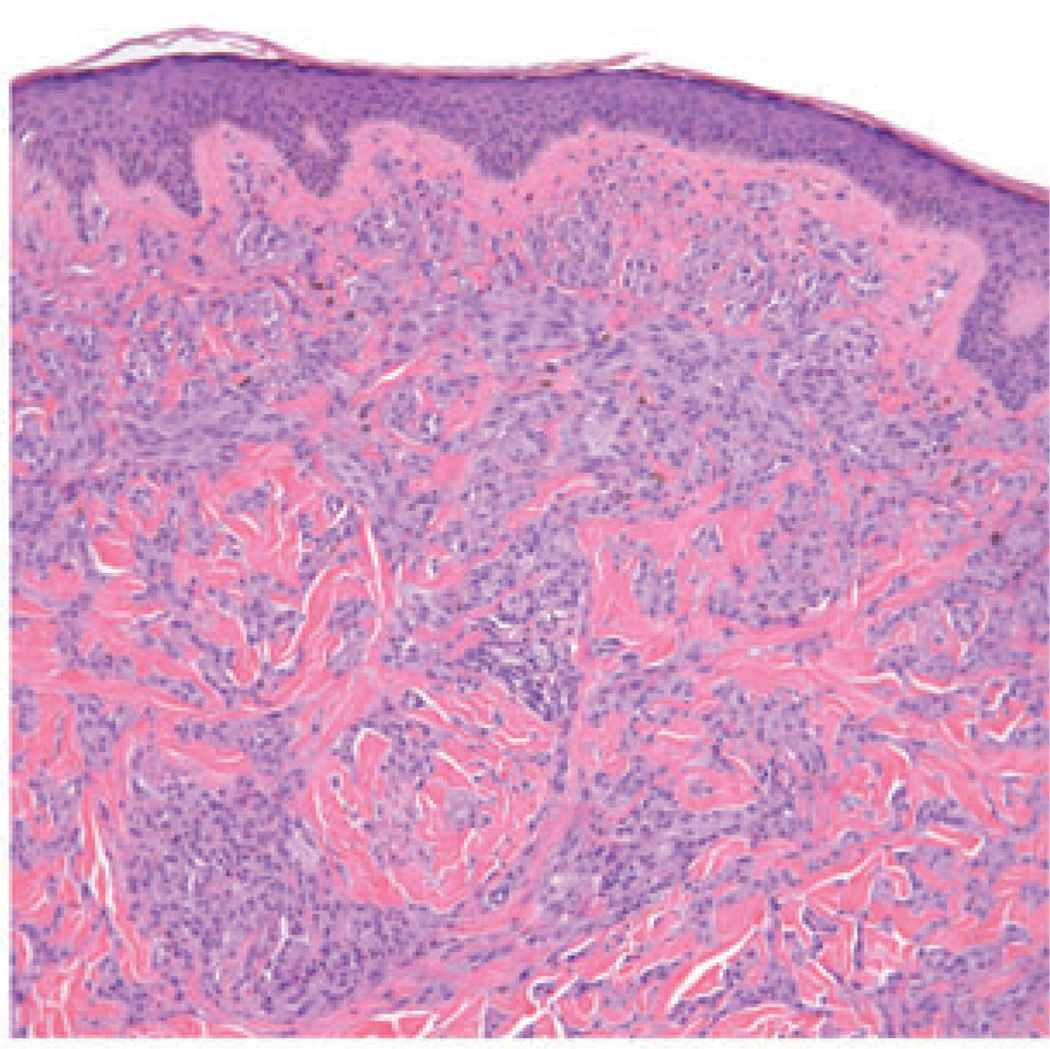
The case illustrated on the cover of this issue of the JCP represents a broad pink plaque that was detected on the back of a 38 year old man. It had been present for at least a six-month period of time and had been clinically judged to be worrisome for basal cell carcinoma. A broad shave biopsy demonstrated a sizable and mostly intradermal proliferation of enlarged melanocytes enmeshed in sclerotic retic-ular dermal collagen (Figures 2–5). Maturation was uneven and imperfect, but the Ki-67 cell proliferation index was very low (not illustrated). Immunohisto-chemical staining for p16 demonstrated preservation of cytoplasmic and nuclear expression by the vast majority of the cells of interest (Figure 6). The proliferation was interpreted descriptively but a large desmoplastic Spitz nevus was strongly favored. As the biopsy was only partial in nature, complete excision of any residuum was recommended.
