Abstract
The precise splicing outcome of a transcribed gene is controlled by complex interactions between cis regulatory splicing signals and trans-acting regulators. In higher eukaryotes, alternative splicing is a prevalent mechanism for generating transcriptome and proteome diversity. Alternative splicing can modulate gene function, affect organismal phenotype and cause disease. Common genetic variation that affects splicing regulation can lead to differences in alternative splicing between human individuals and consequently impact expression level or protein function. In several well-documented examples, such natural variation of alternative splicing has indeed been shown to influence disease susceptibility and drug response. With new microarray- and sequencing-based genomic technologies that can analyze eukaryotic transcriptomes at the exon- or nucleotide-level, it has become possible to globally compare the alternative splicing profiles across human individuals in any tissue or cell type of interest. Recent large-scale transcriptome studies using high-density splicing-sensitive microarray and deep RNA sequencing (RNA-Seq) have revealed widespread genetic variation of alternative splicing in humans. In the future, an extensive catalogue of alternative splicing variation in human populations will help elucidate the molecular underpinnings of complex traits and human diseases, and shed light on the mechanisms of splicing regulation in human cells.
Introduction
Splicing of precursor mRNA (pre-mRNA) is an essential step of eukaryotic gene expression. During splicing, introns are removed from the pre-mRNA and exons are ligated to produce the mature mRNA product. This process is tightly regulated by cis elements within exons and surrounding introns as well as trans acting factors that bind to these cis elements 1. In alternative splicing (AS), this basic process is variable, so that multiple mRNA and protein isoforms can arise from a single gene 2. Alternative splicing of a single gene can generate a variety of mRNA and protein products, each with distinct properties in stability, subcellular localization and function 3. Alternative splicing can be modulated by variation both in the cis genomic splicing signals and in the cellular pathways that regulate splicing 2. In humans, the frequency of alternative splicing has been the subject of scrutiny. During the past 15 years, increasingly powerful technologies have been developed that can detect alternatively spliced transcripts at the global level. As the technology advanced so did estimates of the frequency of alternative splicing in humans 2, 4-6. The most recent estimate by high-throughput RNA sequencing (RNA-Seq) is that more than 90% of multi-exon genes in the human genome are alternatively spliced 5, 6, revealing the extent to which alternative splicing expands the regulatory and functional complexity of higher eukaryotes.
Alternative splicing is not only important for normal cellular functions but also frequently is involved in disease pathogenesis 7. Disrupting the normal splicing pattern can cause disease; and sometimes even a modest shift in the relative proportions of mRNA isoforms from a single gene can be pathogenic 7, 8. The majority of disease-causing splicing mutations affect critical splicing regulatory signals in cis (e.g., mutating consensus splice site sequences at exon-intron boundaries or splicing enhancer/silencer elements within exons or introns) 9. Nevertheless, disease-causing splicing mutations can also act in trans, by disrupting the expression or RNA binding activity of splicing regulators. While cis mutations affect splicing of a single transcript, trans mutations can compromise regulated splicing of many downstream gene targets simultaneously. As such, trans splicing mutations are known to cause a broad spectrum of diseases, such as neurodegenerative disease, muscular dystrophy, heart failure, and cancer 7. The classic example is myotonic dystrophy type 1 (DM1), in which the trinucleotide repeat CUG is expanded in the 3’ UTR of the gene DMPK. This expanded trinucleotide repeat sequesters proteins in the muscleblind family of splicing regulators and affects many muscleblind-dependent splicing events 10.
A wealth of information has demonstrated that pathogenic mutations can disrupt splicing in patient tissues; however less well explored is how complex traits or disease susceptibility can be influenced by normal genetic variation of alternative splicing. In recent years, thanks to new technologies for exon- or nucleotide-level profiling of eukaryotic transcriptomes, growing evidence reveals widespread natural variation of alternative splicing in humans 11. A number of studies have sought to identify splicing variation among people that correlates with single nucleotide polymorphisms (SNPs) in surrounding genomic regions 12-23. In this review, we describe molecular mechanisms that underlie genetic variation of alternative splicing. We illustrate genomic tools for global analysis of alternatively spliced transcripts, and summarize recent studies using these tools to survey alternative splicing differences among human individuals. We discuss the functional impact and disease association of alternative splicing variation. Finally, we discuss how a comprehensive catalogue of splicing variation in human populations will provide important insights into the mechanisms of alternative splicing regulation in human cells.
Mechanisms of Alternative Splicing Regulation
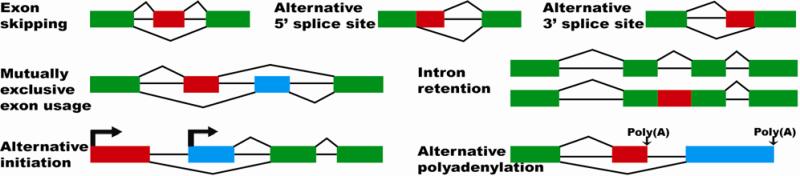
There are five basic types of alternative splicing (Figure 1): exon skipping, alternative 5’ (donor) splice sites, alternative 3’ (acceptor) splice sites, mutually exclusive exon usage, and intron retention. In addition, alternative initiation and alternative polyadenylation provide two other common mechanisms for generating various transcript isoforms. Different types of alternative splicing and transcript isoform variation can occur in a combinatorial manner, sometimes yielding an enormous number of distinct mRNA and protein isoforms from a single gene 24. A famous example is the Drosophila gene Dscam, which produces 38,016 distinct isoforms through alternative splicing of several exon clusters, each containing a large number of exons with mutually exclusive usage 25.
