Abstract
The kingdom of fungi provides model organisms for biotechnology, cell biology, genetics, and life sciences in general. Only when their phylogenetic relationships are stably resolved, can individual results from fungal research be integrated into a holistic picture of biology. However, and despite recent progress, many deep relationships within the fungi remain unclear. Here, we present the first phylogenomic study of an entire eukaryotic kingdom that uses a consistency criterion to strengthen phylogenetic conclusions. We reason that branches (splits) recovered with independent data and different tree reconstruction methods are likely to reflect true evolutionary relationships. Two complementary phylogenomic data sets based on 99 fungal genomes and 109 fungal expressed sequence tag (EST) sets analyzed with four different tree reconstruction methods shed light from different angles on the fungal tree of life. Eleven additional data sets address specifically the phylogenetic position of Blastocladiomycota, Ustilaginomycotina, and Dothideomycetes, respectively. The combined evidence from the resulting trees supports the deep-level stability of the fungal groups toward a comprehensive natural system of the fungi. In addition, our analysis reveals methodologically interesting aspects. Enrichment for EST encoded data—a common practice in phylogenomic analyses—introduces a strong bias toward slowly evolving and functionally correlated genes. Consequently, the generalization of phylogenomic data sets as collections of randomly selected genes cannot be taken for granted. A thorough characterization of the data to assess possible influences on the tree reconstruction should therefore become a standard in phylogenomic analyses.
Keywords: phylogenomics, fungi, opisthokonts, ESTs, consistency, dothideomycetes, selection bias
Introduction
Fungi are abundant in the entire biosphere. They have fascinated mankind since the beginning of written history and have considerably influenced our culture. Edible and medicinal mushrooms are widely used to serve our needs, whereas we have to struggle with fungal pathogens in agriculture and forestry and as causative agents of lethal diseases in humans and animals. In biotechnology, the ability of many fungi to grow easily under controlled conditions implicates fungi as model organisms for cell biology, development, and genetics of eukaryotes (e.g., Gavin et al. 2006; Liti et al. 2009). Many fungal species are of essential importance for their physiological and symbiotic abilities. More than 90% of all land plants are forming mycorrhizal associations that are often crucial for plant growth, development, and fruiting (Wang and Qiu 2006). Furthermore, fungi play an important role as degraders in most ecosystems.
Fungi appear in a vast variety of forms and shapes, and the main strategy to assess the various potentials of an unknown fungus is by comparison with known species. Therefore, reliable phylogenetic information is necessary to facilitate pathogen control or biotechnological applications. Within the domain of Eukarya, the kingdom of fungi, the Mycota, is the sister taxon to the multicellular animals (Metazoa) (Wainright et al. 1993; Liu, Steenkamp, et al. 2009). However, within the fungi, their relationships to the Microsporidia (Keeling et al. 2000; Lee et al. 2008; Koestler and Ebersberger 2011), the split of the arbuscular endomycorrhizal fungi from the former Zygomycota (Schüßler et al. 2001), and the order or below-order relationships within the ascomycetes remained uncertain. Although new taxonomic insights were drawn from recent attempts to resolve the phylogeny of fungi (e.g., Lutzoni et al. 2004; Fitzpatrick et al. 2006; Hibbett 2006; James, Kauff, et al. 2006; Liu, Leigh, et al. 2009; Liu, Steenkamp, et al. 2009; Marcet-Houben and Gabaldon 2009), the backbone of the fungal phylogeny is not yet fully resolved.
One of the reasons for low backbone support is linked to limited gene and taxon sampling. A handful of phylogenetic markers are commonly used (e.g., James, Kauff, et al. 2006; Schoch et al. 2009), bearing the potential drawback of a biased view on evolutionary relationships (Rokas et al. 2003). Moreover, the combined phylogenetic signal of only few genes does not suffice to unequivocally resolve the phylogeny of an entire kingdom (e.g., Jeffroy et al. 2006).
Phylogenomic analyses (Delsuc et al. 2005; Telford 2007) use the phylogenetic signal integrated over many genes as a proxy for the species phylogeny (Gatesy and Baker 2005; Comas et al. 2007). It is hoped that this approach reduces the influence of gene-specific signals (noise) and accentuates the phylogenetic signal generated by the evolutionary relationships of the species. Correspondingly, more recent fungal phylogenies were based on larger set of genes, but the analyses were confined to relatively few taxa (e.g., Robbertse et al. 2006; Cornell et al. 2007; Liu, Steenkamp, et al. 2009; Marcet-Houben and Gabaldon 2009). This substantially increased branch support values but was at the cost of potentially misleading conclusions on phylogenetic relationships due to insufficient taxon sampling (Philippe et al. 2005). Recently, expressed sequence tag (EST) data were proven useful for phylogenomic studies (Hughes et al. 2006; Roeding et al. 2007; Sanderson and McMahon 2007; Dunn et al. 2008; Ebersberger et al. 2009; Meusemann et al. 2010). This wealth of data has only begun to be tapped for fungi (Liu, Leigh, et al. 2009; Liu, Steenkamp, et al. 2009), even though it bears tremendous potential for resolving the fungal part of the tree of life by maximizing both taxon and gene sampling.
Phylogenomic studies result, in many cases, in resolved and well-supported trees (Jeffroy et al. 2006). Unfortunately, these trees not necessarily reflect the true species tree. This is particularly true for notoriously difficult phylogenetic questions usually related to short internal branches or to the placement of rapidly evolving taxa (Jeffroy et al. 2006). For example, three different phylogenetic studies focusing on the early evolutionary relationships of the metazoa (Dunn et al. 2008; Philippe et al. 2009; Schierwater et al. 2009) resulted in three mutually exclusive reconstructions of the early metazoan phylogenies. Using these three studies as an example, Philippe et al. (2011) summarized why merely increasing the amount of data provides no guarantee of arriving at the true tree. Data sets may suffer from the consideration of nonorthologous genes, of genes whose phylogenetic information content has been severely compromised by multiple substitutions, and from sampling of too few taxa or taxa that are evolving too quickly. Moreover, using inappropriate models of sequence evolution can lead to an incorrect interpretation of the phylogenetic signal that remains in the data. Despite many obvious sources of error, there are no clear-cut thresholds to assess a priori the validity of a given data set (Philippe et al. 2011). Problems are usually identified afterward when the reconstructed tree is either incompatible with accepted phylogenetic relationships or when different data sets or different tree reconstruction methods obtain incongruous results. If no a priori knowledge exists, a consistency criterion for evaluating a tree is the only way to assess the credibility of the resulting phylogenetic hypothesis.
In the present study, we maximized taxon and gene sampling by merging data from 99 completely sequenced fungi and 109 fungal EST projects. We extracted and characterized different subsets from these data to 1) assess the influence of data selection procedures in the compilation of phylogenomic data sets and 2) provide independent strategies for reconstructing the fungal phylogeny at different levels of resolution. From the consistent splits, we deduce a comprehensive and refined phylogeny for the fungi. Complementary to recent efforts to resolve the animal phylogeny (Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2011), we provide the second of three pillars to understand the evolution of the multicellular eukaryotic kingdoms, fungi, metazoa, and plants in the past 1.6 billion years.
Materials and Methods
Data Overview and Data Sources
A list of the analyzed taxa and the corresponding data sources is provided in supplementary table S1 (Supplementary Material online).
Preprocessing of EST Data
All ESTs have been screened for vector contaminations with CROSS_MATCH (http://www.phrap.org/phredphrapconsed.html [date last accessed 28 November 2011]) (-minmatch 10 -minscore 20) and with SEQCLEAN (http://compbio.dfci.harvard.edu/tgi/software/ [date last accessed 28 November 2011]). In both cases, the search was performed against the entire UniVec database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html [date last accessed 28 November 2011]).
PolyA tails in the ESTs were removed with SEQCLEAN and sequences with less than 100 nt remaining were discarded. Repetitive elements according to Repbase (Jurka et al. 2005) were soft masked with REPEATMASKER (http://www.repeatmasker.org [date last accessed 28 November 2011]). Eventually, ESTs from a single species were clustered and assembled into contigs with TGICL (Pertea et al. 2003) and subsequently translated in all six reading frames.
Ortholog Search
Definition of the Core Ortholog Set
For our reconstruction of the fungal phylogeny, we compiled three different sets of genes (core orthologs) using the InParanoid-TC approach described in (Ebersberger et al. 2009). In brief, we selected a set of completely sequenced species representing the phylogenetic (sub)-tree of interest as so-called primer taxa for the initial ortholog search (table 1). Only orthologous genes that were present in all primer taxa were considered for further analysis. We named these three core ortholog sets fungi, basidiomycota, and pezizomycotina, respectively, and each of these sets was designed to analyze the corresponding part of the fungal tree. The core ortholog sets are available for download from the HaMStR home page at http://www.deep-phylogeny.org/hamstr (date last accessed 28 November 2011). The names of the core ortholog sets are italicized throughout the manuscript to avoid confusion with the homonymic systematic groups.
