Abstract
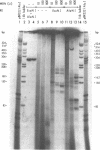
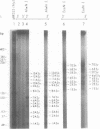
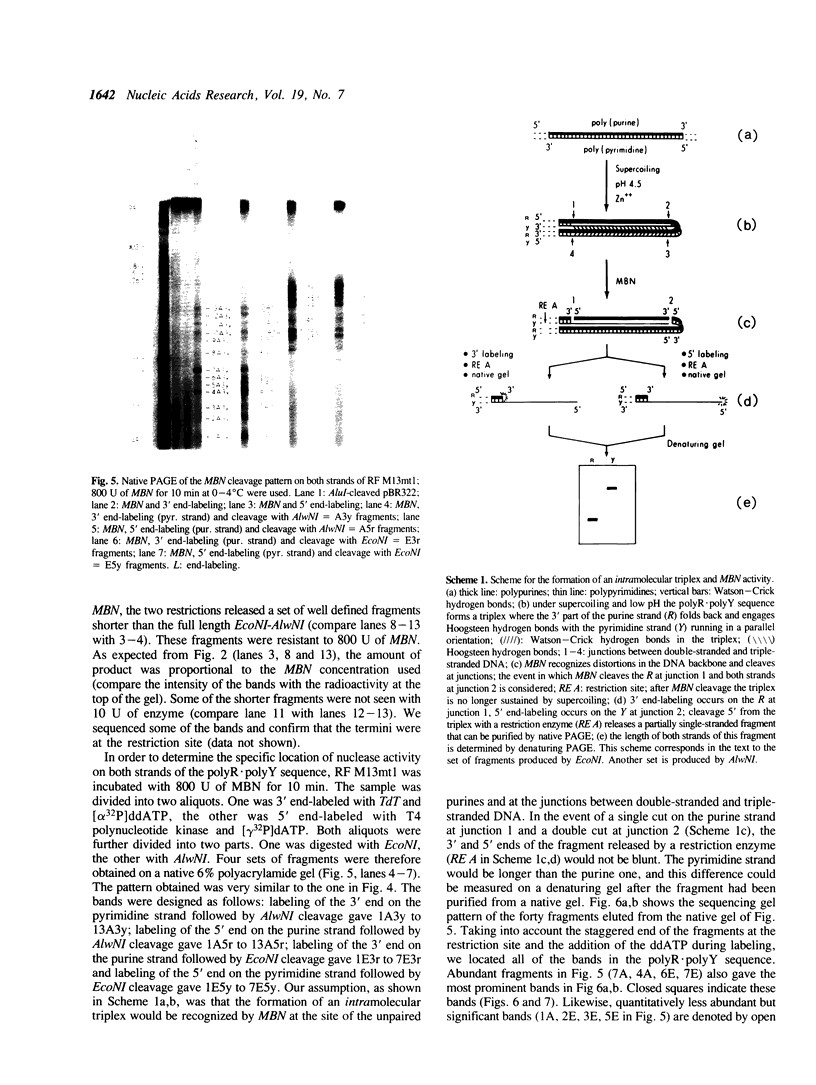
Mung bean nuclease, an enzyme specific for single-stranded DNA, was used to probe a non-B DNA structure present in the mouse metallothionein-I gene. The region sensitive to the enzyme was constituted by a 128 base-pair long polypurine.polypyrimidine sequence located at 1.2-kb from the start of transcription. A detailed analysis of the mung bean nuclease cleavage pattern revealed that: (i) under conditions of supercoiling and low pH a triplex structure was formed, (ii) the triplex was flanked by a sequence with the potential of forming a Z-DNA structure, (iii) most of the enzymatic activity was localized at some of the junctions between double-stranded and triple-stranded DNA and at mismatches in the triplex, (iv) no unpaired bases were observed in the loop or outside the triplex, and (v) the triplex was present in more than one configuration.