Figure 1.
Basic types of alternative splicing and transcript isoform variation.
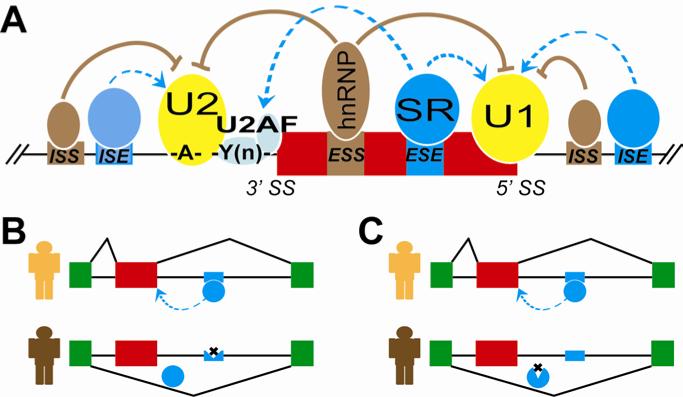
Alternative exon selection and splice site choice are controlled by an intricate regulatory network involving cis splicing elements and trans splicing regulators (Figure 2A) 1. The most essential core splicing elements include the 5’ splice site (5’ ss or donor site) and the 3’ splice site (3’ ss or acceptor site), which define the exon-intron boundary; also important are the branch site and polypyrimidine tract, which lie upstream of the 3’ splice site. These core cis splicing signals are recognized by the spliceosome, a large complex of protein and RNA subunits that are ubiquitously expressed and assemble on the pre-mRNA during splicing. In addition to core cis splicing signals, other auxiliary cis elements in exons and flanking introns can promote or inhibit exon splicing 1. The locations of these elements and their effects on exon splicing categorize these elements as exonic splicing enhancer (ESE), intronic splicing enhancer (ISE), exonic splicing silencer (ESS), or intronic splicing silencer (ISS). Such auxiliary elements are recognized by sequence-specific RNA-binding proteins, collectively known as trans-acting splicing regulators. These regulators determine whether an exon is selected by the spliceosome. For example, SR proteins typically promote exon selection by binding to exonic splicing enhancer elements; conversely, the hnRNP proteins typically inhibit exon selection by binding to exonic splicing silencer elements 26 (Figure 2A). The expression of many splicing regulators is restricted to specific tissues and cell types, thereby controlling networks of tissue-specific alternative splicing events. For example, the splicing regulators Nova-1 and Nova-2 are specifically expressed in the brain and regulate the production of neuronal-specific alternatively spliced transcripts 27, 28. In another example, the epithelial-specific splicing regulators ESRP1 and ESRP2 are transcriptionally repressed during the epithelial-to-mesenchymal transition (EMT) 29, 30. This event flips the switch off a genome-wide epithelial splicing network 29, 30. In addition alternative splicing can be regulated by other RNA and epigenetic features, such as RNA secondary structure 31, RNA polymerase II elongation rate 32, chromatin organization and histone modifications 33.
Figure 2.
Mechanisms of alternative splicing regulation and variation in humans. (A) Alternative splicing is controlled by an intricate regulatory network involving cis splicing elements and trans splicing regulators. The essential core splicing signals include the 5’ splice site (5’ SS), 3’ splice site (3’ SS), branch site (A) and polypyrimidine tract (Y(n)). Other auxiliary cis elements in exons and flanking introns include the exonic splicing enhancer (ESE), intronic splicing enhancer (ISE), exonic splicing silencer (ESS), and intronic splicing silencer (ISS). These auxiliary elements are recognized by transacting splicing regulators that positively or negatively regulate exon inclusion. Positive trans regulators are depicted in blue and negative trans regulators are depicted in brown. (B) A cis polymorphism disrupts an intronic splicing enhancer element and abolishes the interaction between the cis element and the trans regulator, resulting in the switch from the exon inclusion isoform to the exon skipping isoform. (C) A trans polymorphism alters the RNA binding activity of the trans splicing regulator and abolishes the interaction between the cis element and the trans regulator, resulting in the switch from the exon inclusion isoform to the exon skipping isoform.
Since the splicing of pre-mRNA is determined by interactions between cis elements and trans regulators, splicing outcomes can differ among human individuals due to genetic variation that alters such interactions in cis or in trans. In a hypothetical example (illustrated in Figure 2B-C), an exon is only included when it contains an intronic splicing enhancer element that is recognized by a splicing regulatory protein. Here, if the enhancer element is disrupted by a cis polymorphism, then it is not recognized by the splicing regulatory protein: without the protein bound to the enhancer, the exon is skipped (Figure 2B). A similar outcome could arise from a trans polymorphism in which the splicing regulatory protein is mutated in a way that disrupts RNA binding or expression (Figure 2C). Both scenarios could cause genetic variation to affect alternative splicing in human populations. It should be noted, however, that while the effect of a cis polymorphism (Figure 2B) is expected to be local (restricted to a specific adjacent exon), the effect of a trans polymorphism (Figure 2C) might be global, because each splicing regulator may control hundreds to thousands of alternative splicing events in the transcriptome 26.
