Abstract
NDRG4 is a member of the N-myc downregulated gene family (NDRG) belonging to the alpha/beta hydrolase superfamily. We have previously documented discrepancy between our analysis of the expression and function of NDRG4 in glioblastoma multiforme (GBM) and a recent publication by Schilling et al., who reported that NDRG4 is upregulated in GBM compared to human cortex tissues and knock down of NDRG4 reduced the viability of GBM cells. In the present study, we found that NDRG4 is indeed downregulated, at both RNA and protein levels, by quantitative RT-PCR and Western blot analysis, in GBM compared to normal tissues, and that over expression of NDRG4 inhibited proliferation of GBM cells. These new observations can inform the selection of lead molecular compounds for drug discovery as well as novel diagnostics for GBM. They also lend evidence to NDRG4 a role of tumor suppressor.
Introduction
Glioblastoma multiforme (GBM) is the most common and most aggressive malignant primary brain tumor in humans. N-myc downregulated gene family (NDRG) belongs to the alpha/beta hydrolase superfamily and includes four members NDRG1–4, which share about 57–65% amino acid identity (Zhou et al., 2001). In adults, they are expressed in distinct tissues: NDRG1 is relatively ubiquitous expressed; NDRG2 is highly expressed in adult skeletal muscle and brain; NDRG3 is highly expressed in brain and testis; and NDRG4 is specifically expressed in brain and heart (Zhou et al., 2001). NDRG family members usually perform the role of tumor suppressors. For example, NDRG4 is downregulated in colorectal cancer compared to normal tissues and is a tumor suppressor for colorectal cancer (Melotte et al., 2009) and NDRG2 inhibits glioblastoma cell proliferation (Deng et al., 2003).
In a previous study, we applied massively parallel sequencing of expressed sequenced tags (MPSS) technology, and identified many differentially expressed genes between GBM and normal brain tissues, including the downregulation of NDRG4 in GBM tissues compared to normal brain tissues (Lin et al., 2010). However, Schilling et al. (2009) recently reported that NDRG4 is upregulated in GBM compared to human cortex tissues, and knock down of NDRG4 reduced the cell viability of GBM cells. To resolve this discrepancy, we performed additional experiments, and found that NDRG4 is indeed downregulated, at both RNA and protein levels by quantitative RT-PCR and Western blot analysis, in GBM compared to normal tissues.
Material and Methods
Tissue samples
Histologically confirmed GBM and histologically normal nontumor brain specimens (temporal lobe white matter from epilepsy resections) were obtained from the University of Iowa Hospital and Swedish Medical Center, Seattle, WA. All patients gave informed consent prior to collection of specimens according to institutional guidelines.
Quantitative Real-time polymerase chain reaction (qRT-PCR)
The GBM and normal tissues were lysed and total RNA was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Purified RNA (1 μg) was reverse transcribed using random primers (Applied Biosystems, Bedford, MA). The resulting cDNAs were diluted 25-fold and used as templates. qRT-PCR was performed using Assay on Demand gene expression reagents (Invitrogen) on ABI PRISM 7900 HT Sequence Detection System. The primer sequences are: 5′-GGCCTTCTGCATGTAGTGATCCG-3′ and 5′-GGTGATCTCCTGCATGTCCTCG-3′. The primers would PCR a 153 bp fragments from all seven cDNA isoforms of NDRG4 as they localized in the shared regions. The expression of human GUS (beta glucuronicase) was used as endogenous control and performed in triplicate. Quantification of the expression abundance of transcripts was calculated using ΔCt.
Western blot analysis
Total tissue proteins were extracted in lysis buffer and then centrifuged at 12,000×g for 10 min at 4°C. The protein concentration of the supernatant was determined by the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). Aliquots (20 μg) of whole protein lysates were loaded onto sodium dodecyl sulfate polyacrylamide (10%) gels for electrophoresis. For Western blot analysis, proteins were transferred to a polyvinylidene difluoride (PVDF) Immobilon-P membrane (Millipore, Bedford, MA). The membranes were blocked with milk for 1 h at the room temperature and incubated with the primary antibodies directed against GAPDH (Abcam, Cambridge, MA) and NDRG4 (H00065009-M01, Abnova, USA) overnight at 4°C. Then the blots were detected using an HRP-conjugated secondary antibody and visualized by ECL Western Blotting KIT (Pierce, Rockford, IL). The band intensity was quantified in triplicate by the IMAGE-J software for each protein and was normalized to the corresponding GAPDH values.