Full text
PDF

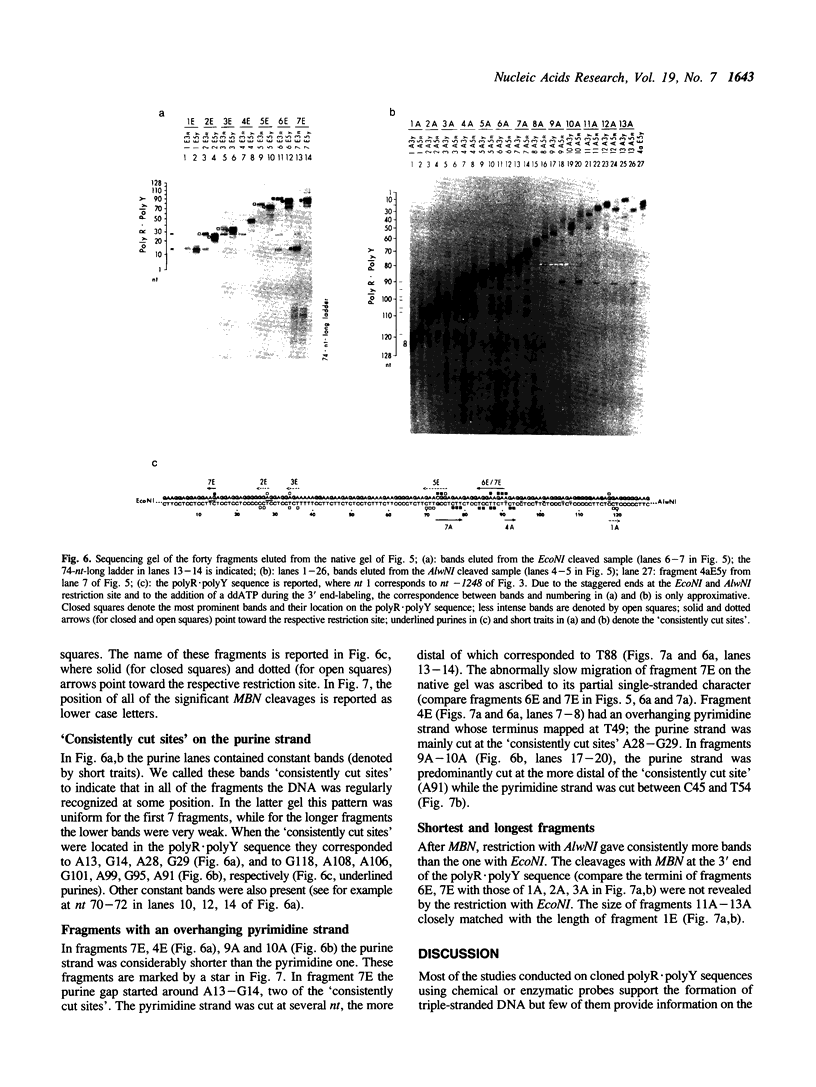
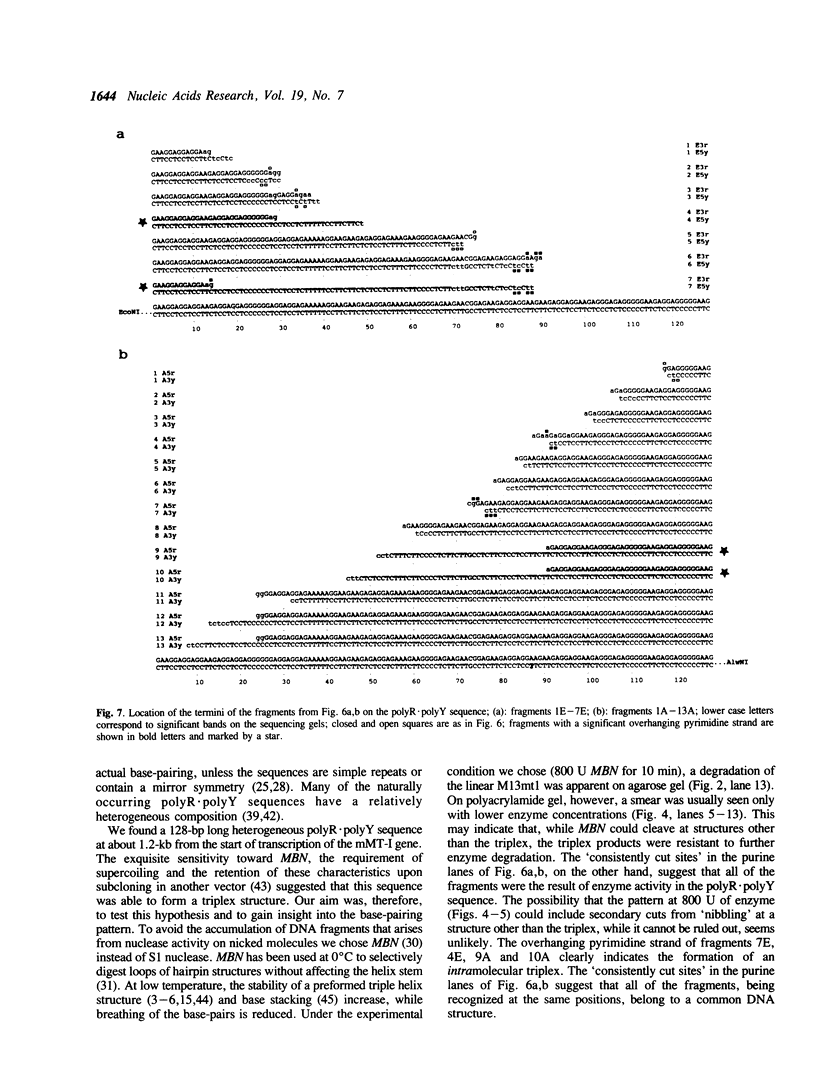


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
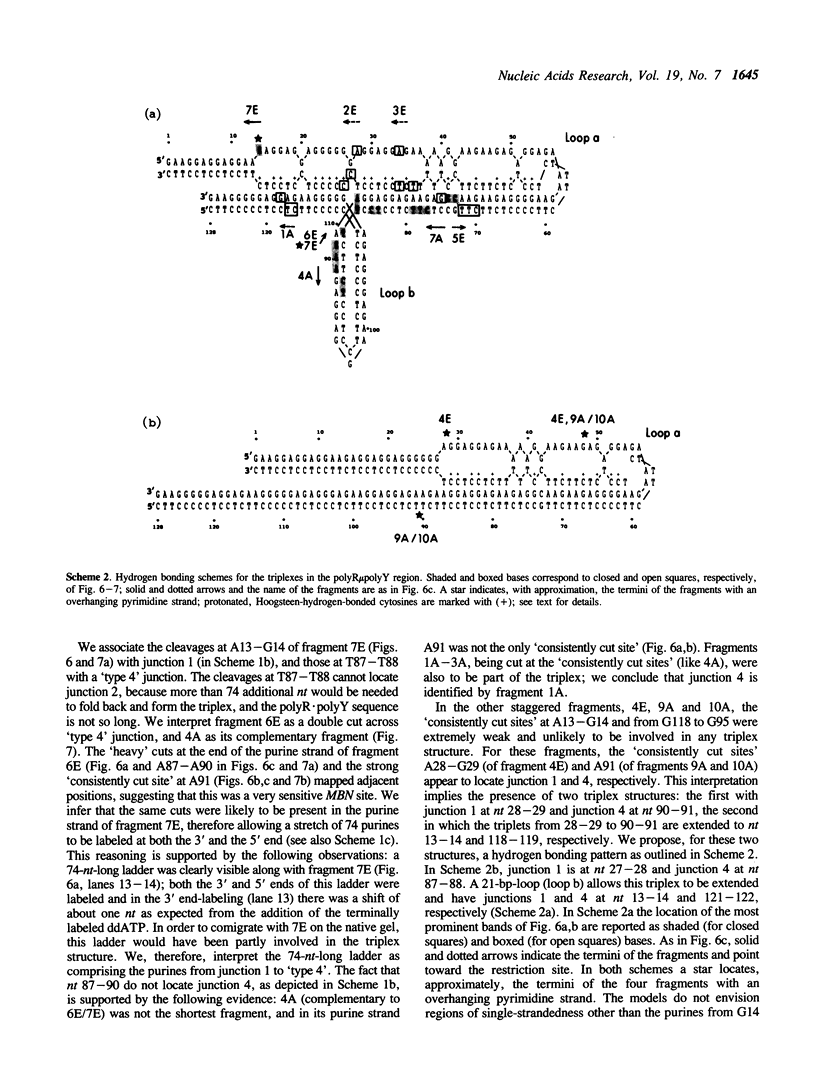
- Antao V. P., Gray D. M., Ratliff R. L. CD of six different conformational rearrangements of poly[d(A-G).d(C-T)] induced by low pH. Nucleic Acids Res. 1988 Jan 25;16(2):719–738. doi: 10.1093/nar/16.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J., Selsing E., Smith P. J. Models of triple-stranded polynucleotides with optimised stereochemistry. Nucleic Acids Res. 1976 Oct;3(10):2459–2470. doi: 10.1093/nar/3.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J. Structures for Poly(U)-poly(A)-poly(U)triple stranded polynucleotides. Nat New Biol. 1973 Jul 25;244(134):99–101. doi: 10.1038/newbio244099a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Bond P. J. Triple-stranded polynucleotide helix containing only purine bases. Science. 1973 Jul 6;181(4094):68–69. doi: 10.1126/science.181.4094.68. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Baumann U., Frank R., Blöcker H. Conformational analysis of hairpin oligodeoxyribonucleotides by a single-strand-specific nuclease. Eur J Biochem. 1986 Dec 1;161(2):409–413. doi: 10.1111/j.1432-1033.1986.tb10460.x. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Christophe D., Cabrer B., Bacolla A., Targovnik H., Pohl V., Vassart G. An unusually long poly(purine)-poly(pyrimidine) sequence is located upstream from the human thyroglobulin gene. Nucleic Acids Res. 1985 Jul 25;13(14):5127–5144. doi: 10.1093/nar/13.14.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobianchi F., Wilson S. H. Enzymes for modifying and labeling DNA and RNA. Methods Enzymol. 1987;152:94–110. doi: 10.1016/0076-6879(87)52013-4. [DOI] [PubMed] [Google Scholar]