Table 1.
Core Ortholog Sets.
| Primer Taxa | Number of Genes | Number of Single-Copy Genesa | |
| fungi | Batrachochytrium dendrobatidis | 1206 | 173 |
| Phycomyces blakesleeanus | |||
| Cryptococcus neoformans | |||
| Schizosaccharomyces pombe | |||
| Yarrowia lipolytica | |||
| Aspergillus fumigatus | |||
| Magnaporthe grisea | |||
| Homo sapiens | |||
| basidiomycota | Ustilago maydis | 559 | 256 |
| Puccinia graminis | |||
| Laccaria bicolor | |||
| Postia placenta | |||
| Schizophyllum commune | |||
| Tremella mesenterica | |||
| pezizomycotina | Sclerotinia sclerotiorum | 2823 | 1226 |
| Fusarium verticillioides | |||
| Stagonospora nodorum | |||
| Mycosphaerella fijiensis | |||
| Coccidioides posadasii | |||
| Aspergillus fumigatus | |||
| Tuber melanosporum |
Core orthologs for which HaMStR (option –strict) detected either no or only a single ortholog in the analyzed completely sequenced fungi (cf. supplementary table S2, Supplementary Material online).
Targeted Search for Orthologs
We extended the core orthologs with sequences from further taxa using HaMStR (Ebersberger et al. 2009). To this end, we aligned the protein sequences for each core ortholog with MAFFT (Katoh et al. 2005) using the options –maxiterate 1000 and –localpair. The resulting multiple sequence alignment, comprising all species from the primer taxa, was converted into a profile hidden Markov model (pHMM) (Durbin et al. 1998) with hmmbuild from the HMMER3 package (http://hmmer.janelia.org [date last accessed 28 November 2011]).
Subsequently, we searched sets of protein sequences or translated EST contigs from taxa not included in the primer taxa for hits with the pHMM. To determine the orthology status of the hmmsearch-hits, HaMStR uses a reciprocity criterion. Each hmmsearch-hit is compared with BLASTP (Altschul et al. 1997) against the proteomes of all primer taxa (HaMStR options –strict and –representative). The reciprocity criterion is only then fulfilled if in all BLASTP searches the protein represented in the core ortholog is identified as highest scoring hit.
Assessing the Copy Number per Gene and Genome
For each gene represented in the three core ortholog sets, we assessed the copy number in the completely sequenced fungi by the following procedure: We ran HaMStR with the option –strict but without the option –representative on the protein set of a given fungal taxon. This resulted in the set of all genes in the search taxon that HaMStR predicted as orthologs. Those cases where HaMStR predicted two or more orthologs are indicative of a gene duplication event that occurred after the split of the search taxon and the closest related primer taxon. The results are summarized in the supplementary table S2 (Supplementary Material online).
Assessing the Evolutionary Rates of the Core Orthologs
We computed for each core ortholog the maximum likelihood (ML) tree from the primer taxon sequences. Sequence alignments and tree reconstruction were performed as outlined in the corresponding paragraphs below. The sum of the branch lengths of the primer taxon tree was then used as a proxy of the evolutionary rate of the gene represented by the core ortholog.
Saturation Plots
Saturation plots were generated as described in Philippe et al. (2011). We computed the pairwise Hamming distances for all sequences in a data set with TREEPUZZLE v5.2 (Schmidt et al. 2002) using the option -weditdist. The ML distances were obtained by summing up the lengths of the branches connecting the corresponding two taxa in the ML tree. We plotted all pairs of corrected–uncorrected distances with R and computed the slope of the linear regression line using the function lm in R.
Data Sets for the Phylogeny Reconstructions
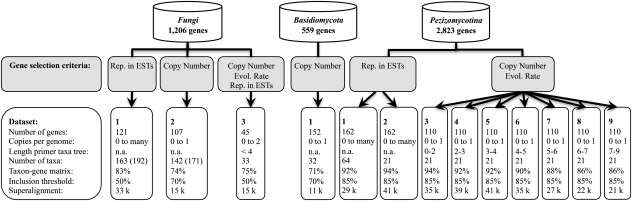
To analyze the phylogeny of the fungi, we compiled 15 different data sets. All data sets were based on the core ortholog sets listed in table 1. An overview of the data sets is given in figure 1 and in supplementary table S2 (Supplementary Material online). Saturation plots for the individual data sets are shown in supplementary fig. S1 (Supplementary Material online).
FIG. 1.
Overview of the data sets used for phylogeny reconstruction. We derived three collections of data sets from the core ortholog collections fungi, basidiomycota, and pezizomycotina, respectively, using different combinations of criteria for gene selection: 1) the abundance of a gene in the EST data (Rep. in ESTs), 2) the maximal number of HaMStR hits for a gene over all analyzed taxa with completely sequenced genomes (copy number), and 3) the evolutionary rate of a gene reflected in the length of the primer taxa tree (“Length primer taxa tree” in substitutions per site). The selection procedure is described in full detail in the Materials and Methods. For the data sets fungi_1 and fungi_2, “Number of taxa” refers to the data sets fungi_1 and fungi_2, respectively. The corresponding numbers for the data sets fungi_1A and fungi_2A (supertree reconstruction) are given in parenthesis. “Taxon–gene matrix” denotes the fraction to which the taxon–gene matrix is filled in the individual data sets. “Inclusion threshold” gives the minimal fraction of ungapped and unambiguous positions in an alignment column to be retained for final analysis. “Superalignment” denotes the length of the final processed superalignment × 1000 amino acid positions that was used for tree reconstruction.
Data Set Fungi_1
We maximized taxon sampling by selecting those genes that are abundant in the EST taxa. To this end, we generated a taxon–gene matrix of all genes in the fungi core ortholog set and all EST taxa. Subsequently, we used an in-house perl script (datamatrix.pl; Simon et al. 2009) to select 121 genes and 57 EST taxa such that each gene is represented in 72% of the EST taxa, and each EST taxon is represented by at least 35% of the genes. The data set was then complemented with sequences from the genome taxa. We chose the following outgroup taxa: Hydra magnipapillata, Nematostella vectensis, Homo sapiens, Capitella sp., Trichoplax adhaerens, Monosiga brevicollis, Capsaspora owczarzaki, and Amoebidium parasiticum. The final data matrix comprised 121 genes and 163 taxa and was filled to 74%.
Data Set Fungi_1A
For the supertree analysis, we complemented data set fungi_1 with fungal EST taxa that were represented by as few as 5 of the 121 genes. This increased the taxon sampling to 195 taxa. Note that we did not consider Cladonia rangifera (12 genes), Pneumocystis carinii (56 genes), and Coniothyrium minitans (5 genes) in this analysis. Initial analyses revealed that neither taxon could be stably placed in the supertree with the current data.
Data Set Fungi_2
This data set is based on genes that occur as single copy in the completely sequenced fungal genomes (table 1). Additionally, we required that each gene must be represented in at least 75 of the 99 analyzed genomes. One hundred and seven genes fulfilled both criteria. To reduce the amount of missing data in the resulting taxon–gene matrix, we included data from all fungal genome taxa but only 27 EST taxa that had at least 25% of the genes represented in their data. Note that this threshold is lower as in data set fungi_1. However, applying the same limit of 35% would have resulted in only a handful of EST taxa to be considered. The following outgroup taxa were chosen: H. magnipapillata, N. vectensis, Gallus gallus, H. sapiens, Oryzias latipes, Danio rerio, Mus musculus, Xenopus tropicalis, Pediculus humanus, Aedes aegypti, Pristionchus pacificus, Lottia gigantea, M. brevicollis, C. owczarzaki, Dictyostelium discoideum, and Dictyostelium purpureum. The final data matrix comprised 107 genes and 142 taxa and was filled to 74%.
Data Set Fungi_2A
For the supertree analysis, we complemented data set fungi_2 with EST taxa that were represented by as few as 5 of 107 genes. This increased taxon sampling to 171 taxa. Note that we did not consider Glomus intraradices (7 genes) and Pisolithus tinctorius (5 genes) in this analysis. Initial analyses revealed that neither taxon could be stably placed in the supertree with the current data.