NDRG4 over expression in GBM cells
GBM cell line U87-MG was transfected with pReceiver-M02-NDRG4 (GeneCopoeia, Rockville, MD), which harbors NDRG4 isoform 1, to generate NDRG4 over expression cells (marked as U87-NDRG4). Cells at 90–95% confluence were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Also, cells were transfected with the empty vector pReceiver-M10 (GeneCopoeia) to generate control cells (marked as U87-Mock). Transfected cells were split and subjected to 400 μg/mL G418 selection (Sigma, St. Louis, MO). After approximately 10 days, monoclonal transfected colonies were picked and a pool of six monoclonal transfectants were combined and cultured for further experiments. The over expression of NDRG4 was confirmed by Western blot analysis.
Cell viability assay
Following infection and selection, 500 U87-NDRG4 and U87-Mock cells were plated in 96-well plates, respectively. Cell viability assays were carried out with the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI) at days 1, 3, 5, 7, and 9, following the manufacturer's instructions.
Results
NDRG4 RNA expression is downregulated in GBM
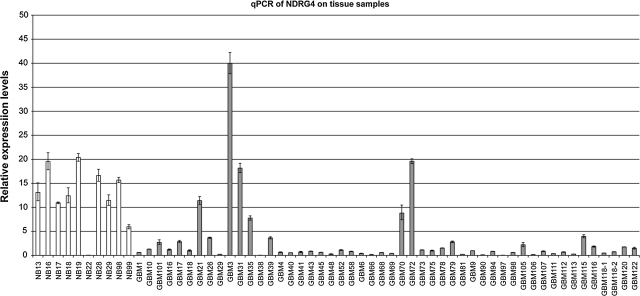
From the MPSS dataset (Lin et al., 2010), we found that the two tags representing NDRG4 expression were both significantly lower in GBM tissues compared with normal tissues (p=7.57E-04 and 3.44E-12) (Table 1). We then performed quantitative RT-PCR on a panel of 49 individual brain tumor samples and 10 individual normal brain tissues, and confirmed that the NDRG4 expression was significantly lower in GBM tissues than in normal tissues (Fig. 1). There are several outliers (Fig. 1), suggesting a heterogeneity nature of the NDRG4 expression, with one case of normal brain tissue with very low NDRG4 expression and about 5 of the 49 GBM cases have expression that of the average normal brain expression levels. We also checked the TCGA (The Cancer Genome Atlas) GBM dataset (http://tcga-data.nci.nih.gov/) for the expression of NDRG4, and found that it is also downregulated in all of the 410 GBM tissues that TCGA profiled (the median AgilentG4502A_07 log2 tumor/normal ratio is −2.15) (Supplementary Table 1).
Table 1.
MPSS Analysis Revealed That NDRG4 is Downregulated in GBM Compared with Normal Brain Tissues
| Tag sequences | GATCCAGGTCATTCCTG | GATCCAGGCCATTCCTG |
|---|---|---|
| Gene symbol | NDRG4 | NDRG4 |
| GBM tissues (TPM) | 110 | 46 |
| GBM tissues (TPM STDEV) | 12 | 0 |
| Normal brains (TPM) | 229 | 269 |
| Normal brains (TPM SD) | 37 | 53 |
| Ratio GBM/normal | 0.48 | 0.17 |
| P-Val: GBM tissues versus normal brains | 7.57E-04 | 3.44E-12 |
TPM, tags per million; SD, standard deviation.
FIG. 1.
NDRG4 mRNA expressions in a panel of 49 individual brain tumor samples (SN series) and 10 individual normal brain tissues (NGRL series). White bars, NGRL series (normal) samples; black, SN series (GBM) samples. Y-axis indicates relative expression levels and X-axis indicates individual samples. Three replicate PCR were performed and the standard errors of the mean were indicated by error bars.
NDRG4 protein expression is downregulated in GBM
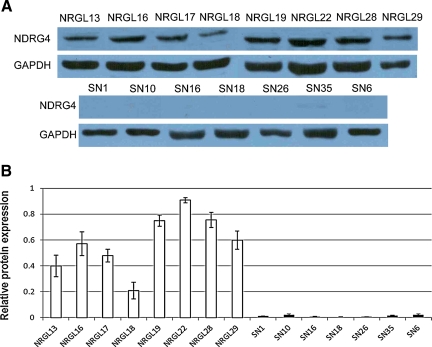
We next investigated the protein expression of NDRG4 by Western blot analysis in seven individual GBM tissues and eight individual normal brain tissues. In agreement with the RNA expression results, NDRG4 protein was expressed in normal brain tissues, but was hardly detectable in GBM tissues (Fig. 2A). A quantitative analysis (Fig. 2B) demonstrated a dramatic downregulation of NDRG4 protein in GBM tissues, ranging from 10–43 times lower than that in normal brain tissues.