- Côté J., Renaud J., Ruiz-Carrillo A. Recognition of (dG)n.(dC)n sequences by endonuclease G. Characterization of the calf thymus nuclease. J Biol Chem. 1989 Feb 25;264(6):3301–3310. [PubMed] [Google Scholar]
- Fitch D. H., Mainone C., Goodman M., Slightom J. L. Molecular history of gene conversions in the primate fetal gamma-globin genes. Nucleotide sequences from the common gibbon, Hylobates lar. J Biol Chem. 1990 Jan 15;265(2):781–793. [PubMed] [Google Scholar]
- Glanville N., Durnam D. M., Palmiter R. D. Structure of mouse metallothionein-I gene and its mRNA. Nature. 1981 Jul 16;292(5820):267–269. doi: 10.1038/292267a0. [DOI] [PubMed] [Google Scholar]
- Greene J. R., Guarente L. Subcloning. Methods Enzymol. 1987;152:512–522. doi: 10.1016/0076-6879(87)52058-4. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Klysik J., Wells R. D. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J Biol Chem. 1988 May 25;263(15):7386–7396. [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. II. Effect of base composition and noncentral interruptions on formation and stability. J Biol Chem. 1989 Apr 5;264(10):5950–5956. [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johnson A. C., Jinno Y., Merlino G. T. Modulation of epidermal growth factor receptor proto-oncogene transcription by a promoter site sensitive to S1 nuclease. Mol Cell Biol. 1988 Oct;8(10):4174–4184. doi: 10.1128/mcb.8.10.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Kohwi Y. Cationic metal-specific structures adopted by the poly(dG) region and the direct repeats in the chicken adult beta A globin gene promoter. Nucleic Acids Res. 1989 Jun 26;17(12):4493–4502. doi: 10.1093/nar/17.12.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker W. D., Kowalski D. Gene-sized pieces produced by digestion of linear duplex DNA with mung bean nuclease. Biochemistry. 1978 Aug 8;17(16):3236–3243. doi: 10.1021/bi00609a010. [DOI] [PubMed] [Google Scholar]
