Abstract
Scientific research has shifted from studies conducted by single investigators to the creation of large consortia. Genetic epidemiologists, for example, now collaborate extensively for genome-wide association studies (GWAS). The effect has been a stream of confirmed disease-gene associations. However, effects on human subjects oversight, data-sharing, publication and authorship practices, research organization and productivity, and intellectual property remain to be examined. The aim of this analysis was to identify all research consortia that had published the results of a GWAS analysis since 2005, characterize them, determine which have publicly accessible guidelines for research practices, and summarize the policies in these guidelines. A review of the National Human Genome Research Institute’s Catalog of Published Genome-Wide Association Studies identified 55 GWAS consortia as of April 1, 2011. These consortia were comprised of individual investigators, research centers, studies, or other consortia and studied 48 different diseases or traits. Only 14 (25%) were found to have publicly accessible research guidelines on consortia websites. The available guidelines provide information on organization, governance, and research protocols; half address institutional review board approval. Details of publication, authorship, data-sharing, and intellectual property vary considerably. Wider access to consortia guidelines is needed to establish appropriate research standards with broad applicability to emerging forms of large-scale collaboration.
Keywords: ethics, research; genome-wide association study; molecular epidemiology
Scientific research has undergone a profound transformation in the last 10–15 years, with a shift from single-laboratory, investigator-initiated research to the creation of large research consortia organized to accomplish strategic goals (1) and to accelerate the translation of basic science discovery to public health benefit (2, 3). The new collaborative arrangements enabled by such consortia are important, both with respect to the scale of scientific output generated (4, 5) and with regard to promoting changes in collaborative research practices and underlying research norms (6). These rapidly changing norms are likely to be especially challenging for investigators trained in the period before such large-scale consortia became commonplace.
In genetic epidemiology, for example, scientists have begun cooperating to an unprecedented degree, often spurred on by the need to accrue sample sizes large enough to allow the identification of common genetic contributions to complex diseases using genome-wide association studies (GWAS) (7). This need for large sample sizes is directly attributable to the widespread realization that risk estimates from GWAS will be modest at best. The effect of such cooperation has been a remarkable stream of confirmed disease-gene associations (8, 9). However, other impacts of these new research arrangements, including their effects on human subjects oversight, data-sharing, publication and authorship practices, research organization and productivity, intellectual property, and possible return of results to study participants, remain to be examined. Comprehensively identifying and understanding such impacts will be important for ensuring the ongoing success of genetic consortia as well as for strengthening public confidence in current and future forms of large-scale collaborative research.
To begin to examine the impact of evolving collaborative research practices on underlying research norms, we conducted a systematic review of one class of emerging large-scale consortia, those that have published findings from a GWAS. Specifically, our aim was to identify all large-scale genomic research consortia that had published the results of at least 1 GWAS analysis since 2005, to characterize these consortia, to determine which of them have publicly accessible policies that govern consortium research practices, and to summarize those policies and procedures. The long-term objective of this work is to develop models and recommendations of research guidelines with broad applicability to facilitate emerging forms of large-scale collaboration.
MATERIALS AND METHODS
For this analysis, GWAS consortia were identified from publications listed in A Catalog of Published Genome-Wide Association Studies (10), compiled by the National Human Genome Research Institute (NHGRI). This catalog, which is continually updated, includes studies that attempt to assay at least 100,000 single nucleotide polymorphisms (11). Studies focused only on candidate genes are excluded. The catalog contains studies collected through PubMed literature searches (National Library of Medicine) that are conducted weekly, daily news and media reports distributed by the National Institutes of Health, and comparison of catalog listings with the GWAS literature in the HuGE Navigator database (12). As of April 1, 2011, a total of 845 GWAS publications were included in the NHGRI catalog. From these, 110 consortia that either explicitly participated in a study from the catalog or had their resources used in a catalog publication (e.g., providing control data) were identified. To enhance comparability of research policies and procedures, we focused on 69 of the 110 consortia that included at least 1 US-based member.