FIG. 2.
Western blot analysis of NDRG4. (A) NDRG4 protein expressions in 8 individual normal brain tissues (NGRL series) and 7 individual GBM samples (SN series). GAPDH was used as a control. (B) Quantification of (a) normalized to GAPDH. White bars, NGRL series (normal) samples; black, SN series (GBM) samples.
NDRG4 acts as a potential tumor suppressor to reduce cell proliferation in GBM
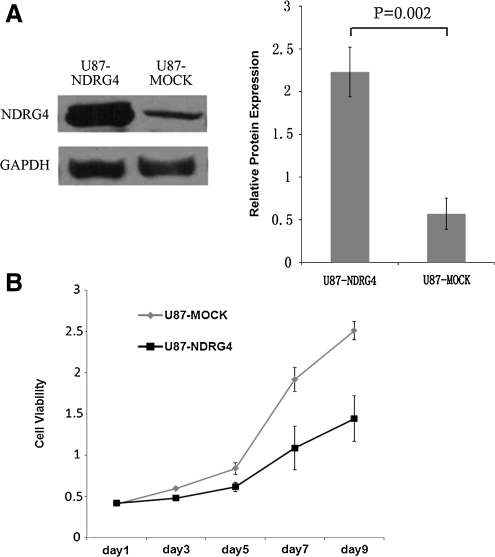
Our data suggest that NDRG4 could play a role of tumor suppressor in GBM, as demonstrated in colorectal cancers (Melotte et al., 2009) where NDRG4 expression is decreased compared to noncancerous colon mucosa, and NDRG4 overexpression suppressed colony formation, cell proliferation, and invasion (Melotte et al., 2009). To test this hypothesis, we overexpressed NDRG4 (isoform 1) in U87-MG glioblastoma cells, and assessed the effect of NDRG4 on cell proliferation. As shown in Figure 3A, NDRG4 protein expression was significantly upregulated in U87 cells that were transfected with NDRG4 (U87-NDRG4) compared to U87-MOCK control. The U87-NDRG4 and U87-MOCK cells with stable NDRG4 expression were then used for cell viability assay. Results showed that NDRG4 overexpression reduced cell viability by about 43% by day 9 (p<0.05) (Fig. 3B).
FIG. 3.
Overexpression of NDRG4 decreases U87-MG cell viability. (A) Western blot analysis of U87-MOCK (control group) and U87-NDRG4 (NDRG4 overexpression) cells. NDRG4 relative protein expression were quantified and showed as ratios of NDRG4/GAPDH (right panel). (B) MTS cell viability analysis of U87-MOCK and U87-NDRG4 cells.
Discussion
Among the four members, three members of the NDRG family were shown to be tumor suppressors in cancers. For example, NDRG1 is identified as a tumor suppressor—its expression could reduce the invasion and metastasis of breast, colon, prostate, and pancreatic cancers by modulating proliferation, differentiation, and angiogenesis of cancer cells (Kehlen et al., 2003; Kovacevic et al., 2008; Liu et al., 2011; Malette et al., 2003; Tschan et al., 2010). NDRG2 is also described as a tumor suppressor. Compared to normal tissues, NDRG2 is downregulated in tumors including thyroid carcinoma (Mordalska et al., 2010), colon cancer (Hwang et al., 2011; Kim et al., 2009; Shi et al., 2009), renal cancer (Ma et al., 2008) and glioblastoma (Deng et al., 2003; Shen et al., 2008; Tepel et al., 2008). NDRG2 overexpression could inhibit glioblastoma cell proliferation (Deng et al., 2003). However, NDRG3 could serve as a tumor promoter for prostate cancer PC3 cell line because overexpression of NDRG3 increased cell growth and migration capability of PC3 cells (Wang et al., 2009). We showed that NDRG4's expression was downregulated at both RNA and protein levels in GBM tissues compared to normal brain tissues, which is in conflict with the findings by Schilling et al. (2009), who showed that NDRG4 is upregulated in GBM compared to human cortex tissues and that knocking down of NDRG4 reduced the cell viability of GBM cells. The discrepancies could be due to several possible reasons. In the report by Schilling et al. (2009), the real-time PCR of NDRG4 was performed by comparing normal primary human astrocytes (NHA 1 and 2) and cultured cells derived from three human GBM xenograft samples (Schilling et al., 2009), whereas in the TCGA as well as in our data (Fig 1 and Supplementary Table 1), the analysis was performed by comparing human GBM tissues with normal brain tissues. The sample size (three cases of GBM) was also very small in Schilling's case but quite large (410 TCGA GBM samples and 49 GBM samples from our laboratory). Considering that there were heterogeneities in the expression of NDRG4 in GBM samples (Fig. 1), conclusions from analysis using small sample size should be interpreted carefully. There also exist possible differences between the GBM tissues derived from the mouse xenografts in Schilling's study versus GBM tissues taken directly from human in our analysis. Additional verification from a third laboratory might be necessary to resolve these discrepancies.