Genomic tools for analysis of alternative splicing
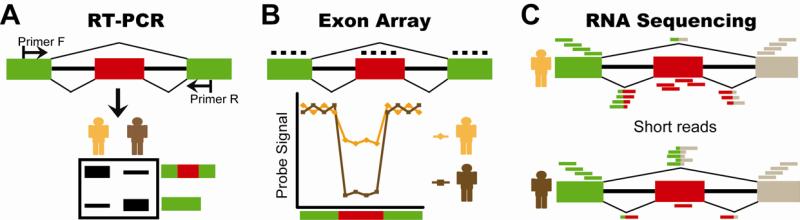
Alternative splicing plays critical roles in development, tissue differentiation, and disease, and so has been a longstanding subject of study. A variety of molecular and genomic tools have been developed to analyze alternative splicing. One widely used molecular approach is reverse transcription PCR (RTPCR) 2. With this method, any alternatively spliced exon(s) of interest can be monitored using a pair of forward and reverse PCR primers that hybridize to flanking exons (Figure 3A). After the RT-PCR reaction, electrophoresis can separate and make visible multiple PCR products of varying sizes, which correspond to distinct mRNA isoforms. The intensities of the different RT-PCR bands can be quantified to estimate the relative proportions of distinct mRNA isoforms 34. Combined with the use of radioactively or fluorescently labeled PCR primers, this approach is highly sensitive and accurately reflects the splicing levels of individual exons. It can also analyze other types of alternative splicing events such as alternative 5’ and 3’ splice sites. The drawback of this approach, however, is that it is relatively labor intensive and time consuming, and cannot be conducted on a genomic scale.
Figure 3.
Molecular and genomic tools for analysis of alternative splicing. (A) RT-PCR. For any alternatively spliced exon of interest, a pair of forward and reverse PCR primers can be designed to target the flanking exons (top panel). After the RT-PCR reaction, PCR products of varying sizes corresponding to distinct mRNA isoforms can be separated and visualized by electrophoresis (bottom panel). The intensities of RT-PCR bands can be quantified to estimate the relative proportions of distinct mRNA isoforms. (B) Exon array. Splicing-sensitive exon arrays use oligonucleotide probes that target specific exons or exon-exon junctions to analyze alternative splicing (top panel). Some platforms only contain probes targeting exon sequences, while others include probes for both exons and exon-exon junctions. The fluorescent intensities of individual probes can be used to infer the splicing activities of alternatively spliced exons (bottom panel). (C) RNA sequencing (“RNA-Seq”). By mapping RNA-Seq reads to exons and exon-exon junctions, one can annotate exon-intron structures and estimate the splicing levels of individual exons. In this diagram, the RNA-Seq data of the individual depicted in the top panel indicates that the alternatively spliced exon is predominantly included. By contrast, the RNA-Seq data of the individual depicted in the bottom panel indicates that the alternatively spliced exon is predominantly skipped.
Alternative splicing was first studied at the genomic level when full-length cDNAs and expressed sequence tags (ESTs) were sequenced on a large scale 2. In this first wave of genome-level investigation, exon-intron structures and alternative splicing events could be identified by aligning cDNA/EST sequences to the reference genome. Despite the utility of this approach, cDNA and EST sequencing have a limited throughput. Currently, there are < 7 million cDNA/EST sequences for human genes in the NCBI UniGene database (http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=9606). It is important to note that these sequences are aggregated over a wide range of tissues, developmental states and diseases. For any given alternative splicing event, the number of EST sequences supporting distinct transcript isoforms in a particular tissue or cell type is usually extremely limited. Thus, cDNA/EST sequencing can depict only a crude picture of alternative splicing in the human transcriptome, but cannot quantify isoform abundance in individual samples. Thus, new genomic technologies are needed for analyzing alternative splicing in any RNA sample of interest in a global and quantitative manner. Over the past few years, two powerful genomic approaches were developed for this purpose: high-density splicing-sensitive microarray 35 and ultra-deep RNA sequencing 36.
High-density splicing-sensitive microarray
Splicing-sensitive microarrays target specific exons or exon-exon junctions with oligonucleotide probes (Figure 3B). In the results of a microarray experiment, the fluorescent intensities of individual probes reflect the usage of alternatively spliced exons. This microarray-based approach was pioneered by Clark and colleagues for a genome-wide analysis of mRNA splicing in Saccharomyces cerevisiae 37. Other research groups and companies subsequently developed microarray platforms for analyzing alternative splicing in a variety of organisms including human, mouse and Drosophila 4, 38-41. Some of these designs only contain probes targeting exon sequences, while others include probes for both exons and exon-exon junctions. For example, the commercially available Affymetrix human exon junction array (HJAY array) has 8 probes per probeset for 315,137 exons and 260,488 exon-exon junctions in human genes; this covers all mRNA/EST-supported exons and alternative splicing events 29, 42.
Ultra-deep RNA sequencing
RNA sequencing (“RNA-Seq”) has emerged as a powerful technology for transcriptome analysis 36. Currently, a single lane of RNA-Seq run on the Illumina HiSeq 2000 sequencer can produce up to 100 million sequence reads. When tens to hundreds of millions of RNA-Seq reads are mapped to the genome, exons, and exon-exon junctions, one can annotate exonintron structures and estimate the relative isoform abundance of individual alternative splicing events 5, 6, 43 (Figure 3C).
