Abstract
Increasing quantitative data generated from transcriptomics and proteomics require integrative strategies for analysis. Here, we present an R package, clusterProfiler that automates the process of biological-term classification and the enrichment analysis of gene clusters. The analysis module and visualization module were combined into a reusable workflow. Currently, clusterProfiler supports three species, including humans, mice, and yeast. Methods provided in this package can be easily extended to other species and ontologies. The clusterProfiler package is released under Artistic-2.0 License within Bioconductor project. The source code and vignette are freely available at http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html
Introduction
In the current postgenomic era, high-throughput experimental techniques including microarray, RNA-Seq, and mass spectrometry were used to detect cellular molecules at the systems level. These kinds of analyses generate huge quantities of data, and thus drive the development of data-mining techniques to capture biological information. A commonly used approach is via clustering in the gene dimension for grouping different genes based on their similarities, such as expression pattern (Yeung et al., 2003), semantic similarity (Yu et al., 2010), and structure of PPI network (Asur et al., 2007). Clustering analysis is widely used to reveal the hidden patterns at the systems level, in particular, to look for shared promoters or regulators, to classify biological processes, to predict poorly characterized or novel genes in coexpression with known functions, and to detect protein communities.
Another common way for searching shared functions among genes is to incorporate the biological knowledge provided by biological ontologies. For instance, Gene Ontology (GO) (Ashburner et al., 2000) annotates genes to biological processes, molecular functions, and cellular components in a directed acyclic graph structure, Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2010) annotates genes to pathways, and Disease Ontology (DO) annotates genes with human disease association (Osborne et al., 2009). Related tools for identifying predominant biological themes in a collection of genes were developed to enhance data interpretation, including GO::TermFinder (Boyle et al., 2004) and GOstats (Falcon and Gentleman, 2007) for GO enrichment analysis, and SubpathwayMiner (Li et al., 2009) for detecting enriched pathways.
Although these tools can automatically calculate the statistically significant categories, the manual step to choose interesting clusters followed by enrichment analysis on each selected cluster is still slow and tedious. Therefore, several new tools such as ClueGO (Bindea et al., 2009) and goProfiles (Sánchez et al., 2007) were developed to partially resolve this problem. However, they only support comparison of two clusters of genes. TM4 MultiExperiment Viewer (MeV) (Saeed et al., 2003), which implements the Tree-EASE (TEASE) algorithm by combining hierarchical clustering with EASE (Hosack et al., 2003), can perform clustering analysis followed by GO enrichment calculation, but without tools designed for comparing and visualizing functional differences among clusters. Here, we present an R package called clusterProfiler for statistical analysis of GO and KEGG, allowing biological theme comparison among gene clusters.
Materials and Methods
The clusterProfiler was implemented in R, an open-source programming environment (Ihaka and Gentleman, 1996), and was released under Artistic License 2.0 within Bioconductor project (Gentleman et al., 2004). The clusterProfiler package depends on the Bioconductor annotation data GO.db and KEGG.db to obtain the maps of the entire GO and KEGG corpus. Bioconductor annotation packages org.Hs.eg.db, org.Mm.eg.db, and org.Sc.sgd.db were imported for genome-wide annotation of mapping Entrez gene identifiers or ORF identifiers for humans, mice, and yeast, respectively.
The clusterProfiler package offers a gene classification method, namely groupGO, to classify genes based on their projection at a specific level of the GO corpus, and provides functions, enrichGO and enrichKEGG, to calculate enrichment test for GO terms and KEGG pathways based on hypergeometric distribution. To prevent high false discovery rate (FDR) in multiple testing, q-values (Storey, 2002) are also estimated for FDR control. Furthermore, clusterProfiler supplies a function, compareCluster, to automatically calculate enriched functional categories of each gene clusters and provides several methods for visualization.
The comparison function was designed as a general package for comparing gene clusters of any kind of gene–ontology associations, not only GO and KEGG, but also other biological and biomedical ontologies, such as DO that annotates the human genome in terms of disease (Osborne et al., 2009). As demonstrated in the online vignette, clusterProfiler can cooperate with R package DOSE and compare gene–disease associations among gene clusters. Comparing gene clusters in the context of disease is a critical step in translating molecular findings from high-throughput methods into clinical relevance.
Results
The clusterProfiler package is implemented for gene cluster comparison. It is not limited to gene clusters obtained from gene expression data, but can also be applied to gene clusters from other approaches, such as PPI modules and miRNA target genes (Yu and He, 2011).
To illustrate how clusterProfiler assesses and compares biological themes among gene clusters, we analyzed the publicly available expression dataset of breast tumour tissues from 200 patients (GSE11121, Gene Expression Omnibus) (Schmidt et al., 2008). We first identified 5230 differentially expressed genes (DEGs) using the significant analysis of microarray (SAM) algorithm (Schwender and Ickstadt, 2008), with cutoff of p-values <0.01 and FDR <0.05, and then identified eight gene clusters from these DEGs, which have different expression patterns across samples, using soft clustering algorithm (Kumar and Futschik, 2007). Finally, we used clusterProfiler to compare these gene clusters by their enriched biological processes, with the strict cutoff of p-values <0.01 and FDR <0.05.