After reviewing all consortia descriptions, we developed a 3-part definition of a “GWAS consortium” in order to focus our analysis on a relatively homogeneous set of collaborative research endeavors. A GWAS consortium was defined as:
1) a team of individual investigators who are associated with a study or a research institution, or a group of studies and research institutions,
2) who assemble large data sets of genome-wide genotypic data and phenotypic information from existing resources, recruitment of new study participants, or both,
3) for the purpose of discovering associations between the genotypic variants and phenotypes, including diseases, risk factors, and other physical characteristics.
A total of 55 consortia met this definition, and 14 did not. Web Table 1, which is posted on the Journal’s website (http://aje.oxfordjournals.org/), lists the 14 consortia that did not meet all criteria, according to our review. Web Table 2 lists the 55 consortia that did meet all 3 criteria.
For the 55 GWAS consortia, we identified key consortia characteristics using Internet searches to locate consortia websites, researcher or institution-hosted websites, and/or references to the consortia in other studies. Using this information, we characterized each consortium by the type of consortium members identified (i.e., individual investigators who form or are invited to join a consortium; research centers, including universities, hospitals, nonprofit organizations, and for-profit corporations; studies, including a cohort or database of participants, or another GWAS study; or other consortia, including GWAS consortia that meet our analysis definition), the disease or trait studied, the date of the first scientific publication from the consortium in the NHGRI catalog, whether or not the consortium had a dedicated website, and whether consortium guidelines were publicly available on that website. As is shown in Web Table 3, the diseases studied in the consortia GWAS were categorized using the International Classification of Diseases, Tenth Revision (13). Risk factors related to those diseases and other traits are also listed in Web Table 3, as well as other traits not listed in the International Classification of Diseases, Tenth Revision.
Of the 55 consortia, 30 were found to have publicly available websites, and of these, 14 had accessible guidelines, at times referred to as “research protocols,” “data-sharing plans,” “consortium details,” and/or “usage policy.” We then undertook a more detailed analysis of these consortia, including determining the number of papers published by the consortia, the number of consortium members, and components of the available consortia guidelines. We searched the PubMed database to determine the approximate number of papers published by searching for the acronym of each consortium and also by searching for the full name of the consortium. Abstracts and, when necessary, articles were reviewed individually to ensure that the consortium was involved. Original research studies authored by the consortium as well as review papers were counted as papers published by the consortium. Consortia websites were also searched for a list of members, and the number of members was counted.
Finally, publicly available guidelines for these 14 consortia websites were characterized using the following categories: 1) organization and governance guidelines that describe the management structure, organizational hierarchy, committee organization, and/or decision-making protocols for the consortium; 2) research protocols that address how to standardize, analyze, and/or contribute data, that describe how to apply to use the consortium’s research resources, that stipulate laboratory technologies and techniques to be used, and/or that provide quality control procedures for consortium projects; 3) institutional review board (IRB) review of research data used by consortium members; and 4) publication and authorship guidelines that present a plan for how the consortium’s work is published, specify who can be included as authors, give rules for citing the consortium in research papers, and/or provide intellectual property guidelines that address the need to respect intellectual property and fairness in collaboration.
RESULTS
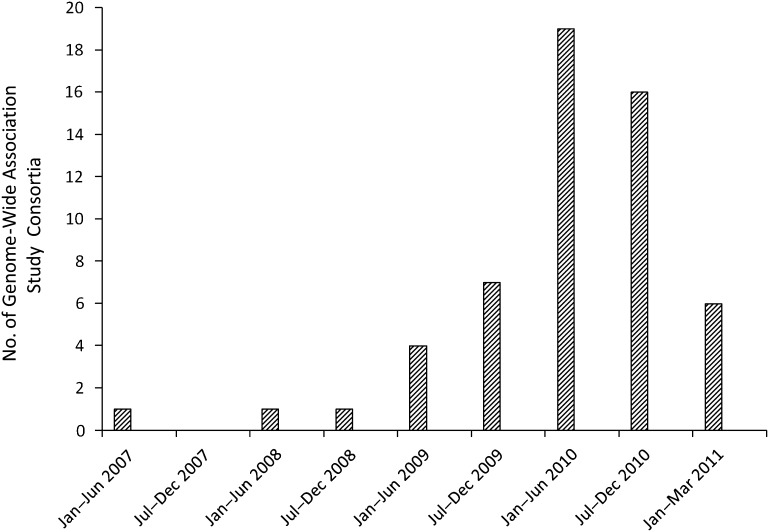
Among the 55 GWAS consortia that met our analysis definition, 23 have members that are individual investigators (42%), 15 have members that are research centers (27%), 16 have members that are studies (29%), and 1 has members that are other consortia (2%). Of the 48 diseases, risk factors, and/or traits studied by these consortia, the 3 most common are related to diseases of the circulatory system (19%), neoplasms (17%), and mental and behavioral disorders (17%). Several other traits are studied, including brain structure, height, and telomere length. As is shown in Figure 1, the number of consortia publishing a first paper in the NHGRI catalog increased dramatically beginning in 2009 and peaked in the first half of 2010, reflecting the increase in publication of GWAS studies during the same time period (9). (Note that the last bar in the figure is for the first 3 months of 2011, while other bars represent 6-month intervals.)