Currently, the strengths of high-density splicing-sensitive microarrays and ultra-deep RNA sequencing in alternative splicing analysis complement each other. High-density splicing sensitive microarrays are a cost-effective way to assay exons and alternative splicing events that are known. Coupled with appropriate analysis algorithms, microarray platforms can detect changes in alternative splicing patterns at a low false positive rate, as demonstrated in numerous studies 29, 42, 44. Unlike microarray analysis, RNA-Seq does not require prior knowledge of gene structures and splicing events and has a nucleotide-level resolution. Once RNA-Seq reaches sufficient coverage for an exon, it can accurately estimate the exon's inclusion levels in mature mRNA transcripts 6, 43, 45. Nevertheless, to obtain quantitative exon-level measurements over the entire transcriptome, especially for less abundant transcripts, RNA-Seq must go very deep, which requires substantial experimental cost 36. We anticipate that, in the near term, microarray- and sequencing-based approaches for alternative splicing analysis will co-exist. However, because the cost of high-throughput RNA sequencing has been dropping precipitously, we expect RNA-Seq will eventually supplant microarrays as the standard tool for transcriptome analysis. Indeed, since the advent of RNA-Seq a number of studies have adopted this technology to characterize multiple layers of regulatory variation in the human transcriptome (reviewed by 46).
It should be noted that genetic polymorphisms could potentially confound the splicing analysis of microarray- and sequencing-based transcriptome profiles. The oligonucleotide probes on splicing-sensitive microarrays are designed using the reference genome sequence. In microarray hybridization, the fluorescent intensity of any given probe depends on the abundance of its target mRNA transcript as well as the binding affinity of the probe to its target. In the absence of any effect on expression and splicing, a genetic polymorphism can decrease probe intensity simply by reducing the binding affinity of a microarray probe to its target, creating a spurious signal for altered expression or splicing. An analysis of Affymetrix exon array data of 57 individuals showed that this confounding issue is a serious source of false positives for detecting genetic variation of alternative splicing 47. A practical solution is to remove all microarray probes overlapping with polymorphic sites from analysis 47. This problem is less severe for RNA-Seq, because RNA-Seq mapping protocols can tolerate a small number of mismatches 5. However, genetic polymorphisms can still complicate the mapping of RNA-Seq reads 48, especially larger-scale polymorphisms such as insertions or deletions. In the future it will be useful to develop computational procedures that can reliably map RNA-Seq reads to polymorphic sites in the genome and transcriptome.
Genome-wide survey of alternative splicing variation in human populations
The key to identifying genetically controlled variation in alternative splicing is to associate alternative splicing differences in people with particular genetic polymorphisms. Differences in alternative splicing between individuals/alleles could be manifested either as all-or-none switches between competing mRNA isoforms, or as shifts in the relative proportions of multiple mRNA isoforms of a single gene.
The first large-scale survey of SNP-associated alternative splicing was conducted by Nembaware and colleagues through the combined analysis of the dbSNP and dbEST databases 49. Using ESTs that map to both SNPs and isoform-specific exon-exon junctions of alternatively spliced transcripts, the authors estimated that 21% of the alternatively spliced isoforms detected from EST data arose from allele-specific splicing, with a conservative lower bound estimate of 6% if one considered only SNPs that caused all-or-none switches between isoforms. This study provides the first genomic evidence that an appreciable percentage of alternative splicing events reported in human genes might be attributed to genetic polymorphisms. In another study, using RT-PCR Hull and colleagues investigated the splicing patterns of 250 exons in 22 individuals of European ancestry 12. Of the 250 exons, six showed considerable splicing differences among individuals. Moreover, for each of the six exons, the splicing differences observed among individuals correlated well with the genotypes of a particular cis SNP in the neighboring genomic region, suggesting alternative splicing was being genetically controlled 12.
Recently, genome-wide patterns of alternative splicing variation in human populations have been investigated by several groups using the HapMap lymphoblastoid cell lines (LCLs) as the model system 13, 14, 16, 17, 20, 21, 50. Across the entire genome, the SNPs in these HapMap LCLs were characterized extensively by the international HapMap project 51, 52, making it convenient to test associations of splicing patterns with genetic polymorphisms. By combining Affymetrix exon 1.0 array analysis of two unrelated European individuals and RT-PCR analysis of splicing patterns within a three-generation family, Kwan and colleagues demonstrated the Mendelian inheritance of alternative splicing patterns of three genes (OAS1, CAST and CRTAP) 14. Subsequently, an exon 1.0 array study by Kwan et al. extended the analysis to HapMap LCLs from 57 unrelated individuals of European ancestry 13. They identified 177 genes whose relative transcript isoform proportions (owing to alternative splicing, alternative initiation and alternative polyadenylation) correlated strongly with surrounding SNPs. The same exon 1.0 array approach was also used to characterize splicing variation among 176 HapMap LCLs of European and African ancestry 17, 50.