- LIPSETT M. N. COMPLEX FORMATION BETWEEN POLYCYTIDYLIC ACID AND GUANINE OLIGONUCLEOTIDES. J Biol Chem. 1964 Apr;239:1256–1260. [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtler A., Stover M. L., Angilly J., Kream B., Rowe D. W. Isolation and characterization of the rat alpha 1(I) collagen promoter. Regulation by 1,25-dihydroxyvitamin D. J Biol Chem. 1989 Feb 25;264(6):3072–3077. [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structure of (dG)n.(dC)n under superhelical stress and acid pH. J Biomol Struct Dyn. 1987 Oct;5(2):275–282. doi: 10.1080/07391102.1987.10506393. [DOI] [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Margot J. B., Demers G. W., Hardison R. C. Complete nucleotide sequence of the rabbit beta-like globin gene cluster. Analysis of intergenic sequences and comparison with the human beta-like globin gene cluster. J Mol Biol. 1989 Jan 5;205(1):15–40. doi: 10.1016/0022-2836(89)90362-8. [DOI] [PubMed] [Google Scholar]
- Miles H. T., Frazier J. Infrared study of helix strandedness in the poly A-poly U system. Biochem Biophys Res Commun. 1964;14:21–28. doi: 10.1016/0006-291x(63)90204-3. [DOI] [PubMed] [Google Scholar]
- Miller H. Practical aspects of preparing phage and plasmid DNA: growth, maintenance, and storage of bacteria and bacteriophage. Methods Enzymol. 1987;152:145–170. doi: 10.1016/0076-6879(87)52016-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Sobell H. M. A molecular model for gene repression. Proc Natl Acad Sci U S A. 1966 May;55(5):1201–1205. doi: 10.1073/pnas.55.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Shapiro B., Lipkin L. E., Maizel J. V., Jr DNAase I hypersensitive sites may be correlated with genomic regions of large structural variation. J Mol Biol. 1984 Aug 25;177(4):591–607. doi: 10.1016/0022-2836(84)90039-1. [DOI] [PubMed] [Google Scholar]