Figure 1.
Distribution of genome-wide association study consortia (n = 55) with published articles in the National Human Genome Research Institute’s Catalog of Published Genome-Wide Association Studies (10) as of April 1, 2011, by date of first publication. (Note that the last bar is for the first 3 months of 2011, while other bars represent 6-month intervals.)
Only 14 of the 55 GWAS consortia (25%) have publicly accessible research guidelines (Table 1). Of these, the number of members of each consortium varies widely. Among the 6 consortia with “individual investigator” members (the Ovarian Cancer Association Consortium (OCAC) (14), the Pancreatic Cancer Case-Control Consortium (PANC4) (15), the International Multiple Sclerosis Genetics Consortium (IMSGC) (16), the Type 1 Diabetes Genetics Consortium (T1DGC) (17), the Alzheimer’s Disease Genetics Consortium (ADGC) (18), and the International Serious Adverse Event Consortium (iSAEC) (19)), 5 have between 30 and 61 members, while 1 has 376 members. Among the 5 consortia with “research centers” that are members (the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) (20), Genetic Markers for Osteoporosis (GENOMOS) (21), the Melanoma Genetics Consortium (GenoMEL) (22), the Inflammatory Bowel Disease Genetics Consortium (IBDGC) (23), and Electronic Medical Records and Genomics (eMERGE) (24)), the number of members ranges between 5 and 44. Finally, the number of members in consortia comprised of “studies” (the Breast Cancer Association Consortium (BCAC) (25), Genetic Factors for Osteoporosis (GEFOS) (26), and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) (27)) ranges from 5 to 50. Most consortia have published between 1 and 10 publications (as of April 1, 2011), while 1 has 84 publications listed in PubMed (data not shown). No apparent relation was seen between the number or type of consortium members and the number of publications (data not shown). Eight of the consortia with accessible guidelines are sponsored by the National Institutes of Health (IBDGC, T1DGC, eMERGE, BCAC, CIMBA, OCAC, PANC4, and GenoMEL), including 5 that are supported by the Epidemiology and Genomics Research Program of the National Cancer Institute (BCAC, CIMBA, OCAC, PANC4, and GenoMEL).
Table 1.