Such genetic studies of alternative splicing are facilitated by the advent of the high-throughput RNA sequencing technology. For example, Pickrell and colleagues generated 1.2 billion single-end RNASeq reads on LCLs of 69 Nigerian individuals from the HapMap project 21. By treating the isoform proportions of alternatively spliced genes as quantitative traits, they identified 187 genes with splicing patterns that correlated with neighboring SNPs, revealing putative splicing quantitative trait loci (sQTLs). In a similar study, 60 HapMap LCLs of European ancestry were analyzed by Montgomery and colleagues through paired-end RNA sequencing for discovery of putative sQTLs 20. Both RNA-Seq studies come with a caveat, however, because the sequencing depth was a modest ~20 million reads per individual, likely generating a high rate of false positives and false negatives in splicing analysis. Clearly, a true set of sQTLs must be confirmed by experimental validation and further replication. In another ultra-deep RNA-Seq analysis of two HapMap European individuals, the authors observed a significant enrichment of detected sQTLs within high-confidence eQTL target genes 16. This result suggests that alternative splicing variation among human individuals can regulate steady-state overall transcript levels, potentially through the effect on mRNA stability and degradation 16.
The genetic information on HapMap LCLs is readily available, making them the cell type of choice for most studies on transcriptome variation in human populations. Nevertheless, alternative splicing regulation can strongly depend on tissue and cell type 1, 26, thus a SNP that affects splicing in LCLs may not necessarily affect splicing in other tissues. Such tissue specificity of sQTLs was examined by Heinzen and colleagues, who combined genome-wide SNP arrays with the Affymetrix exon 1.0 arrays to compare genetic control of alternative splicing in 93 cortical brain samples and 80 peripheral blood mononuclear cell (PBMC) samples 19. Indeed, although 80 high-confidence sQTLs were identified, only 49% were shared between the two tissue types. In a similar study, LCLs and osteoblasts were compared by Kwan and colleagues, who estimated that 78% of sQTLs overlap in the two tissues 15. Both studies confirmed that genetic regulation of alternative splicing can be both tissue-specific and tissue-independent.
Although most studies to date have focused on cis-acting polymorphisms, multiple lines of evidence suggest that trans-acting polymorphic sites could broadly affect splicing regulation in human populations. A recent study on cis and trans regulation of gene expression variation indicates that the majority of polymorphic sites with influence on gene expression act in trans to target genes 53. In a variety of human diseases, genetic mutations that disrupt trans-acting splicing regulators can trigger disease pathogenesis or modify disease severity 7. Thus, it is entirely conceivable that common genetic variation of splicing regulators can act as modifiers of complex traits or diseases, by altering the splicing of downstream gene targets. Importantly, variation in a brain-specific splicing regulator FOX1 has been recently implicated in autism 54, 55. The discovery and functional characterization of trans-acting splicing polymorphisms is expected to accelerate in the future.
Functional impact and disease association of alternative splicing variation in human populations
High-throughput transcriptome studies are producing a fast-growing catalogue of splicing variation in human populations, but so far information on the functional impact of such splicing variation is limited. Currently, our ability to detect alternative splicing events greatly exceeds our ability to characterize the functions of alternatively spliced gene products. Nonetheless, for several genes, genetic variation of alternative splicing has well-established functions (Table 1). For example, in HMSD, an intronic SNP (rs9945924) causes pronounced skipping of exon 2 by weakening the strength of the 5’ splice site 22, 56. This HMSD exon-skipping isoform generates a novel minor histocompatibility antigen, which affects the immune response and could serve as a potential target for immunotherapy 56. In another example, an allele of ERAP2 (a gene involved in MHC class I antigen presentation) harbors an A-to-G SNP (rs2248374) at the canonical 5’ splice site of exon 10. This SNP activates a downstream cryptic splice site, subjecting the resultant transcript to nonsense mediated decay and significantly reducing the steady-state mRNA level of ERAP2 18, 57. Consequently, primary lymphocytes homozygous for the G allele express less MHC Class I at the B cell surface, suggesting this alternative splicing event affects MHC antigen presentation. Interestingly, genetic analyses of this SNP in six human populations revealed a strong signature of balancing selection 57, suggesting this alternative splicing polymorphism confers an unknown adaptive benefit. A similar example was found in OAS1, a gene important for the innate immune response to virus infection. Here, a G-to-A SNP (rs10774671) at the 3’ splice site of exon 7 abolishes splice site activity, resulting in the usage of an internal 3’ splice site and the production of a protein isoform with reduced enzymatic activity 58. This SNP also affects the response to interferon (IFN) therapy in hepatitis C patients in that HCV patients homozygous for the A allele do not respond to interferon and exhibit a higher degree of liver fibrosis 59. In SCN1A, a gene encoding a neuronal sodium-channel alpha subunit, an intronic SNP (rs3812718) was found that modulates the alternative splicing of exon 5 and influences the dose response to antiepileptic drugs 60.
Table 1.