Data Set Fungi_3
We constructed a third data set to zoom in on deep fungal relationships. The genes were selected according to the following criteria: 1) each gene had to be represented by maximally two co-orthologs in the 99 fungal genomes, 2) the length of the core ortholog tree has to be smaller than four substitutions per site, and 3) each gene must be represented in at least 5 of the 11 basal fungal EST taxa. We chose the following outgroup taxa: M. brevicollis, H. sapiens, N. vectensis, H. magnipapillata, and C. owczarzaki. Moreover, we limited taxon sampling for the Basidiomycota and the Ascomycota to one representative each for the major clades within the two phyla, Basidiomycota: Cryptococcus neoformans (Tremellomycetes), Laccaria bicolor (Agaricomycetes), Sporobolomyces roseus (Pucciniomycotina), and Ustilago maydis (Ustilaginomycotina) and Ascomycota: Neurospora crassa (Sordariales), Trichoderma virens (Hypocreales), Sclerotinia sclerotiorum (Leotiomycetes), Stagonospora nodorum (Dothideomycetes), Aspergillus niger (Eurotiomycetes), Tuber melanosporum (Pezizomycetes), Yarrowia lipolytica (Saccharomycetes), and Schizosaccharomyces pombe (Taphrinomycetes). The Microsporidia were excluded from this analysis. This was done to avoid potentially incorrect inferences in the tree reconstruction due to their high evolutionary rates and the resulting problem of long-branch attraction (cf. Liu, Steenkamp, et al. 2009). The final data set comprised 45 genes and 33 taxa, and the taxon–gene matrix was filled to 75%.
Data Set Basidiomycota_1
To resolve the early splits within the basidiomycetes, we compiled a fourth data set based on the basidiomycota core orthologs. From the 559 genes in the set, we selected 152 single-copy genes that are represented in at least 17 of the 20 basidiomycete and closely related ascomycete genome taxa. The data were complemented with sequences from 12 EST taxa that had at least 36 genes (24%) represented in their data. The final data matrix comprised 152 genes and 32 taxa and was filled to 71%.
Data Sets Pezizomycotina_1–9
Our data sets to resolve the evolutionary relationships within the Pezizomycotina were based on the genes represented in the pezizomycotina core ortholog set. For data set pezizomycotina_1, we used the perl script datamatrix.pl (Simon et al. 2009) to selected 162 genes and 16 EST taxa such that each gene is represented in 70% of the EST taxa, and each EST taxon is represented by at least 30% of the genes. The final data matrix comprised 162 genes and 64 taxa and was filled to 88%.
For data set pezizomycotina_2, we used the same 162 genes as in pezizomycotina_1 but limited the taxon sampling to 20 pezizomycotina (18 genome taxa and 2 EST taxa) and Y. lipolytica. We reduced the taxon set for two reasons: 1) many of the taxa are very closely related. A consideration of all taxa inflates tree space and hence increases the complexity of the tree search without any obvious benefit. 2) Taxa represented only by small EST projects increase the amounts of missing data. Where we had the choice between a genome taxon and a closely related EST taxon, we selected the genome taxon. Aureobasidion pullulans (13,000 ESTs) and Geomyces pannorum (11,000 ESTs) were used to complement the genome taxon sampling for the Dothideomycetes and the Leotiomycetes, respectively.
To assess the effect of the copy number and of the evolutionary rates of the proteins on the outcome of the tree reconstruction, we generated the data sets pezizomycotina_3–9. First, we categorized the 1,226 single-copy genes according to the length of the corresponding primer taxa tree into seven bins: [0-2[, [2–3[, [3–4[, [4–5[, [5–6[, [6–7[, and [7–9[ expected substitutions per site. From each bin, we then randomly chose 110 genes without replacement and collected the corresponding orthologs from the same taxa as in data set pezizomycotina_2.
Protein Sequence Alignments
Protein sequence alignments were generated individually for each core ortholog with MAFFT (Katoh et al. 2005) using the options –maxiterate 1000 and –localpair.
Generation of the Supermatrices for the Tree Reconstruction
We first concatenated the individual protein sequence alignments for a data set. In the resulting supermatrices, we denoted missing data by an X. Subsequently, we processed the alignments by retaining only those columns where more than a given fraction of the taxa were represented by an amino acid (cf. Marcet-Houben and Gabaldon 2009). The inclusion threshold was set to 50% for data sets fungi_1 and fungi_3 and to 70% for data sets fungi_2 and basidiomycota_1. Note that the high proportion of EST taxa in data sets fungi_1 and fungi_3 required a less stringent inclusion threshold. For the data sets, pezizomycotina_1–9 we used the most stringent inclusion threshold of 85%. This ensured that missing data do not interfere with the accurate placement of the Dothideomycetes (see below). For data sets fungi_1 and fungi_2, we alternatively processed the alignments with Gblocks (Talavera and Castresana 2007). We used the Gblocks server at http://molevol.cmima.csic.es/castresana/Gblocks_server.html (date last accessed 28 November 2011) with reduced stringency settings by allowing gaps within final blocks and less strict flanking positions. ML tree reconstruction was repeated for both alignments processed with Gblocks obtaining the same trees as with the standard processed alignments. Thus, we conclude that our results and conclusions are not significantly influenced by the alignment processing strategy.
Phylogenetic Tree Reconstruction
ML Trees
ML trees were reconstructed from each of the supermatrices with RAxML-HPC v7.2.2 (Stamatakis 2006) using the PROTGAMMAILGF model of sequence evolution. The LG model (Le and Gascuel 2008) was identified to give the best fit to the data by running ProtTest v2.4 (Abascal et al. 2005) on the concatenated fungi_1 and fungi_2 alignments, respectively. Moreover, it was identified as the best model in 217 of 228 cases (96%) when ProtTest was run on the individual alignments in the two data sets. We computed 100 bootstrap trees for each data set and combined the individual trees into a strict consensus tree with TREEPUZZLE v5.2 (Schmidt et al. 2002).
Bayesian Tree Search
Bayesian tree searches were conducted with PhyloBayes 3.2b and the CAT + Gamma model (Lartillot and Philippe 2004). For each data set or partition of a data set, we performed three independent runs. The runs were pairwise checked for convergence with bpcomp discarding the first 1,000 trees as burn-in and then sampling every second tree. The consensus tree was built from the two runs with the smallest discrepancy observed across all bipartitions (maxdiff in the bpcomp output).
Maximum Parsimony Tree Reconstruction
Maximum parsimony (MP) trees were reconstructed with PAUP* using equal weighting for all characters and treating gaps as missing data. We assessed branch support by performing 100 nonparametric bootstrap replicates.
MRP Supertree Approach
We reconstructed for each aligned gene in the data sets fungi_1A and fungi_2A ten ML trees with RAxML-HPC (Stamatakis 2006) and the PROTGAMMAILGF model of sequence evolution. All ten trees were based on a different starting tree. The resulting 10 × 121 (fungi_1A) and 10 × 107 (fungi_2A) ML trees were used for matrix representation with parsimony (MRP) supertree construction based on Baum/Ragan coding of the trees (Baum 1992; Ragan 1992). Parsimony analysis was done with PAUP* performing a heuristic search with stepwise addition of taxa, ten random starting points, and tree bisection and reconnection (TBR) branch swapping. If more than one most parsimonious tree was obtained, the strict consensus tree was taken. To estimate the branch support of the supertree, we drew 100 bootstrap samples from the input trees (Burleigh et al. 2006). Each sample was treated as the original data. Support for a split in the original MRP supertree was then estimated as the number of bootstrapped trees that contain the split.
Supernetwork Reconstruction
Clades in the eight fungal backbone trees (data sets fungi_1/1A and fungi_2/2A) were collapsed at the displayed taxonomic level with FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/ [date last accessed 28 November 2011]). From the collapsed input trees, we constructed the supernetwork with SplitsTree v4.11.3 (Huson 1998) setting the minimum number of trees to consider a given split to 3. All splits were given equal weight irrespective of their support in the individual trees.
Gene Ontology Overrepresentation Analysis
The gene ontology (GO) (Ashburner et al. 2000) annotations for human genes were retrieved from Ensembl (build 52) using the biomaRt package (Durinck et al. 2005) from Bioconductor (Gentleman et al. 2004). The GO annotations for Saccharomyces cerevisiae genes were retrieved from GO (version 10/8/2010). The pipeline for the GO enrichment analysis was implemented using the topGO R package (Alexa et al. 2006) from Bioconductor in R version R-2.10.1. The significance level of the enrichment for a GO term was determined by a hypergeometric test (one-sided Fisher exact test). For graphical display, we arranged the significant terms for the “Biological Process” GO subontology using the following procedure: A GO graph object was built containing the set of significant terms using the GOgraph function of the GOstat R package (Beissbarth and Speed 2004). To reduce the size of the resulting graph, we iteratively deleted all nonsignificant parental terms from the graph and reconnected the significant child terms of a deleted node to its parents. The resulting GO graphs were imported to the Cytoscape graph visualization tool (Shannon et al. 2003) to perform the graph layout. The GO terms were subsequently manually organized into a map of functional groups based on their shared roles in biological processes.