Genome-wide Association Study Consortia With Publicly Available Guidelines Accessible From Websites and Components of Those Guidelines as of April 1, 2011
| Consortium (Reference No.) | Abbreviation | Website | Component of Guidelines |
|||
| Organization/Governance | Research Protocols/Methodology | Institutional Review Board Approval | Data-Sharing/Publication/Authorship/Intellectual Property | |||
| Breast Cancer Association Consortium (25) | BCAC | http://www.srl.cam.ac.uk/consortia/bcac/ | X | X | X | |
| Consortium of Investigators of Modifiers of BRCA1/2 (20) | CIMBA | http://www.srl.cam.ac.uk/consortia/cimba/index.html | X | X | X | |
| Ovarian Cancer Association Consortium (14) | OCAC | http://www.srl.cam.ac.uk/consortia/ocac/index.html | X | X | X | |
| Pancreatic Cancer Case-Control Consortium (15) | PANC4 | www.panc4.org/ | X | X | ||
| Genetic Factors for Osteoporosis Consortium (26) | GEFOS | http://www.gefos.org/ | X | X | X | |
| Genetic Markers for Osteoporosis (21) | GENOMOS | http://www.genomos.eu/ | X | X | ||
| Melanoma Genetics Consortium (22) | GenoMEL | http://www.genomel.org/ | X | X | X | X |
| International Multiple Sclerosis Genetics Consortium (16) | IMSGC | https://www.imsgc.org | X | X | X | X |
| Inflammatory Bowel Disease Genetics Consortium (23) | IBDGC | http://medicine.yale.edu/intmed/ibdgc | X | X | X | X |
| Type 1 Diabetes Genetics Consortium (17) | T1DGC | https://www.t1dgc.org/home.cfm | X | X | X | X |
| Alzheimer’s Disease Genetics Consortium (18) | ADGC | http://alois.med.upenn.edu/adgc | X | X | X | X |
| Cohorts for Heart and Aging Research in Genomic Epidemiology (27) | CHARGE | http://web.chargeconsortium.com/ | X | X | X | |
| Electronic Medical Records and Genomics (24) | eMERGE | https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page | X | X | X | X |
| International Serious Adverse Event Consortium (19) | iSAEC | www.saeconsortium.org/ | X | X | X | X |
All of the publicly accessible Web-based guidelines for GWAS consortia provide information about the organization and governance of the consortium, and a few include an organizational diagram (Table 1 and Web Table 4). These guidelines typically describe a steering committee, an executive or management committee, and/or a scientific advisory board, and most include an analysis committee, study groups, or a data coordinating center. Many of the guidelines list individual or research center consortia members and working groups or subcommittees for data analysis.
Four of the GWAS consortia that make use of individual samples and/or data for analyses indicate in their website guidelines that IRB approval is required before project decisions are made (Table 1 and Web Table 4). These include GenoMEL, IMSGC, IBDGC, and T1DGC. The ADGC requires a “memorandum of understanding” for analysis proposals, but specific guidelines for these are available only to ADGC members. In contrast, the CHARGE consortium, which performs primarily meta-analyses and does not share individual subject data, does not mention IRB approvals on its website. Unlike the GWAS consortia that are focused on specific diseases, eMERGE was formed “to develop, disseminate, and apply approaches to research that combine DNA biorepositories with electronic medical record systems for large-scale, high-throughput genetic research” (24). One of the 3 eMERGE work groups, the Consent and Community Consultation group, deals with IRB issues, including model language for consent forms (28). The iSAEC, whose mission is to “identify and validate DNA-variants useful in predicting the risk of drug-induced serious adverse events” (19), requires that researchers who qualify for data access comply with a series of restrictions through a signed agreement. However, a review of the first publication from each of these consortia demonstrated that 3 of the consortia that did not mention IRB approvals on their website did describe such procedures in those publications (CIMBA, PANC4, and GEFOS), illustrating that IRB approvals were obtained for these analyses.
Although most of the consortia guidelines include policies for data-sharing, publication, authorship, and intellectual property, these vary considerably in content and the level of detail provided (Table 1 and Web Table 4). For example, GenoMEL and IMSGC require that data generated for consortium projects be submitted to the consortium data repository, while T1DGC and CHARGE have specific guidelines for data confidentiality and data-sharing among working group members. Guidelines for 5 consortia—CIMBA, IMSGC, T1DGC, ADGC, and CHARGE—have explicit authorship and publication policies that are publicly accessible. Among the consortia focused on particular diseases, only T1DGC requires a certain acknowledgement statement on all presentations and publications, and only the CHARGE guidelines mention intellectual property issues. An objective of the iSAEC is to “manage [intellectual property] relating to [pharmacogenetic] markers useful in predicting [serious adverse events] to ensure broad and open access” (19).