Fig. 2.
A scanning magnification view demonstrates a broad melanocytic proliferation with its center of gravity in the reticular dermis.
Fig. 5.
Large fascicles of enlarged spitzoid melanocytes are apparent throughout the reticular dermis. Occasional melanocytes in mitosis could be found.
Fig. 6.
Immunohistochemical staining for p16 demonstrates preservation of expression, both nuclear and cytoplasmic, by most of the cells of interest. (Loss of p16 expression, as can be seen with melanoma, was not observed.)
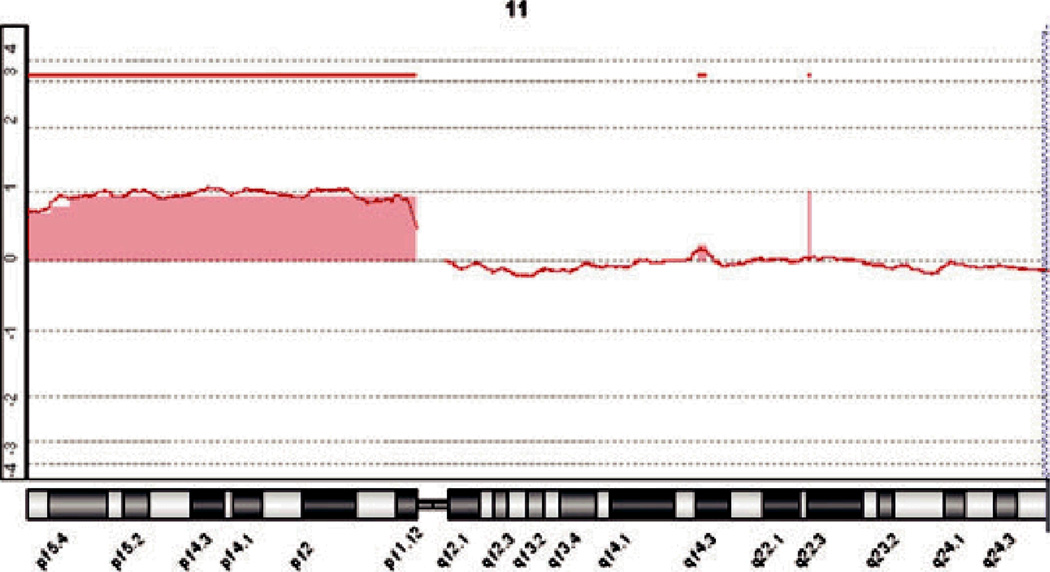
A subsequent excisional specimen demonstrated a significant residuum that was microscopically sim-ilar to what has been illustrated in Figures 2–5. The dermatopathologist who received that specimen acknowledged that a large desmoplastic Spitz nevus seemed likely but found a peculiar presentation of melanoma difficult to dismiss from the differen-tial diagnosis. At the second dermatopathologist’s request, molecular assessment of the original tumor was pursued. After completion of aCGH, a clear-cut gain in chromosome 11p was documented (Figure 1). No other chromosomal copy number aberrations were noted. A methodological notation regarding our aCGH protocol is included at the end of this manuscript.
Fig. 1.
A tracing from aCGH analysis shows clear gain in signal at the position of chromosome 11p. The remainder of the genome showed little variation from baseline. (The above graph shows the log2 of the ratios of the fluorescence intensity of the hybridized tumor-derived DNA and that of a differentially-labeled normal reference genome along chromosome 11 on the x-axis. A log2 ratio of 0 represents normal copy number.)
The documentation of 11p gain is not uncommon in the context of large desmoplastic Spitz nevi. Such sizable proliferations often have marked associated desmoplasia, an infiltrative periphery, and harbor HRAS mutations.5 – 8 Chromosome 11p gain is relatively tightly associated with HRAS mutancy. (HRAS mutations with resultant copy number increase of the mutant allele can also be seen in other tumors, such as bladder or thyroid cancer.) Although the clinical presentation of such tumors may not be distinctive, the attributes observed via conventional microscopy and molecular assessment are relatively characteristic. Extended follow-up of patients who have undergone excision of HRAS-mutated Spitz nevi has not demonstrated clinically malignant behavior.7 For this reason, we refer to such lesions as Spitz nevi rather than Spitz tumors.5
We posit that molecular-microscopical correla-tion has become increasingly important in dermatopathology and its future is now. Historically speaking, clinicopathological correlation has been a pillar in the practice of dermatopathology and its importance has been a point of emphasis in the recent writings of one of us.9,10 However, as the discipline of dermatopathology advances, simply mastering parallels between what is seen on the patient and what is viewed through our oculars and objectives will be insufficient. Increasingly, being able to infer the presence of key molecular scenarios by means of careful microscopy will permit narrow, specific diagnoses to be rendered or will facilitate focused molecular assessment. Excellence in molecular-microscopical correlation will be especially prized if demanding fiscal straits impede the widening application of molecular techniques.