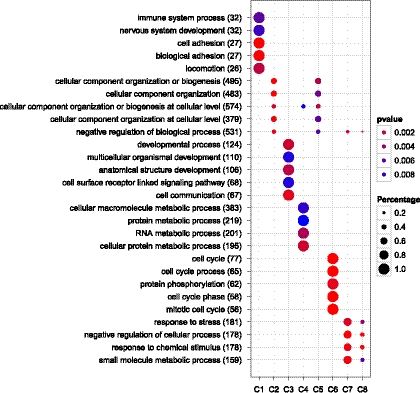
As illustrated in Figure 1, clusterProfiler pinpointed different clusters of DEGs related to different biological processes, such as cluster 2 and cluster 5 related to cellular component organization, cluster 3 related to developmental process, and cluster 6 related to cell cycle.
FIG. 1.
Comparison of GO enrichment of gene clusters.
Dots represent term enrichment with color coding: red indicates high enrichment, blue indicates low enrichment. The sizes of the dots represent the percentage of each row (GO category).
These results are consistent with the main findings of the previous studies, showing that high expression of proliferation-associated genes confers a good prognosis in breast cancer (Schmidt et al., 2008). This analysis provides valuable information for further investigations.
Conclusion and Discussion
In this work, we present a new ontology-based tool, clusterProfiler, that offers three methods, groupGO, enrichGO and enrichKEGG, for gene classification and enrichment analyses. Most importantly, clusterProfiler applies biological term classification and enrichment analyses to gene cluster comparison, helping to better understand higher order functions of biological system. We also adopted a model design with more flexibility. The comparison function of clusterProfiler is designed as a general framework for comparing any kind of biological and biomedical ontologies as demonstrated in the online vignette. In addition, clusterProfiler provides a visualization module for displaying analysis results.
This package is a simple-to-use tool, specifically designed for biologists who wish to analyze high-throughput data obtained from transcriptomics or proteomics. It can be easily extended to support new organisms, and integrated into pipelines for high-throughput data analysis, especially in cooperating with other Bioconductor packages.
We plan to employ three strategies to improve this package in the future. First, we will use semantic similarity among KEGG pathways and GO terms to aggregate closely related categories. This can be anticipated to result in more interpretable outcomes. Second, we will rank gene similarities in each cluster and then correlate them to search for enriched categories. This will provide more sensibility for finding active gene modules. Third, we will develop a statistical model based on the induced directed acyclic graph to compare functional profiles as a whole rather than a set of unrelated categories. These strategies will increase the versatility of clusterProfiler.
Acknowledgments
This work was partially supported by National “973” Projects of China (2011CB910700), the 2007 Chang-Jiang Scholars Program, “211” Projects, National Natural Science Foundation of China (20871057), Guangdong Natural Science Research Grant (32209003), and the Fundamental Research Funds for the Central Universities (21611303 to G. Yu and 11610101 to Q.-Y. He).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Ashburner M. Ball C.A. Blake J.A. Botstein D. Butler H. Cherry J.M., et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asur S. Ucar D. Parthasarathy S. An ensemble framework for clustering protein–protein interaction networks. Bioinformatics. 2007;23:i29–i40. doi: 10.1093/bioinformatics/btm212. [DOI] [PubMed] [Google Scholar]
- Bindea G. Mlecnik B. Hackl H. Charoentong P. Tosolini M. Kirilovsky A., et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.I. Weng S. Gollub J. Jin H. Botstein D. Cherry J.M., et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S. Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C. Carey V.J. Bates D.M. Bolstad B. Dettling M. Dudoit S., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack D.A. Dennis G., Jr. Sherman B.T. Lane H.C. Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R. Gentleman R. R: a language for data analysis and graphics. J Comp Graph Stat. 1996;5:299–314. [Google Scholar]
- Kanehisa M. Goto S. Furumichi M. Tanabe M. Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L. Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Li X. Miao Y. Wang Q. Jiang W. Xu C., et al. SubpathwayMiner: a software package for flexible identification of pathways. Nucleic Acids Res. 2009;37:e131. doi: 10.1093/nar/gkp667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J. Flatow J. Holko M. Lin S. Kibbe W. Zhu L., et al. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10:S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I. Sharov V. White J. Li J. Liang W. Bhagabati N., et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sánchez A. Salicrú M. Ocaña J. Statistical methods for the analysis of high-throughput data based on functional profiles derived from the Gene Ontology. J Stat Plann Inference. 2007;137:3975–3989. [Google Scholar]
- Schmidt M. Böhm D. von Törne C. Steiner E. Puhl A. Pilch H., et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- Schwender H. Ickstadt K. Empirical Bayes analysis of single nucleotide polymorphisms. BMC Bioinformatics. 2008;9:144. doi: 10.1186/1471-2105-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498. [Google Scholar]
- Yeung K.Y. Medvedovic M. Bumgarner R.E. Clustering gene-expression data with repeated measurements. Genome Biol. 2003;4:R34. doi: 10.1186/gb-2003-4-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. He Q.-Y. Functional similarity analysis of human virus-encoded miRNAs. J Clin Bioinformatics. 2011;1:15. doi: 10.1186/2043-9113-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. Li F. Qin Y. Bo X. Wu Y. Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]