DISCUSSION
This analysis of 845 papers from the NHGRI catalog of GWAS identified 55 GWAS consortia with at least 1 US member that met the GWAS consortium definition developed for this analysis. Among these 55 consortia, 30 (55%) had a dedicated website for the consortium, and only 14 (25%) had publicly accessible guidelines from that website. Thus, only one-fourth of the identified GWAS consortia had publicly accessible policies and procedures relevant to the operation of those consortia. Furthermore, although the guidelines that are available generally provide information on the organization and governance of each consortium and on research procedures and protocols, only half of the guidelines address IRB approval procedures, and the level of detail on publication, authorship, data-sharing, and intellectual property policies varies considerably. Although a full comparison of 41 consortia without US members identified in the NHGRI catalog was beyond the scope of this analysis, only 4 (9.8%) have publicly accessible guidelines on their websites, demonstrating a similar trend among international consortia.
Many GWAS consortia are formed for the purpose of enlarging a study’s sample size to increase statistical power, while others are broader collaborations with a GWAS component. Thus, by definition, such consortia consist of large numbers of investigators, research centers, or studies that must collaborate effectively to successfully complete and publish results from a GWAS analysis. Most consortia with guidelines identified in this analysis were formed to study 1 disease or 1 class of diseases, but they differ fundamentally from the traditional model of a single investigator undertaking a defined research study. As a result, guidelines to effectively and fairly govern these efforts are essential, including guidelines for phenotype harmonization (29, 30). However, most recommendations for the responsible conduct of collaborative research were developed before these large-scale consortia became commonplace (31–33). Our analysis demonstrates that only a fraction of GWAS consortia have publicly available guidelines, although some well-developed examples exist (16, 17, 22–24, 27). Without additional information from specific consortia, it is impossible to know whether the lack of publicly accessible guidelines reflects a lack of formally adopted guidelines or whether consortia have simply chosen to keep their guidelines accessible to members only. To the extent that consortia are choosing to keep their guiding principles out of public view, this represents a lost opportunity, both with regard to educating others about the successful pursuit of collaborative research and with regard to demonstrating responsible practice to a wider range of interested stakeholders (research participants, funders, etc.).
Recently, Knoppers et al. (34), from the Public Population Project in Genomics, the European Network for Genetics and Genomic Epidemiology, and the Centre for Health, Law, and Emerging Technologies, proposed a “Code of Conduct” for international genomic research, consisting of 7 principles and procedures: quality, accessibility, responsibility, security, transparency, accountability, and integrity. This guidance is based on the values of “ (i) mutual respect and trust between scientists, stakeholders and participants; and (ii) a commitment to safeguarding public trust, participation and investment” (34, p. 46). Based on the analysis presented here, the limited number of guidelines that are currently publicly available reduces the ability to determine whether GWAS consortia procedures are consistent with these proposed principles. Further, greater transparency of guidelines would allow new consortia to build on the prior experience of existing consortia. For example, recommendations about effective organization structures, recommendations about the level of detail needed for research protocols, and guidelines for when IRB approvals are needed for consortium projects would facilitate establishment of new collaborative arrangements.
There were several limitations of this analysis. First, the NHGRI catalog captures only a subset of all active consortia undertaking large-scale genomic research. For example, the Epidemiology and Genomics Research Program of the National Cancer Institute supports 44 consortia that are investigating a variety of types of cancer (35). Thirty-five of these consortia meet the GWAS consortium definition used in this analysis. Of these, 8 consortia had published results from a GWAS as of April 1, 2011, and thus were included in the NHGRI catalog and in our analysis. Among the remaining 27 consortia, 17 (63%) were found to have websites, a somewhat larger proportion than the consortia identified from the NHGRI database with websites (30 of 55; 55%). Still other consortia did not yet have GWAS publications in the NHGRI catalog as of April 1, 2011—for example, the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis Study, which published the design of the study in 2010 (36).
Second, there were limitations to our method for identifying consortia publications because of long consortium names, because of consortium abbreviations with other meanings, and because the names of consortia do not appear in consistent form in author lists or abstracts. In addition, although some consortia were formed specifically for the purpose of performing a GWAS, others are part of broader research collaborations. The latter type of consortia may have guidelines embedded within a more general framework of consortium policies, rather than having specific policies for the GWAS component, and thus may not have been included in this analysis.