Correlation between precise molecular attributes and exacting histomorphology is in its infancy. Recognizing gain of chromosome 11p and HRAS mutation in large desmoplastic Spitz nevi represents but one example. Loss of chromosome 3p in childhood-type spitzoid melanomas may be another.8 Our collective ability to infer specific, narrow genetic findings based upon simple conventional microscopy holds the potential to improve the power of dermatopathology as a tool in patient care.
Methodological notation: Manual micro-dissection of the tumor was performed using a stereo-microscope (Leica MZ12) to avoid significant normal cell contamination. DNA was extracted using the chloroform phenol extraction. The resultant DNA was labeled using a Bioprime Array CGH Genomic Labeling Kit according to the manufacturer’s instructions (Invitrogen, Carlsberg, CA). Briefly, 500 to 1000ng of test DNA and reference DNA (Promega, Madison, WI) were labeled with dCTP-Cy5 and dCTP-Cy3, respectively. The labeled DNA was mixed and hybridized onto an Agilent CGH microarray according to the manufacturer’s protocol version 6.2.1 (Agilent Technologies, Santa Clara, CA). The array was scanned using the Agilent microarray scanner G2505C and analyzed using Genomic Workbench Lite software 6.0.
Fig. 3.
Melanocytes in fascicles and cords are arrayed throughout much of the reticular dermis.
Fig. 4.
The melanocytes comprising the dermal component do not clearly mature with descent and are encompassed by sclerotic collagen.
References
- 1.Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009;33:1146. doi: 10.1097/PAS.0b013e3181a1ef36. Erratum in: Am J Surg Pathol 2010; 34: 688. [DOI] [PubMed] [Google Scholar]
- 2.McCalmont T. Fillet of FISH. J Cutan Pathol. 2011;38:327. doi: 10.1111/j.1600-0560.2011.01675.x. [DOI] [PubMed] [Google Scholar]
- 3.Bauer J, Bastian BC. Distinguishing melanocytic nevi from melanoma by DNA copy number changes: comparative genomic hybridization as a research and diagnostic tool. Dermatol Ther. 2006;19:40. doi: 10.1111/j.1529-8019.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 4.Bastian BC. Molecular cytogenetics as a diagnostic tool for typing melanocytic tumors. Recent Results Cancer Res. 2002;160:92. doi: 10.1007/978-3-642-59410-6_13. [DOI] [PubMed] [Google Scholar]
- 5.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000;157:967. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokx WA, van Dijk MC, Ruiter DJ. Molecular cytogenetics of cutaneous melanocytic lesions – diagnostic, prognostic and therapeutic aspects. Histopathology. 2010;56:121. doi: 10.1111/j.1365-2559.2009.03452.x. [DOI] [PubMed] [Google Scholar]
- 7.van Engen-van Grunsven AC, van Dijk MC, Ruiter DJ, Klaasen A, Mooi WJ, Blokx WA. HRAS-mutated Spitz tumors: a subtype of Spitz tumors with distinct features. Am J Surg Pathol. 2010;34:1436. doi: 10.1097/PAS.0b013e3181f0a749. [DOI] [PubMed] [Google Scholar]
- 8.McCalmont TH. In my opinion. J Cutan Pathol. 2011;38 doi: 10.1111/j.1600-0560.2010.01661.x. [DOI] [PubMed] [Google Scholar]
- 9.McCalmont TH. Fact or fiction? J Cutan Pathol. 2010;37:1130. doi: 10.1111/j.1600-0560.2010.01616.x. [DOI] [PubMed] [Google Scholar]
- 10.McCalmont TH. Baldy. J Cutan Pathol. 2010;37:1030. doi: 10.1111/j.1600-0560.2010.01588.x. [DOI] [PubMed] [Google Scholar]