- RICH A. Formation of two- and three-stranded helical molecules by polyinosinic acid and polyadenylic acid. Nature. 1958 Feb 22;181(4608):521–525. doi: 10.1038/181521a0. [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 1989 Sep 19;28(19):7859–7870. doi: 10.1021/bi00445a048. [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989 Jun 22;339(6226):637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- Sheardy R. D., Winkle S. A. Temperature-dependent CD and NMR studies on a synthetic oligonucleotide containing a B-Z junction at high salt. Biochemistry. 1989 Jan 24;28(2):720–725. doi: 10.1021/bi00428a046. [DOI] [PubMed] [Google Scholar]
- Shehee W. R., Loeb D. D., Adey N. B., Burton F. H., Casavant N. C., Cole P., Davies C. J., McGraw R. A., Schichman S. A., Severynse D. M. Nucleotide sequence of the BALB/c mouse beta-globin complex. J Mol Biol. 1989 Jan 5;205(1):41–62. doi: 10.1016/0022-2836(89)90363-x. [DOI] [PubMed] [Google Scholar]
- Todd Miles H., Frazier J. A strand disproportionation reaction in a helical polynucleotide system. Biochem Biophys Res Commun. 1964;14:129–136. doi: 10.1016/0006-291x(64)90242-6. [DOI] [PubMed] [Google Scholar]
- Usdin K., Furano A. V. The structure of the guanine-rich polypurine:polypyrimidine sequence at the right end of the rat L1 (LINE) element. J Biol Chem. 1989 Sep 15;264(26):15681–15687. [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Weinreb A., Collier D. A., Birshtein B. K., Wells R. D. Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J Biol Chem. 1990 Jan 25;265(3):1352–1359. [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. Methods Enzymol. 1987;154:329–350. doi: 10.1016/0076-6879(87)54083-6. [DOI] [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]