Despite these limitations, the ascertainment scheme used here was sufficient to begin broadly describing collaborative practices in this quickly evolving scientific area. Next steps in this research will include a survey of existing GWAS consortia members to provide a more thorough evaluation of the available guidelines and to begin an iterative process for the development of models and recommendations to facilitate large-scale collaborative research. Such a process will need to involve all relevant stakeholders, including investigators, IRB members, and study participants. For example, in GWAS studies funded by the National Institutes of Health, IRB approvals and data-sharing plans are already required, providing one mechanism with which to begin standardizing policies.
As innovative genomic technologies develop, especially DNA sequencing, it is clear that large-scale research collaborations will continue. Some of these consortia will build on those that were developed for GWAS, and others will be formed to address new scientific goals. For example, participants in a Human Genome Epidemiology Network workshop presented a vision for GWAS collaboration “to create a sustainable, credible knowledge base on genetic variation and human diseases… [that] involves [the collaboration of] research investigators, systematic reviewers, online publishers, and database developers” (37, p. 275). Thus, the need for well-founded guidelines to address collaborative practices and define norms for responsible conduct in this leading-edge research domain will only increase. Wider availability of consortia guidelines would allow for review of the current policies on data-sharing, authorship, publication, intellectual property, and return of results to participants and the development of consensus on appropriate standards that could be uniformly implemented among large-scale consortia. This would not only assist in developing a consistent set of “best practices” to guide genomic research practice but also provide a sound basis from which interested stakeholders (e.g., funders, investigators, research participants) might judge the relative effectiveness of specific collaborative undertakings.
In summary, 55 distinct GWAS consortia were identified from 845 GWAS publications for this analysis, yet only 14 of these have publicly accessible research guidelines on consortia websites. Most of the available guidelines include information on the organization and governance of the consortium, as well as research protocols, but only half address IRB approval procedures. Importantly, the documentation of policies on publication, authorship, data-sharing, and intellectual property varies considerably. Wider availability of consortia guidelines is needed to identify and implement appropriate research standards with broad applicability to emerging forms of large-scale collaboration.
Supplementary Material
Acknowledgments
Author affiliations: Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington (Melissa A. Austin); Department of Environmental and Occupational Health Sciences, School of Public Health, University of Washington, Seattle, Washington (Marilyn S. Hair); and Department of Bioethics and Humanities, School of Medicine, University of Washington, Seattle, Washington (Stephanie M. Fullerton).
This work was supported by the National Human Genome Research Institute of the National Institutes of Health (grant P50 HG003374) and the National Center for Research Resources of the National Institutes of Health (grant UL1 RR025014).
The authors thank Dr. Bruce Psaty, whose seminar presentation at the University of Washington Institute for Public Health Genetics in 2010 gave them the idea for this work.
Conflict of interest: none declared.
Glossary
Abbreviations
- ADGC
Alzheimer’s Disease Genetics Consortium
- BCAC
Breast Cancer Association Consortium
- CHARGE
Cohorts for Heart and Aging Research in Genomic Epidemiology
- CIMBA
Consortium of Investigators of Modifiers of BRCA1/2
- eMERGE
Electronic Medical Records and Genomics
- GEFOS
Genetic Factors for Osteoporosis
- GenoMEL
Melanoma Genetics Consortium
- GENOMOS