Examples of alternative splicing variation in human populations and their functional or pathological consequences.
| Gene symbol | Effect of SNP on splicing | Functional or pathological consequence |
|---|---|---|
| HMSD | An intronic SNP (rs9945924) causes pronounced skipping of exon 2 by weakening the 5′ splice site. | Generates a novel minor histocompatibility antigen, which affects the immune response and could serve as a potential target for immunotherapy56. |
| ERAP2 | A SNP (rs2248374) residing within the canonical 5′ splice site of exon 10 triggers activation of a downstream cryptic splice site, leading to mRNA nonsense-mediated decay and a significant reduction in the steady-state mRNA level of ERAP218, 57. | Primary lymphocytes homozygous for the G allele express less MHC Class I at the B cell surface, suggesting that this alternative splicing event affects MHC antigen presentation18, 51. |
| OAS1 | A SNP (rs10774671) at the 3′ splice site of exon 7 abolishes splice site activity, resulting in the usage of an internal 3′ splice site. | Produces a protein isoform with reduced enzymatic activity58. Affects the response to interferon (IFN) therapy in hepatitis C patients59, and influences susceptibility to multiple sclerosis65. |
| SCN1A | An intronic SNP (rs3812718) modulates the alternative splicing of exon 5. | Influences the dose response to antiepileptic drugs60. |
| IRF5 | An intronic SNP (rs2004640) generates a consensus GT 5′ splice site of the alternative first exon 1B, producing an alternative transcript isoform initiated at exon 1B. | Associated with increased risk of systemic lupus erythematosus63, 64. |
| CD45 (PTPRC) | An exonic SNP (rs17612648) within exon 4 disrupts an exonic splicing silencer, leading to increased inclusion of exon 4. | Associated with multiple sclerosis61. |
| LDLR | An exonic SNP (rs688) promotes skipping of its exon 12. | Associated with total and LDL-cholesterol levels in females especially in pre-menopausal women67. |
An alternative to gene-specific functional experiments is to examine whether SNPs that cause splicing differences can be linked to phenotypic traits or diseases. Common genetic variants that affect the alternative splicing of IRF5, OAS1, CTLA4, and PTPRC (also known as CD45) were linked to several autoimmune diseases 61-65. Genetic polymorphisms that affect the alternative splicing of NPSR1 (also known as GPRA) were implicated in asthma 66. Another example was found in the low-density lipoprotein receptor (LDLR), in which a SNP (rs688) promotes skipping of exon 12 and is strongly associated with total and LDL-cholesterol levels in females especially in pre-menopausal women 67. Similarly, in cortical brain and PBMC samples, the 80 identified associations between SNPs and splicing were systematically compared to 41 published genome-wide association studies (GWAS) for 50 different traits 19. Here, up to 13 SNP-splicing associations appeared to be responsible for previously reported GWAS signals of human traits (out of a total of 84 reported). Likewise, splicing QTL data of human osteoblasts were compared with a GWAS of bone mineral density, showing ~20% of GWAS signals may be attributed to alternative splicing variation 15. Such studies illustrate that intersecting sQTLs with GWAS signals provides an effective means to identify the causal regulatory effects of GWAS hits.
Genetic variation of alternative splicing and the splicing code
Despite the importance of alternative splicing, the rules that govern splicing regulation (i.e. the “splicing code”) in individual tissues and cell types remain poorly understood. Many cis splicing elements are degenerate, and their effects on splicing usually depend on the surrounding sequence context as well as the abundance and activity of trans acting regulators 1. Although experimental and computational approaches have made considerable progress in elucidating the splicing code of mammalian cells 1, 68, 69, currently it is still challenging to predict the precise alternative splicing pattern of a gene from the primary genomic sequence.
Natural variation of alternative splicing provides a rich resource for dissecting the mechanisms of splicing regulation. When a given cis SNP can be linked to the splicing pattern of a particular exon across human populations, this suggests that a critical cis splicing regulatory element is created or disrupted by that genetic variant. Intuitively, this is analogous to conventional minigene studies of splicing regulation, in which the role of a putative splicing regulatory element can be assessed by mutational analysis of the element in an artificial minigene splicing reporter construct 70, 71. Here, the genetic variation of alternative splicing that exists in natural human populations can be viewed as the results of “whole-genome mutagenesis experiments” during human evolution. By globally correlating alternative splicing variation in the human transcriptome to genetic polymorphisms in the human genome, we can gain significant insights into the genomic signals of splicing regulation and identify novel regulatory elements.
It must be emphasized however, that specific SNPs found to be associated with alternative splicing are often not causal but rather are in linkage disequilibrium with the causal polymorphisms. Indeed, most of the significantly associated cis SNPs identified by sQTL analysis are far from the exons of interest 13; in contrast, cis splicing elements are typically in close proximity to the exons they regulate 1, 69. Therefore, after an sQTL is identified, the exon and flanking intronic regions must be resequenced to identify potential causal SNPs that might regulate splicing 12, 18. The causal effect of SNPs identified by such resequencing can be confirmed by minigene experiments, in which the genomic sequences corresponding to distinct alleles are cloned into a splicing reporter construct and the splicing efficiencies are compared. This approach has been used in several studies to pinpoint the causal SNPs responsible for sQTL signals as well as the affected cis splicing regulatory elements 12, 18.