Systematic Classification
Unless otherwise noted, we derived the systematic classification of fungal taxa from the Index Fungorum (http://www.indexfungorum.org [date last accessed 28 November 2011]), which is connected with Species Fungorum (http://www.speciesfungorum.org [date last accessed 28 November 2011]), and MycoBank (http://www.mycobank.org/ [date last accessed 28 November 2011]).
Data Availability
All sequence data as well as the alignments are available at http://www.deep-phylogeny.org/fungi (date last accessed 28 November 2011).
Results
The Characteristics of Genes in Phylogenomic Data Sets
We screened the annotated protein sets from 99 completely sequenced fungi as well as ESTs from further 106 fungal taxa for the presence of orthologs to 1,206 evolutionary conserved protein coding nuclear genes with well-supported orthology from animals to fungi. Subsequently, we pursued two approaches to reduce this raw taxon–gene matrix into a phylogenomic data set suitable for tree reconstruction. Data set fungi_1 was compiled according to common procedures of phylogenomic analyses focusing on taxa with limited sequence data (e.g., Roeding et al. 2007; Simon et al. 2009; Meusemann et al. 2010). To maximize both taxon and gene sampling and to minimize the amount of missing data, we chose those genes that are prevalent in the EST sets. This produced a final taxon–gene matrix comprising 121 genes and 163 taxa (eight outgroup taxa) with 25% missing data (fungi_1). For data set fungi_2, we preselected 107 single-copy genes for phylogeny reconstruction irrespective from their representation in EST data. We then sacrificed most EST taxa and considered only those 27 that had at least one quarter of the preselected genes represented. The resulting data matrix comprised 107 genes and 143 taxa (16 outgroup taxa) and had 26% missing data.
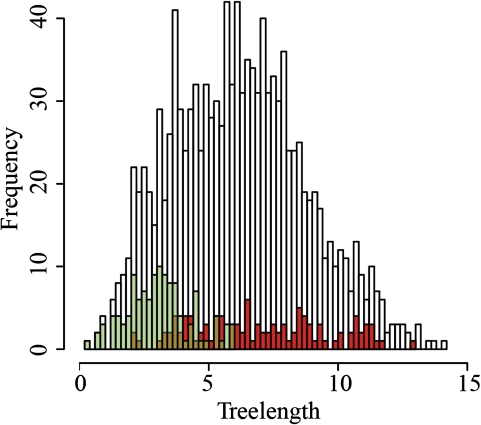
In the next step, we characterized the two data sets in detail. We first compared the copy numbers of the genes (supplementary table S2A, Supplementary Material online). By definition, all genes in fungi_2 are single copy in all analyzed fungal genomes. In contrast, this applied only to 4 of the 121 genes in data set fungi_1. The remaining 117 genes were represented with up to 19 copies in the genomes (supplementary fig. S3, Supplementary Material online). Next, we compared the evolutionary rates of the proteins in the two data sets (fig. 2). This revealed that the selection for genes that are represented in many EST sets (fungi_1) introduced a strong bias toward slowly evolving genes (cf. supplementary fig. S2, Supplementary Material online for a further analysis of this bias). In contrast, fungi_2 covers a broad spectrum from slowly to quickly evolving genes. As a consequence, the discrepancy between observed and corrected pairwise sequence distance—frequently referred to as saturation (Philippe et al. 2011)—was less pronounced in the fungi_1 set as compared with fungi_2 (supplementary fig. S1, Supplementary Material online). Subsequently, we analyzed the influence of the selection procedure on the function of the chosen genes. A GO (Ashburner et al. 2000) overrepresentation analysis unveiled a marked functional correlation between the genes in fungi_1. The genes mainly participate in carbohydrate and energy metabolism as well as in protein synthesis and ribosome function (supplementary fig. S4A, Supplementary Material online). No such pronounced functional correlation was seen for the genes in data set 2 (supplementary fig. S4B, Supplementary Material online). Finally, we determined the overlap between the genes in the data sets 1 and 2. Only two genes were represented in both sets (not shown). In summary, the two data sets differ in all aspects and represent two independent and complementary roads toward reconstructing the evolutionary relationships of fungi.
FIG. 2.
EST guided compilation of phylogenomic data sets selects for slowly evolving genes. The histogram shows the lengths of the primer taxa trees for the 1,206 genes in the fungi core ortholog set. The values for the subset of genes in fungi_1 that have been selected due to their abundance in the EST data are colored in green. The values for the single-copy genes in the data set fungi_2 are colored in red. Numbers of genes with agreeing tree lengths in both sets are colored in brown. Note that this does not imply that the same genes are present in both sets. Only two genes are shared between fungi_1 and fungi_2 (see text).
Overlap to the AFTOL2 Gene Set
Recently, the fungal tree of life initiative has suggested a collection of 74 genes for analyzing the fungal phylogeny (http://aftol.org/pages/AFTOL2_locib.html [date last accessed 28 November 2011]). We determined the overlap between these loci, and the genes represented in our data sets fungi_1 and fungi_2. Of the 74 loci, 21 are not represented in the fungi core ortholog set. For these genes, the transitive circle of pairwise orthology predictions could not be closed. Of the remaining 53 genes, only three are represented to a sufficient extent in the EST data to be considered in data set fungi_1, and only five genes met the criteria to be considered in the data set fungi_2 (supplementary table S2A, Supplementary Material online).
Reconstructing the Backbone of the Fungal Tree of Life
Data set fungi_1 comprised, after alignment processing, 121 genes, 164 taxa, and 33,199 aa. We computed the ML tree and the MP tree for this superalignment (supplementary fig. S5A and B, Supplementary Material online). A complementary Bayesian tree search showed no sign of convergence. We followed the suggestions by the Phylobayes developers and split the data into three nonoverlapping subsets of 41, 41, and 39 genes in size where the assignments of individual genes to a subset were random. The Bayesian tree searches were then performed individually for each partition for a minimum of 46,000 and a maximum of 57,000 generations. Two runs from partition 3 converged (maxdiff: 0.09), and their consensus tree is shown in supplementary fig. S5C (Supplementary Material online). The remaining runs again showed no tendency for convergence (maxdiff > 0.3) and were not further considered. Eventually, we computed a supertree from data set fungi_1A (121 genes, 192 taxa; supplementary fig. S5D, Supplementary Material online). The same procedures were then repeated for data sets fungi_2 and fungi_2A (supplementary fig. S6, Supplementary Material online). Again, the individual runs for the Bayesian tree search using the full data set fungi_2 did not converge. After dividing the data set into two partitions, two runs for partition 2 showed tendency to converge (maxdiff: 0.18 after 20,000 generations).
Combined Evidence From Fungi_1 and Fungi_2: The Supernetwork
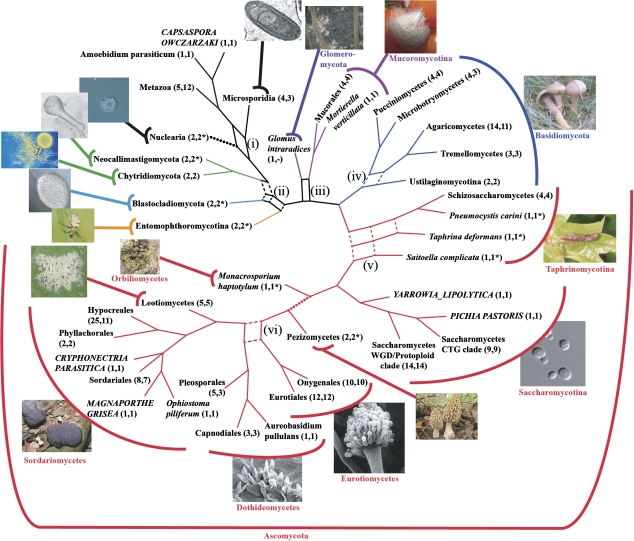
Our analysis of two complementary data sets with four different tree reconstruction methods has resulted in eight trees (supplementary figs. S5 and S6, Supplementary Material online). Each of these trees sheds light on the relationships between the major fungal clades from a different angle. We reason that splits that were consistently recovered in most or all of the trees have a good chance to reflect the true evolutionary relationships of the corresponding fungal taxa. To identify these stable parts in the fungal backbone phylogeny, we used SplitsTree (Huson 1998) to combine the eight trees into a supernetwork (fig. 3). On the chosen level of resolution, most taxa could be placed consistently. This directs attention to reticulate areas in the fungal backbone where the branching pattern differs between the individual trees. Specifically, this concerns the positions of (i) Nuclearia and Microsporidia relative to each other, (ii) the Entomophthoromycotina and the Blastocladiomycetes, (iii) G. intraradices as sole representative of the Glomeromycota, (iv) the smut fungi (Ustilaginomycotina) at the base of the Basidiomycota, (v) the Taphrinomycetes (Taphrina deformans and Saitoella complicata), and (vi) the Dothideomycetes (Pleosporales, Capnodiales, and A. pullulans). To elaborate one example (ii): a clade comprising the Blastocladiomycetes and the Entomophthoromycotina received high support in the ML and MP analysis of the fungi_1 data set (ML bootstrap support [MLBS]: 100; MP bootstrap support [MPBS]: 90; supplementary fig. S5A and B, Supplementary Material online). The position of the two taxa was not resolved in the Bayesian consensus tree (supplementary fig. S5C, Supplementary Material online), and the supertree inferred from fungi_1A suggested a sister group relationship of the Blastocladiomycetes and the Chytridiomycetes + Neocallimastigomycetes (supplementary fig. S5D, Supplementary Material online). Interestingly, the latter topology was also seen in the ML tree of the fungi_2 set (MLBS: 80, supplementary fig. S6A, Supplementary Material online). However, the remaining three trees for this data set provided either no or discrepant information for the position of the two taxa (supplementary fig. S6B–D, Supplementary Material online).