Genetic Markers for Osteoporosis
- GWAS
genome-wide association study(ies)
- IBDGC
Inflammatory Bowel Disease Genetics Consortium
- IMSGC
International Multiple Sclerosis Genetics Consortium
- IRB
institutional review board
- NHGRI
National Human Genome Research Institute
- OCAC
Ovarian Cancer Association Consortium
- PANC4
Pancreatic Cancer Case-Control Consortium
- iSAEC
International Serious Adverse Event Consortium
- T1DGC
Type 1 Diabetes Genetics Consortium
References
- 1.Wuchty S, Jones BF, Uzzi B. The increasing dominance of teams in production of knowledge. Science. 2007;316(5827):1036–1039. doi: 10.1126/science.1136099. [DOI] [PubMed] [Google Scholar]
- 2.Hall KL, Feng AX, Moser RP, et al. Moving the science of team science forward: collaboration and creativity. Am J Prev Med. 2008;35(suppl 2):S243–S249. doi: 10.1016/j.amepre.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Disis ML, Slattery JT. The road we must take: multidisciplinary team science. Sci Transl Med. 2010;2(22):22cm9. doi: 10.1126/scitranslmed.3000421. (doi:10.1126/scitranslmed.3000421) [DOI] [PubMed] [Google Scholar]
- 4.Stvilia B, Hinnant CC, Schindler K, et al. Composition of scientific teams and publication productivity at a national science lab. J Am Soc Inform Sci Technol. 2011;62(2):270–283. [Google Scholar]
- 5.Lee S, Bozeman B. The impact of research collaboration on scientific productivity. Soc Sci Med. 2005;35(5):673–702. [Google Scholar]
- 6.Anderson MS, Ronning EA, Devries R, et al. Extending the Mertonian norms: scientists’ subscription to norms of research. J Higher Educ. 2010;81(3):366–393. doi: 10.1353/jhe.0.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 8.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 9.Visscher PM, Montgomery GW. Genome-wide association studies and human disease: from trickle to flood. JAMA. 2009;302(18):2028–2029. doi: 10.1001/jama.2009.1643. [DOI] [PubMed] [Google Scholar]
- 10.Hindorff LA, MacArthur J, Wise A, et al. A Catalog of Published Genome-Wide Association Studies. Bethesda, MD: National Human Genome Research Institute; 2011. ( http://www.genome.gov/gwastudies/). (Accessed April 1, 2011) [Google Scholar]
- 11.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Human Genome Epidemiology Network, Centers for Disease Control and Prevention. HuGE Navigator (Version 2.0). GWAS Integrator. Atlanta, GA: Centers for Disease Control and Prevention; 2011. ( http://hugenavigator.net/HuGENavigator/gWAHitStartPage.do). (Accessed April 1, 2011) [Google Scholar]
- 13.National Center for Health Statistics, Centers for Disease Control and Prevention. International Classification of Diseases, Tenth Revision (ICD-10) Atlanta, GA: Centers for Disease Control and Prevention; 2011. ( http://www.cdc.gov/nchs/icd/icd10.htm). (Accessed April 1, 2011) [Google Scholar]
- 14.Ovarian Cancer Association Consortium. OCAC: The Ovarian Cancer Association Consortium. Cambridge, United Kingdom: Genetic Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge; 2011. ( http://www.srl.cam.ac.uk/consortia/ocac/index.html). (Accessed August 25, 2011) [Google Scholar]
- 15.Pancreatic Cancer Case-Control Consortium. Pancreatic Cancer Case-Control Consortium. Rochester, MN: Mayo Clinic; 2011. ( http://www.panc4.org/). (Accessed April 1, 2011) [Google Scholar]
- 16.International Multiple Sclerosis Genetics Consortium. International Multiple Sclerosis Genetics Consortium. Boston, MA: Harvard Medical School; 2011. ( https://www.imsgc.org). (Accessed April 1, 2011) [Google Scholar]
- 17.Type 1 Diabetes Genetics Consortium. Type 1 Diabetes Genetics Consortium. Winston-Salem, NC: School of Medicine, Wake Forest University; 2011. ( https://www.t1dgc.org/home.cfm). (Accessed April 1, 2011) [Google Scholar]
- 18.Alzheimer’s Disease Genetics Consortium. Alzheimer’s Disease Genetics Consortium. Philadelphia, PA: School of Medicine, University of Pennsylvania; 2009. ( http://alois.med.upenn.edu/adgc). (Accessed April 1, 2011) [Google Scholar]
- 19.International SAE Consortium. International SAE Consortium. Chicago, IL: International SAE Consortium Ltd; 2011. ( www.saeconsortium.org/). (Accessed March 21, 2012) [Google Scholar]