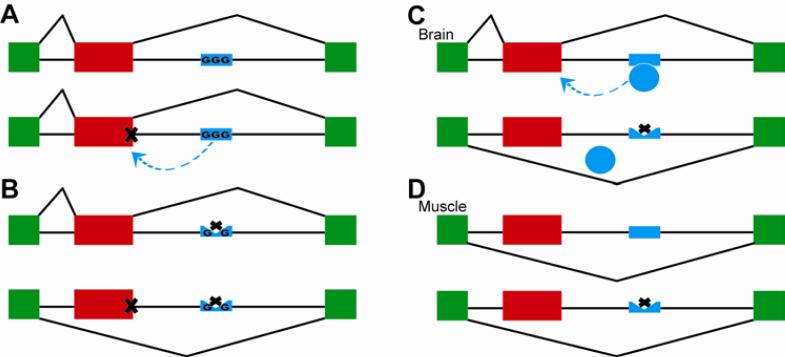
Genetic variation of alternative splicing can also reveal higher-order interactions among distinct classes of cis splicing regulatory elements. This was aptly demonstrated by a recent study on 5’ splice site SNPs and their context-dependent splicing effects 22. In this work, Lu et al. analyzed the HapMap LCLs of 7 individuals using RT-PCR to examine the splicing patterns of 129 exons containing 5’ splice site SNPs. Surprisingly, despite the critical role of the 5’ splice site in exon recognition, only a small fraction of the tested 5’ splice site SNPs affected splicing patterns. By comparing exons affected by 5’ splice site SNPs to exons that were unaffected, Lu et al. discovered that the effects of 5’ splice site SNPs were buffered by adjacent sequence signals that promote exon recognition. For example, the GGG motif was the most enriched trinucleotide sequence downstream of exons unaffected by 5’ splice site SNPs, consistent with previous reports on the GGG motif as an intronic splicing enhancer that promotes the recognition of weak 5’ splice sites 72. Using a similar approach, Fu and colleagues showed that the effect of single nucleotide mutations at the 5’ most nucleotide of an exon can be buffered by a strong polypyrimidine tract upstream of the 3’ splice site 73. These results indicate that the effect of a given SNP on splicing can be buffered by surrounding sequence elements (see Figure 4A-B for a schematic illustration). Consequently, the splicing of exons lacking such compensatory mechanisms would be more susceptible to genomic mutations.
Figure 4.
Genetic variation of alternative splicing provides insights into the mechanisms of splicing regulation. (A-B) The splicing effect of a SNP can be buffered by other splicing regulatory elements in the surrounding region. (A) The intronic poly-G runs act as splicing enhancer elements when located downstream of weak 5’ splice sites. A SNP that significantly weakens the 5’ splice site is buffered by the downstream poly-G run and so it does not affect inclusion of the exon. (B) In the absence of the downstream intronic poly-G run, the same 5’ splice site SNP results in the switch from the exon inclusion isoform to the exon skipping isoform. (C-D) The tissue specificity of splicing QTLs suggests tissue-specific cis splicing elements. A cis intronic SNP results in the switch from the exon inclusion isoform to the exon skipping isoform in the brain (C), while in the muscle both alleles undergo complete skipping of the exon (D). These patterns suggest that the SNP disrupts a brain-specific cis splicing enhancer element.
Conclusion
Inheritable differences in splicing add an important layer to the molecular underpinnings of complex traits and human diseases. Although the current knowledge about genetic variation of alternative splicing in humans is strongly biased towards a single cell type (lymphoblastoid cell line), the rapid improvement in the capacity and cost structure of high-throughput sequencing technologies will enable genome-scale analyses of alternative splicing variation in many other tissues in the near future. For example, the recently launched NIH Genotype-Tissue Expression Program aims to analyze genotype-transcriptome correlations in 30 to 50 tissues collected from ~160 deceased donors (https://commonfund.nih.gov/GTEx/overview.aspx). A comprehensive catalogue of expression and splicing QTLs across diverse tissues and cell types will facilitate a variety of important investigations. It will provide a powerful resource for human geneticists to infer the functional relevance of GWAS results, by mapping significant GWAS signals to eQTLs/sQTLs of disease-related tissues (for example GWAS signals of psychiatric diseases to eQTLs/sQTLs of brain tissues). Additionally, the tissue-specificity of splicing QTLs will inform the mechanisms of splicing regulation and reveal the creation or loss of tissue-specific cis splicing elements. For example, if a particular SNP decreases splicing of an adjacent exon in the brain but not in the muscle, then hypothetically, the SNP disrupts a splicing regulatory element that is brain specific (Figure 4C-D). Likewise, systematic analyses of trans splicing QTLs across multiple tissues should identify common genetic variation in master regulators of tissue-specific splicing that globally contributes to splicing variation of downstream target exons.
Another important future direction is to develop new computational tools that reliably identify pathogenic mutations that influence splicing. Many exonic and intronic mutations can disrupt splicing 9. For example, a systematic analysis of synonymous mutations within exon 12 of CFTR, the disease gene of cystic fibrosis, revealed that approximately one quarter of synonymous mutations would result in aberrant splicing and non-functional protein products 74. Such splicing mutations constitute a major class of human disease mutations. However, our current ability to predict whether a given genomic mutation would alter splicing is very limited. A number of computational tools are available for this purpose (reviewed by 75); however, these tools generally are inaccurate or designed to analyze SNPs in specific types of splicing signals (e.g., splice sites). To accurately predict the splicing effect of genomic variants, we need a comprehensive understanding of cis splicing regulatory elements and how these elements interact with each other as well as with trans splicing regulators. Recently, several studies demonstrated that advanced machine learning techniques can computationally predict tissue-specific alternative splicing patterns by integrating hundreds of genomic and RNA sequence features 69, 76. A major goal for the future is the development of computational approaches that accurately predict common and rare genetic variants that alter splicing. This will provide the tools human geneticists need to follow up on exome sequencing and whole genome sequencing studies of human diseases, and facilitate using alternative splicing variants as disease markers for personalized medicine.