FIG. 3.
Supernetwork summarizing the eight fungal backbone trees inferred from data sets fungi_1 and fungi_2. The numbers of species represented by each leaf are given in parenthesis for the data sets fungi_1 and fungi fungi_2, respectively. An * denotes those instances where either one or both species are absent from data set fungi_2 and are represented only in the supertree based on fungi_2A. A “-” indicates that a taxon is entirely missing in a data set. (i)–(vi) identify reticulate regions in the backbone phylogeny (see text). Colors highlight major systematic groups of the fungi (Ascomycota: red; Basidiomycota: blue; Mucoromycotina: magenta; Glomeromycota: purple; Entomophthoromycotina: yellow; Blastocladiomycota: marine; and Chytriodiomycota/Neocallimastigomycota: green). Contractions of dashed branches in the network result in the topologies that are supported by our refined analyses of fungal subtrees or in the case of the Taphrinomycotina by the study of Liu, Leigh, et al. (2009). The two dotted branches are supported only by one data set (fungi_1). Note that the involved taxa, that is, the nucleariids and Monacrosporium haptotylum are absent from the data sets fungi_2. The full trees are given as supplementary figs. S5 and S6 (Supplementary Material online). A list of the species depicted on the individual photos is provided as supplementary table S3 (Supplementary Material online).
In the following, we concentrated on the problematic splits in figure 3. For two of the six reticulate regions (i and iii), no further data were available for a more refined analysis. A further region (v) has been recently thoroughly investigated by Liu, Leigh, et al. (2009), who convincingly recovered the Taphrinomycotina as a monophyletic taxon. We therefore narrowed our focus to the remaining reticulate regions (ii), (iv), and (vi).
Focusing on Fungal Subtrees
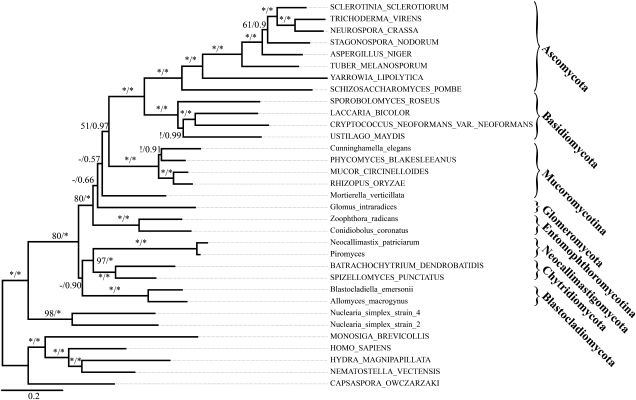
The phylogenetic signals in the data sets fungi_1/1A and fungi_2/2A are insufficient for confidently resolving certain splits in the fungal backbone phylogeny. We, therefore, first compiled a third set of genes from the fungi core ortholog set to reinvestigate split (ii). This time, we selected particularly slowly evolving genes that occur with no more than two copies in the available fungal genome. A stricter approach selecting only single-copy genes was not possible since too few of these genes are represented in the basal fungal taxa for which only ESTs are available. To reduce also the complexity of the tree search, we limited the taxon sampling for the Basidiomycota and Ascomycota to four and eight taxa, respectively, and did not consider the problematic Microsporidia. The final data set to resolve the reticulate region (ii) comprised 45 genes, 33 taxa, and 15,093 aa (fungi_3), and the extent of saturation in the data was substantially lower compared with the data sets fungi_1 and fungi_2 (cf. supplementary fig. S1, Supplementary Material online). We thus did the best to minimize the confounding effects of saturation, inclusion of paralogs, long-branch attraction, and missing data on the tree reconstruction. The results are summarized in figure 4. In essence, the ML tree provides no further information on the deep fungal relationships. In contrast, the Bayesian consensus tree (two independent runs, 22,000 generations, maxdiff: 0.09) supports again an evolutionarily early split of the Blastocladiomycetes, Chytridiomycetes, and Neocallimastigomycetes (BPP: 0.9) and a later branching of the Entomophthoromycotina.
FIG. 4.
The deep-level relationships of the fungi inferred from 46 slowly evolving single-copy genes (data set fungi_3). Shown is the Bayesian consensus tree. The Blastocladiomycota are placed into a monophyletic clade together with the core chytrids. Branch support values represent ML bootstrap support and Bayesian posterior probabilities, respectively. “-” and “!” represent unresolved and conflictingly resolved splits in the ML tree, respectively. An * denotes 100% bootstrap support or a Bayesian posterior probability of 1. Names of taxa represented by genome sequences are written in capital letters, and names of taxa represented by ESTs are written in lower case.
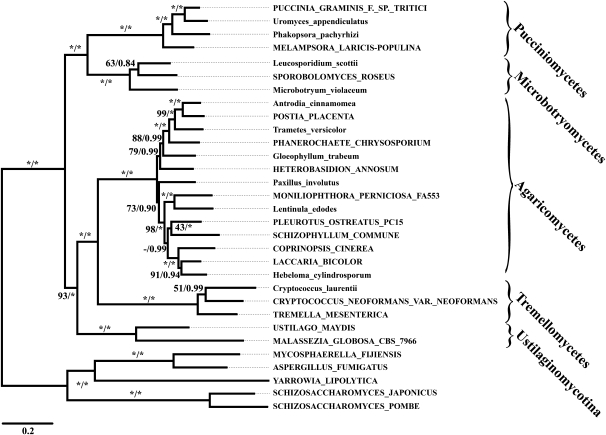
We pursued an extended procedure to address the problematic splits within the Basidiomycota (region (iv)) and within the Pezizomycotina (region (vi)), respectively. We first adapted the core ortholog sets to the particular phylogenetic problem by choosing the primer taxa only from the subtree of interest (table 1). From the two resulting core ortholog collections, we derived the data sets basidiomycota_1 and pezizomycotina_1, respectively, and used them for phylogeny reconstruction. The subtree of the Basidiomycota (fig. 5) recovers the five major basidiomycete taxa represented in our data as monophyletic clades. In addition, it now confidently resolves also the phylogenetic position of the smut fungi as sister to the Agaricomycetes/Tremellomycetes (MLBS: 93; Bayesian posterior probability [BPP]: 1). The situation proved to be more difficult for the Pezizomycotina. The major clades are again resolved in congruency to the backbone phylogeny. However, the position of the Dothideomycetes still remained unclear. In the ML tree, they are placed as sister to the Eurotiomycetes (MLBS: 56; supplementary fig. S7A, Supplementary Material online), whereas in the corresponding Bayesian analysis (two independent runs, 70,000 generations, maxdiff: 0.01), a grouping of the Dothideomycetes with Agaricomycetes and Leotiomycetes is seen (BPP: 0.8; supplementary fig. S7B, Supplementary Material online). Thus, even with this refined and comprehensive data set (162 genes and 64 taxa), the exact position of this group is not resolvable.
FIG. 5.
Phylogenetic relationships of the Basidiomycota based on the data set basidiomycota_1. The Ustilaginomycotina are stably resolved as sister to the Agaricomycotina. Branch support values represent ML bootstrap support and Bayesian posterior probabilities, respectively, where a “-” denotes an unresolved split. An * denotes 100% bootstrap support or a Bayesian posterior probability of 1. Names of taxa represented by genome sequences are written in capital letters, and names of taxa represented by ESTs are written in lower case.