- 20.Consortium of Investigators of Modifiers of BRCA1/2. CIMBA: The Consortium of Investigators of Modifiers of BRCA1/2. Cambridge, United Kingdom: Genetic Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge; 2007. ( http://www.srl.cam.ac.uk/consortia/cimba/index.html). (Accessed August 25, 2011) [Google Scholar]
- 21.Genetic Markers for Osteoporosis Consortium. GENMOS. Rotterdam, the Netherlands: Genetic Laboratory, Department of Internal Medicine, Erasmus MC; 2011. ( http://www.genomos.eu/). (Accessed August 25, 2011) [Google Scholar]
- 22.Melanoma Genetics Consortium. GenoMEL: the Melanoma Genetics Consortium. Leeds, United Kingdom: University of Leeds; 2011. ( http://www.genomel.org/). (Accessed April 1, 2011) [Google Scholar]
- 23.Inflammatory Bowel Disease Genetics Consortium. Inflammatory Bowel Disease Genetics Consortium. New Haven, CT: Department of Internal Medicine, Yale School of Medicine; 2011. ( http://medicine.yale.edu/intmed/ibdgc). (Accessed April 1, 2011) [Google Scholar]
- 24.Electronic Medical Records and Genomics Consortium. The eMERGE Network: electronic Medical Records & Genomics. Nashville, TN: Department of Biomedical Informatics, Vanderbilt University School of Medicine; 2011. ( https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page). (Accessed April 1, 2011) [Google Scholar]
- 25.Breast Cancer Association Consortium. BCAC: Breast Cancer Association Consortium. Cambridge, United Kingdom: Genetic Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge; 2011. ( http://www.srl.cam.ac.uk/consortia/bcac/). (Accessed August 25, 2011) [Google Scholar]
- 26.Genetic Factors for Osteoporosis Consortium. GEFOS. Rotterdam, the Netherlands: Genetic Laboratory, Department of Internal Medicine, Erasmus MC; 2011. ( http://www.gefos.org/). (Accessed April 1, 2011) [Google Scholar]
- 27.CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology: CHARGE Consortium. Seattle, WA: University of Washington; 2011. ( http://web.chargeconsortium.com/). (Accessed April 1, 2011) [Google Scholar]
- 28.Clayton EW, Smith M, Fullerton SM, et al. Confronting real time ethical, legal, and social issues in the Electronic Medical Records and Genomics (eMERGE) consortium. Genet Med. 2010;12(10):616–620. doi: 10.1097/GIM.0b013e3181efdbd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett SN, Caporaso N, Fitzpatrick AL, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol. 2011;35(3):159–173. doi: 10.1002/gepi.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macrina FL. Dynamic Issues in Scientific Integrity: Collaborative Research, 1995. Washington, DC: American Academy of Microbiology; 1995. ( http://academy.asm.org/index.php/colloquium-program/general-microbiology-/199-dynamic-issues-in-scientific-integrity-collaborative-research-1995-a). (Accessed July 22, 2011) [PubMed] [Google Scholar]
- 32.National Academy of Sciences. Overcoming Barriers to Collaborative Research: Report of a Workshop. Washington, DC: National Academy Press; 1999. ( http://books.nap.edu/books/0309067847/html/index.html). (Accessed July 22, 2011) [PubMed] [Google Scholar]
- 33.Shamoo AE, Resnik DB. Responsible Conduct of Research. 2nd ed. New York, NY: Oxford University Press; 2009. [Google Scholar]
- 34.Knoppers BM, Harris JR, Tassé AM, et al. Towards a data sharing Code of Conduct for international genomic research [letter] Genome Med. 2011;3(7):46. doi: 10.1186/gm262. (doi:10.1186/gm262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epidemiology and Genomics Research Program, Division of Cancer Control and Population Sciences, National Cancer Institute. EGRP-Supported Epidemiology Consortia. Bethesda, MD: National Cancer Institute; 2010. ( http://epi.grants.cancer.gov/Consortia/single/genomel.html). (Accessed April 1, 2011) [Google Scholar]
- 36.Preuss M, König IR, Thompson JR, et al. Design of the Coronary Artery Disease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3(5):475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khoury MJ, Bertram L, Boffetta P, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170(3):269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.