Acknowledgements
We thank Peter Stoilov and Keyan Zhao for helpful comments on this manuscript. This work was supported by National Institutes of Health grant R01GM088342 and a junior faculty grant from the Edward Mallinckrodt Jr Foundation.
Contributor Information
Zhi-xiang Lu, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242.
Peng Jiang, Department of Internal Medicine, University of Iowa, Iowa City, IA 52242.
Yi Xing, Department of Internal Medicine and Department of Biomedical Engineering, University of Iowa, IA 52242, yi-xing@uiowa.edu.
References
- 1.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 3.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 10.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:R162–169. doi: 10.1093/hmg/ddl181. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 11.Graveley BR. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet. 2008;24:5–7. doi: 10.1016/j.tig.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 14.Kwan T, Benovoy D, Dias C, Gurd S, Serre D, Zuzan H, Clark TA, Schweitzer A, Staples MK, Wang H, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwan T, Grundberg E, Koka V, Ge B, Lam KC, Dias C, Kindmark A, Mallmin H, Ljunggren O, Rivadeneira F, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lalonde E, Ha KC, Wang Z, Bemmo A, Kleinman CL, Kwan T, Pastinen T, Majewski J. RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res. 2011;21:545–554. doi: 10.1101/gr.111211.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Duan S, Bleibel WK, Wisel SA, Huang RS, Wu X, He L, Clark TA, Chen TX, Schweitzer AC, et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum Genet. 2009;125:81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulombe-Huntington J, Lam KC, Dias C, Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS Genet. 2009;5:e1000766. doi: 10.1371/journal.pgen.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu ZX, Jiang P, Cai JJ, Xing Y. Context-dependent robustness to 5' splice site polymorphisms in human populations. Hum Mol Genet. 2011;20:1084–1096. doi: 10.1093/hmg/ddq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am J Hum Genet. 2006;78:291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 25.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. DrosophilaDscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 28.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. Embo J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 35.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 38.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto ML, Clark TA, Gee SL, Kang JA, Schweitzer AC, Wickrema A, Conboy JG. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 2009;113:3363–3370. doi: 10.1182/blood-2008-05-160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing Y, Stoilov P, Kapur K, Han A, Jiang H, Shen S, Black DL, Wong WH. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. Rna. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci U S A. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Benovoy D, Kwan T, Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008;36:4417–4423. doi: 10.1093/nar/gkn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nembaware V, Wolfe KH, Bettoni F, Kelso J, Seoighe C. Allele-specific transcript isoforms in human. FEBS Lett. 2004;577:233–238. doi: 10.1016/j.febslet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 50.Fraser HB, Xie X. Common polymorphic transcript variation in human disease. Genome Res. 2009;19:567–575. doi: 10.1101/gr.083477.108. [DOI] [PubMed] [Google Scholar]
- 51.International-HapMap-Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 52.International-HapMap-Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung VG, Nayak RR, Wang IX, Elwyn S, Cousins SM, Morley M, Spielman RS. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 56.Kawase T, Akatsuka Y, Torikai H, Morishima S, Oka A, Tsujimura A, Miyazaki M, Tsujimura K, Miyamura K, Ogawa S, et al. Alternative splicing due to an intronic SNP in HMSD generates a novel minor histocompatibility antigen. Blood. 2007;110:1055–1063. doi: 10.1182/blood-2007-02-075911. [DOI] [PubMed] [Google Scholar]
- 57.Andres AM, Dennis MY, Kretzschmar WW, Cannons JL, Lee-Lin SQ, Hurle B, Schwartzberg PL, Williamson SH, Bustamante CD, Nielsen R, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F. Variation in antiviral 2',5'-oligoadenylate synthetase (2'5'AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El Awady MK, Anany MA, Esmat G, Zayed N, Tabll AA, Helmy A, El Zayady AR, Abdalla MS, Sharada HM, El Raziky M, et al. Single nucleotide polymorphism at exon 7 splice acceptor site of OAS1 gene determines response of hepatitis C virus patients to interferon therapy. J Gastroenterol Hepatol. 2011;26:843–850. doi: 10.1111/j.1440-1746.2010.06605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinzen EL, Yoon W, Tate SK, Sen A, Wood NW, Sisodiya SM, Goldstein DB. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–883. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch KW, Weiss A. A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J Biol Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- 62.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 63.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Pons-Estel B, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 64.Kozyrev SV, Lewen S, Reddy PM, Pons-Estel B, Witte T, Junker P, Laustrup H, Gutierrez C, Suarez A, Francisca Gonzalez-Escribano M, et al. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis Rheum. 2007;56:1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- 65.Fedetz M, Matesanz F, Caro-Maldonado A, Fernandez O, Tamayo JA, Guerrero M, Delgado C, Lopez-Guerrero JA, Alcina A. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens. 2006;68:446–449. doi: 10.1111/j.1399-0039.2006.00694.x. [DOI] [PubMed] [Google Scholar]
- 66.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, Makela S, Rehn M, Pirskanen A, Rautanen A, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–304. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 67.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 70.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 71.Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–181. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 72.Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol. 2009;16:1094–1100. doi: 10.1038/nsmb.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu Y, Masuda A, Ito M, Shinmi J, Ohno K. AG-dependent 3'-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res. 2011;39:4396–4404. doi: 10.1093/nar/gkr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM. Prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat. 2008;29:1304–1313. doi: 10.1002/humu.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]