Revisiting the Position of the Dothideomycetes (Region vi)
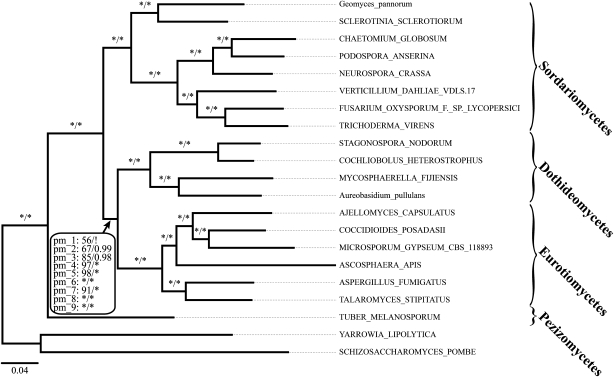
The internal branch determining the position of the Dothideomycetes within the Pezizomycotina is extremely short (∼1 substitution per 100 sites; cf. supplementary figs. S5–S7, Supplementary Material online). This suggests that the diversification of the Leotiomycetes/Sordariomycetes, the Dothideomycetes, and the Eurotiomycetes from their shared common ancestor occurred in close temporal succession. Slowly evolving proteins, which are prevalent in the pezizomycotina_1 set (supplementary fig. S8, Supplementary Material online), may lack the phylogenetic signal to confidently resolve this branch. We investigated this possibility by considering all 1,226 single-copy genes in the pezizomycotina core ortholog set distinguishing seven categories from slowly (pezizomycotina_3) to quickly evolving proteins (pezizomycotina_9). The taxon sampling was restricted to 20 Pezizomycotina (18 genome taxa + 2 EST taxa) and 1 outgroup taxon to level out the extent of missing data between the individual data sets. To judge the effect of the reduced taxon sampling, we also computed trees with the original set of 162 genes (pezizomycotina_2). When comparing these trees to the trees based on the same 162 genes but using the full taxon set, we observed only a single difference: In the Bayesian consensus tree (two independent runs, 14,000 generations each, maxdiff < 0.1), the Dothideomycetes are now confidently resolved as sister group to the Eurotiomycetes (BPP: 0.99). This grouping has already been seen in the Bayesian trees based on data sets fungi_1 and fungi_2 (supplementary figs. S5C and S6C, Supplementary Material online). We conclude that our reduction in taxon sampling introduced no artifacts. Next, we analyzed the trees reconstructed with the binned data sets. All 16 trees unanimously place the Dothideomycetes as sister group to the Eurotiomycetes (fig. 6). Moreover, the statistical support of this split increases with the evolutionary rate of the genes in the data set.
FIG. 6.
Phylogenetic relationships of the Pezizomycotina. The Dothideomycetes are consistently placed as sister to the Eurotiomycetes. The depicted tree is based on the pezizomycotina_2 data set. Branch support values represent ML bootstrap support and Bayesian posterior probabilities, respectively. For the split defining the monophyletic group of Dothideomycetes and Eurotiomycetes, the support values (pm_1–9) from all 18 pezizomycotina trees (9 ML and 9 Bayesian trees, respectively, for the data sets pezizomycotina_1–9) are displayed. Notably, only the Bayesian tree of pezizomycotina_1 resolves the position of the Dothideomycetes differently (marked by a “!”), albeit with weak support (cf. supplementary fig. S7B, Supplementary Material online). An * denotes 100% bootstrap support or a Bayesian posterior probability of 1. Names of taxa represented by genome sequences are written in capital letters, and names of taxa represented by ESTs are written in lower case.
Discussion
Saccharomyces cerevisiae was the first eukaryote species whose genome sequence was fully determined (Goffeau et al. 1996). In recent years, the number of completely sequenced fungal genomes has substantially increased, and to date, roughly one-third of the annotated eukaryotic whole-genome sequences originate from fungi (http://www.diark.org, as of 1 July 2011 [date last accessed 28 November 2011]). This reflects the relevance of the fungal kingdom for evolutionary, medical, and biotechnological research. A robust phylogeny for the fungi is now required to form the scaffold of such studies (Martin et al. 2011). Due to the wealth of data, phylogenomic approaches are the obvious method of choice to reconstruct a new fungal tree of life (e.g., Fitzpatrick et al. 2006; Liu, Steenkamp, et al. 2009).
Phylogenomic Data Are Not a Black Box
Phylogenomic data sets usually represent a collection of ten to hundreds of genes. Gene choice is commonly considered random conditioned only by the requirement that orthologs are identified. This presumably random gene selection procedure is claimed one of the major advantages of phylogenomic studies as it renders gene-specific influences in the tree reconstruction negligible (reviewed in Philippe et al. (2011)). The inclusion of taxa represented only by ESTs, however, complicates this issue by adding a further selection criterion. It is common practice to preferentially choose such genes that are abundant over the various EST sets to avoid too gappy taxon–gene matrices (e.g., Simon et al. 2009; Meusemann et al. 2010). The effect of this procedure on the presumed randomness of the gene selection has never been investigated. Here, we have shown that an EST-directed selection of genes for phylogenomic analyses brings along a number of issues whose influence on the tree reconstruction require careful consideration. First, it bears a high risk of including data from multicopy genes (cf. supplementary fig. S3, Supplementary Material online). This has the potential of blurring the phylogenetic signal in the data by including paralogs. Second, it introduces a bias toward slowly evolving genes (cf. fig. 2 and supplementary figs. S2 and S8, Supplementary Material online). Evolutionary rates of genes are heavily dependent on the respective expression rates (e.g., Drummond et al. 2005), where highly expressed genes have a strong tendency to evolve slowly. In turn, genes that are highly expressed over a broad phylogenetic range have a higher probability of being represented in many and particularly in small EST sets. This explains why phylogenomic data sets with maximized EST sampling are highly enriched for slowly evolving genes. Still the strength of this bias is astounding. Third, it creates a bias of preferentially selecting genes that are involved in the primary metabolism. These genes are likely to act as housekeeping genes and are therefore expressed in all tissues from which mRNA for EST library construction was extracted. The high prevalence of metabolic genes in our EST-directed gene selections therefore is not surprising.
Our analysis reveals that the general assumption that phylogenomic data sets are random collections of genes cannot be taken as granted. Our difficulties in placing the Dothideomycetes in the fungal tree with data set fungi_1 comprising only slowly evolving genes illustrate how this deviation from randomness can influence phylogenomic analyses. Thus, a thorough characterization of phylogenomic data sets with respect to copy number, evolutionary rate, and function of the individual genes should become a standard.
The Criterion of Consistency in Phylogeny Reconstructions
We present a phylogenomic analysis of an entire kingdom whose backbone tree is based on two complementary data sets. We use different gene sets, differently composed ingroup and outgroup as well as different thresholds for alignment postprocessing. Four different tree reconstruction methods (ML, MP, Bayesian, and MRP supertree) were employed to thoroughly explore the phylogenetic signals in the data. Moreover, for those splits that remained unresolved in the first round of analyses, we consulted further data to address the particular phylogenetic problems in greater detail. The comprehensiveness of our approach is informed by the fact that reconstructing evolutionary relationships spanning more than a billion years with a single set of data can only represent a first rough approximation. Although a multiplicity of selection criteria for genes and alignment postprocessing procedures exist to enrich the phylogenetic signal in a data set (e.g., Talavera and Castresana 2007; Marcet-Houben and Gabaldon 2009; Meusemann et al. 2010; Philippe et al. 2011), it is almost impossible to assess a priori whether or not a given data set is suitable to resolve a certain phylogeny. Branch support values commonly associated with the reconstructed trees also do not help. They reflect the stability of a tree given the analyzed data. However, they provide no information on how closely the reconstructed tree resembles the true evolutionary relationships of the analyzed species. Currently, the only way to reenforce phylogenetic hypotheses is the analysis of complementary data (e.g., Comas et al. 2007). If the phylogenetic placement of a taxon remains stable over different data sets and different tree reconstruction methods, it is likely to reflect the true evolutionary relationships of this taxon. However, the criterion of consistency can only be adopted when several trees are available from independent approaches. Generally, this happens only over time when more and more trees are published. In the present study, we have taken a shortcut by basing our conclusions on several independent analyses performed at the same time. We are, therefore, confident that splits that have been consistently resolved in our analysis have a very good chance of being confirmed by future studies based on amino acid sequence alignments. Ultimately, however, our phylogenetic hypotheses have to be confirmed with data of a different type, such as rare genomic changes or morphological characteristics. In turn, those taxa that could not be stably placed by us are indicative of either current shortage in data, methodological problems, or both. This may direct future studies to identify the improvements required to decisively attach these taxa to the fungal tree.
The Phylogenetic Backbone of the Fungi
Phylogeny of the Early Branching Fungi
Our trees consistently support the monophyly of the fungal kingdom. The Nuclearia and the Microsporidia are placed as closest relatives to the fungi. The relative branching order of the two outgroups, however, remains unresolved (fig. 3, region (i)). An accurate placement of the Microsporidia has already proven hard due to their extraordinary high evolutionary rates (Corradi and Keeling 2009; Koestler and Ebersberger 2011). For the Nuclearia, on the other hand, only few ESTs exist and this lack of data makes it currently impossible to address this issue in greater detail.
Within the fungi, the monophyletic group of Neocallimastigomycetes and Chytridiomycetes, sometimes referred to as core chytrids (James, Letcher, et al. 2006; Hibbett et al. 2007), splits first from the backbone. This underpins the common ancestry of the two taxa forming the most basal lineage among the fungi (cf. James, Letcher, et al. 2006; Liu, Steenkamp, et al. 2009) but see Jones et al. (2011). More interesting is the position of the Blastocladiomycetes. Morphological and ecological similarities initially suggested a common ancestry with the core chytrids. However, a tree based on ribosomal DNA sequences suggested that the Blastocladiomycetes split more recently from the fungal backbone than the chytrids (James, Letcher, et al. 2006). Accordingly, the systematic classification of the Blastocladiomycetes was revised, and they were given their own phylum Blastocladiomycota (James, Letcher, et al. 2006; Hibbett et al. 2007). The only phylogenomic analysis of the early fungal relationships reproduced the new placement of the Blastocladiomycota, albeit with low support (Liu, Steenkamp, et al. 2009). Our analysis emphasizes that the evolutionary relationships of the Blastocladiomycota are not yet decisively resolved (fig. 3, region (ii) and fig. 4). The position of this phylum in the fungal tree varies depending on the data set and the tree reconstruction method. Thus, nonphylogenetic signal in the data seems to confound tree reconstruction in our study but presumably also in previous studies. From this perspective, our analysis of 45 slowly evolving single-copy genes (fungi_3) using the CAT model of sequence evolution (Lartillot and Philippe 2004) reflects most closely the suggested approaches to resolve difficult phylogenetic questions (Philippe et al. 2011). Interestingly, the corresponding tree is congruent with the initial classification of the Blastocladiomycota as sister to the chytrids (BPP: 0.9; fig. 4). Thus, revising the systematic classification of this taxon deserves consideration.
The Entomophthoromycotina (Hibbett et al. 2007) are well separated from the earlier branching Chytridiomycota and are placed outside of the remaining fungi. Only their relationships to the Blastocladiomycota discussed above require further analysis. The position of the Glomeromycota relative to the monophyletic Mucoromycotina remains unclear (fig. 3, region (iii)). The supertree analysis of data set fungi_1A places the two taxa within one clade, whereas the remaining trees either suggest that the Glomeromycota split first or failed to resolve the branching order. It is, however, noteworthy that in data sets 1 and 3, the Glomeromycota are represented only by G. intraradices, whereas in data set 2, they are not represented at all. We need to accept that at the moment, available data do not allow to confidently attach glomeromycetes to the phylogenetic backbone of the fungi.
In summary, current reconstructions of early fungal relationships clearly suffer from insufficient data. Meaningful phylogenomic analyses require a careful selection of genes and taxa for tree reconstruction. However, entire phyla or large taxonomic groups, such as the Blastocladiomycota, the Entomophthoromycotina, or the Glomeromycota, are represented only by one or two taxa and even then only by few ESTs in some cases. Notably, our analysis entirely misses the recently proposed Cryptomycota (Jones et al. 2011) including the Rozellida that may comprise the earliest branching lineage of true fungi. Thus, more genomic or transcriptomic data from the early branching fungi and the closest relatives of fungi, such as the nucleariids is necessary to facilitate a profound circumscription of the fungal kingdom.
Phylogeny of the Dikarya
The well-supported Dikarya comprise the monophyletic Basidiomycota and Ascomycota, respectively. Within the Basidiomycota, the rust fungi (Pucciniomycotina), the smut fungi (Ustilaginomycotina), and the Agaricomycotina are each recovered as monophyletic clades. However, the order in which the three taxa emerged is not consistently resolved (fig. 3, region (iv)). We solved this tie in favor of the clade Ustilaginomycotina + Agaricomycotina using a third data set tailored to address the evolutionary relationships within the Basidiomycota (fig. 5). The Basidiomycota serve as an illustrative example that there is most likely no universal data set suitable to resolve each and every split in a kingdoms’ phylogeny (cf. Fong and Fujita 2011). Lineage-specific events, such as duplications or losses of individual genes, or even whole-genome duplications (WGDs) require adapting phylogenomic data sets to the phylogenetic (sub-) tree of interest. There is one indication that this is relevant for the Basidiomycota in particular, as will be discussed below.
Our analysis of all fungi is based on a set of 1,205 genes for which orthologs could be identified in representatives of seven major fungal lineages and humans as outgroup. As expected, the pezizomycotina core ortholog set—whose primer taxa span a considerably smaller evolutionary distance—contains most of these evolutionarily conserved genes from the fungi set (942; 78%). The situation is different for the basidiomycota core ortholog set. Although its primer taxa were also closely related, only 235 (20%) of the genes represented in the fungi set survive the core ortholog selection procedure. For the remainder, the transitive circle of pairwise orthology predictions cannot be closed. This strongly suggests that factors specific to the Basidiomycota have a strong impact on the core ortholog selection. Evidence has emerged that the development of large and diverse structures of fruiting bodies with most basidiomycetes being dependent on sexual propagation coincides with an expansion of gene families involved in signaling pathways, for example, small GTPases or pathways involved in sexual reproduction (Martin et al. 2008; Ohm et al. 2010; Raudaskoski and Kothe 2010). Other basidiomycetes, like the ectomycorrhizal fungus L. bicolor, show extensive spread of transposable elements (Martin et al. 2008). These observations are indicative of a generally high genomic flexibility of the Basidiomycota, which may affect the orthology prediction.
The Ascomycota subdivide into three subphyla: Taphrinomycotina, Saccharomycotina, and Pezizomycotina. The monophyly of the early branching Taphrinomycotina is supported by five of our eight backbone trees, of which unfortunately, only two contain the three EST taxa S. complicata, T. deformans, and P. carinii. However, in concordance to our findings has a recent and extensive study already convincingly resolved the Taphrinomycotina as a monophyletic clade (Liu, Leigh, et al. 2009). We therefore did not follow up on this issue with an extra analysis. The Saccharomycotina are consistently resolved as a monophyletic clade with Y. lipolytica at its base. The remaining species belong to the family of Saccharomycetaceae and are arranged in two major clades. The first clade comprises the species that share a modification in the genetic code. These species translate the codon CTG into a serine rather than into a leucine. Pichia pastoris is consistently placed at the base of the CTG clade. However, this species adheres to the standard genetic code. Thus, the split of P. pastoris delimits the upper bound for the age of this modification in the genetic code. The second major clade harbors the monophyletic species complex around S. cerevisiae, which experienced a WGD (Kellis et al. 2004) (WGD clade), together with its protoploid allies (Souciet et al. 2009). The whole family of Saccharomycetaceae has experienced difficulties in morphological classification in the past. Our data are in line with previous studies suggesting a revision of that group (supplementary figs. S5 and S6, Supplementary Material online).
Within the Pezizomycotina, all classes are monophyletic, and their phylogeny is well resolved. On the level of orders, the phylogenetic position of the Diaporthales is represented only by Cryphonectria parasitica, and its relationship relative to the Sordariales, Ophiostomatales, and Magnaporthe grisea remains ambiguous. Further phylogenomic studies to resolve this issue will require a substantially improved taxon sampling, especially for the Diaporthales, the Ophiostomatales, and the Magnaporthales. Limitation in data availability is, however, not of relevance for our problem in resolving the position of the Dothideomycetes. The fungal classes of interest are represented by ten or more species each, and for most of them, whole-genome sequences are available. Lineage-specific effects interfering with the orthology assignments, as encountered with the Basidiomycota, most likely play no role. Most of the genes in the fungi core ortholog set are represented in the pezizomycotina core ortholog set as well. Moreover, a collection of 164 genes from this set (pezizomycotina_1) did not consistently resolve the issue (supplementary fig. S7, Supplementary Material online). Instead, a situation emerges that is commonly neglected in phylogenomic studies. The genes in the data sets fungi_1 and pezizomycotina_1 may be too slowly evolving to resolve this rather bush-like part in the fungal tree of life (Rokas and Carroll 2006; Fong and Fujita 2011). This interpretation is in line with our results. The four trees based on data set fungi_2 consistently place the Dothideomycetes as sister to the Eurotiomycetes. The same branching pattern is consistently seen with the data sets pezizomycotina_2–9 (fig. 6). Notably, statistical support for this branching increased with the evolutionary rate of the genes in the seven data sets. This provides strong indication that, indeed, the accumulation of slowly evolving genes in the data sets fungi_1 and pezizomycotina_1, as a consequence of EST guided gene selection, interfered with resolving the phylogenetic position of the Dothideomycetes. In summary, our analysis of nine independent data sets (data sets fungi_2 and pezizomycotina_2–9) with varying taxon samplings covering a broad spectrum from very slowly to very quickly evolving genes consistently identified the Eurotiomycetes as sister taxon of the Dothideomycetes.
Conclusions
Here, we have presented the to date most comprehensive phylogeny for the kingdom of fungi. As a major advance over existing studies, we have used the congruency between results from different and complementary data to assess the stability of our phylogenetic conclusions. Splits that consistently occur in our trees have good chances to be recovered also with other amino acid sequence data. In our final backbone phylogeny (supplementary fig. S9, Supplementary Material online), we could decisively place the Ustilaginomycotina as sister to the Agaricomycotina and the Dothideomycetes as sister to the Eurotiomycetes. Moreover, we observed recurring evidence—particularly from slowly evolving genes analyzed with the most sensitive methods—suggesting the alliance of Blastocladiomycota with the core chytrids. This placement reflects the traditional fungal classification where the Blastocladiomycota represented a subphylum within the former Chytridiomycota. A reclassification of the basal fungal lineages deserves therefore consideration. In summary, we provide a stable basis for future studies on fungi assessing the evolution of their peculiar features, for example, fruiting body development, ecological impact, or even to allow new insights into the evolution of multicellular organization. The placement of some taxa like Microsporidia or chytrids will require more genome-wide data, detailed analyses, and careful consideration. Moreover, it will be interesting to see to what extent alternative phylogenetically informative data, such as rare genomic changes or even morphological characters confirm or challenge our results. From the methodological point of view, kingdom-wide phylogeny reconstructions should adhere to some aspects highlighted in our study. There is no globally optimal data set capable in resolving every split in a kingdom’s phylogeny. Instead, individual subtrees may require revisiting with refined phylogenomic data sets adapted to resolve the problematic splits. These data sets then have to cope with, for example, lineage-specific events interfering with the orthology prediction, as seen with the Basidiomycota, or with the problems imposed by bushy parts in the kingdoms phylogeny, as seen with the Dothideomycetes. Together with our finding that the randomness of phylogenomic data sets with respect to gene selection cannot be taken for granted, this strongly suggests that a comprehensive characterization of phylogenomic data sets should become a standard.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/ [date last accessed 28 November 2011]).
Acknowledgments
We gratefully acknowledge the following institutions for providing fully sequenced fungal genomes: the Department of Energy Joint Genome Institute, the Broad Institute, the Baylor College of Medicine, Genolevures, the Podospora anserina genome project, as well as M. Sogin and H. Morrison from the Josephine Bay Paul Center for generously providing us with data from Antonospora locustae. We thank Sascha Strauss for help with the data processing, Martin Eckert for helpful discussion, and Tina Koestler and Dannie Durand for critically reading the manuscript. E.K. and K.V. acknowledge support by the Deutsche Forschungsgemeinschaft (DFG). This work was supported by the Wiener Wissenschafts-, Forschungs- und Technologie Fonds (WWTF) and from the DFG priority program SPP 1174 Deep Metazoan Phylogeny (grant HA 1628/9-2 to A.v.H.).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. (20 co-authors) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum BR. Combining trees as a way of combining data sets phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Burleigh JG, Driskell AC, Sanderson MJ. Supertree bootstrapping methods for assessing phylogenetic variation among genes in genome-scale data sets. Syst Biol. 2006;55:426–440. doi: 10.1080/10635150500541722. [DOI] [PubMed] [Google Scholar]
- Comas I, Moya A, Gonzalez-Candelas F. From phylogenetics to phylogenomics: the evolutionary relationships of insect endosymbiotic gamma-Proteobacteria as a test case. Syst Biol. 2007;56:1–16. doi: 10.1080/10635150601109759. [DOI] [PubMed] [Google Scholar]
- Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Keeling PJ. Microsporidia: a journey through radical taxonomical revisions. Fung Biol Rev. 2009;23:1–8. [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, et al. (18 co-authors) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Durbin R, Eddy S, Krogh A. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JJ, Fujita MK. Evaluating phylogenetic informativeness and data-type usage for new protein-coding genes across Vertebrata. Mol Phylogenet Evol. 2011;61:300–307. doi: 10.1016/j.ympev.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Baker RH. Hidden likelihood support in genomic data: can forty-five wrongs make a right? Syst Biol. 2005;54:483–492. doi: 10.1080/10635150590945368. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, et al. (32 co-authors) Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. (25 co-authors) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. (16 co-authors) Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, et al. (17 co-authors) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS. A phylogenetic overview of the Agaricomycotina. Mycologia. 2006;98:917–925. doi: 10.3852/mycologia.98.6.917. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, et al. (67 co-authors) A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hughes J, Longhorn SJ, Papadopoulou A, Theodorides K, de Riva A, Mejia-Chang M, Foster PG, Vogler AP. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles) Mol Biol Evol. 2006;23:268–278. doi: 10.1093/molbev/msj041. [DOI] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. (70 co-authors) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota) Mycologia. 2006;98:860–871. doi: 10.3852/mycologia.98.6.860. [DOI] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Jones MD, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards TA. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–203. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Koestler T, Ebersberger I. Zygomycetes, microsporidia, and the evolutionary ancestry of sex determination. Genome Biol Evol. 2011;3:186–194. doi: 10.1093/gbe/evr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. (26 co-authors) Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol Biol Evol. 2009;26:27–34. doi: 10.1093/molbev/msn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Steenkamp ET, Brinkmann H, Forget L, Philippe H, Lang BF. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzoni F, Kauff F, Cox C, et al. (46 co-authors) Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am J Bot. 2004;91:1446–1480. doi: 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T. The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS One. 2009;4:e4357. doi: 10.1371/journal.pone.0004357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, et al. (68 co-authors) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Martin F, Cullen D, Hibbett D, Pisabarro A, Spatafora JW, Baker SE, Grigoriev IV. Sequencing the fungal tree of life. New Phytol. 2011;190:818–821. doi: 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- Meusemann K, von Reumont BM, Simon S, et al. (16 co-authors) A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol. 2010;27:2451–2464. doi: 10.1093/molbev/msq130. [DOI] [PubMed] [Google Scholar]
- Ohm RA, de Jong JF, Lugones LG, et al. (32 co-authors) Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, et al. (12 co-authors) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DT, Manuel M, Worheide G, Baurain D. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, et al. (20 co-authors) Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9:847–859. doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbertse B, Reeves JB, Schoch CL, Spatafora JW. A phylogenomic analysis of the Ascomycota. Fungal Genet Biol. 2006;43:715–725. doi: 10.1016/j.fgb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Roeding F, Hagner-Holler S, Ruhberg H, Ebersberger I, von Haeseler A, Kube M, Reinhardt R, Burmester T. EST sequencing of Onychophora and phylogenomic analysis of Metazoa. Mol Phylogenet Evol. 2007;45:942–951. doi: 10.1016/j.ympev.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, McMahon MM. Inferring angiosperm phylogeny from EST data with widespread gene duplication. BMC Evol Biol. 2007;7(Suppl 1):S3. doi: 10.1186/1471-2148-7-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Sung GH, Lopez-Giraldez F, et al. (64 co-authors) The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Schüßler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res. 2001;105:1413–1421. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Strauss S, von Haeseler A, Hadrys H. A phylogenomic approach to resolve the basal pterygote divergence. Mol Biol Evol. 2009;26:2719–2730. doi: 10.1093/molbev/msp191. [DOI] [PubMed] [Google Scholar]
- Souciet JL, Dujon B, Gaillardin C, et al. (54 co-authors) Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009;19:1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Telford MJ. Phylogenomics. Curr Biol. 2007;17:R945–R946. doi: 10.1016/j.cub.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- Wang B, Qiu YL. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. doi: 10.1007/s00572-005-0033-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data as well as the alignments are available at http://www.deep-phylogeny.org/fungi (date last accessed 28 November 2011).