We did attempt to do IHC analysis for glioma tissues from Chinese patients with a tissue microarray consisting of 35 GBM tissues and 5 normal brain tissues. However, getting a good titration proved difficult for the antibody in the IHC analysis. The background was high and it was hard to obtain accurate quantification results. It seemed that the staining intensities were slightly higher in GBM tissues comparing to normal brain tissues (data not shown), which would contradict with our Western blot and RT-PCR results. Our interpretation is that the seemly higher expression of NDRG4 expression in IHC was due to nonspecific staining of the antibody. Although this antibody detected a right sized band of about 41 kDa in Western blot analysis, it could change its specificity in IHC due to different ways of denaturing the NDRG4 antigens between IHC and Western blot analysis. The discrepancies between immunohistochemistry and Western blotting for certain antibodies are not unexpected, as antigenic epitopes could change in different ways dependent on denaturing conditions, for example, formalin and SDS-PAGE, resulting in changes of antibody specificity between Western blot and IHC for certain antibodies. For example, Gibault et al. (2011) assessed 57 sarcomas by Western blot analysis and analyzed their correlation with array comparative genomic hybridization and immunohistochemistry results. They found that the Western blot and immunohistochemistry results were concordant in 23 out of 43 cases (53%), with discrepancies in 20 cases (47%) for the 46 samples with good data in both cases, and that Western blot results were more correlated with array comparative genomic hybridization status than immunohistochemistry (Gibault et al., 2011). Furthermore, they found that some tumors like T19, that are homozygously deleted for PTEN and lack mRNA or protein expression on Affymetrix and Western blot data, displayed a strong signal on immunohistochemistry, in several independent experiments (Gibault et al., 2011). Therefore, when there is a conflict between the Western blot analysis and IHC, the IHC data should be used with caution before ruling out technical bias in IHC such as nonspecific staining.
The differences between our data and that of Schilling et al. (2009) could also be due to different isoforms detected by different antibodies used in the two studies. We used the NDRG4 antibody from Abnova Inc., which is a mouse monoclonal antibody against the full-length recombinant NDRG4 protein. However, the epitope is not known and we do not know if this antibody will detect a specific isoform or all isoforms of NDRG4 considering multiple isoforms (H, B, and Bvar isoforms) exist (Zhou et al., 2001). Schilling et al. (2009) used sigma's NDRG4 antibody, which is a rabbit polyclonal antibody for immunogen RQQIGNVVNQANLQLFWNMYNSRRDLDINRPGTVPNAKTLRCPVMLVVGDNAPAEDGVVECNSKLDPTTTTFLKMADSGGLP. This immunogen is common to all isoforms of NDRG4 (isoforms 1–6 and isoforms H, B, and Bvar). A ClustalW alignment revealed that this immunogen has significant homology to other members of NDRG family including NDRG1, 2, and 3 (over 60% identities in the homologous egions for NDRG1-3) (Supplementary Fig. 1). Melotte et al. (2009) used the same anti-NDRG4 antibody from Abnova Inc. as us, and their conclusion of the role of NDRG4 in colorectal cancer as a tumor suppressor is similar to what we derived for gliomas.
Conclusion
NDRG4's expression was downregulated at both RNA and protein levels in GBM tissues compared to normal brain tissues. These new observations can inform the selection of lead molecular compounds for drug discovery as well as novel diagnostics for GBM. They also lend evidence to NDRG4 a role of tumor suppressor.
Supplementary Material
Acknowledgements
This work was supported by a grant (81072060/H1618) from the National Science Foundation of China, and by grants from the Swedish Science Foundation and The Ben and Catherine Ivy Foundation.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Deng Y. Yao L. Chau L. Ng S.S. Peng Y. Liu X., et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–347. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- Gibault L. Ferreira C. Perot G. Audebourg A. Chibon F. Bonnin S., et al. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.163. [DOI] [PubMed] [Google Scholar]
- Hwang J. Kim Y. Kang H.B. Jaroszewski L. Deacon A.M. Lee H., et al. Crystal structure of the human N-Myc downstream-regulated gene 2 protein provides insight into its role as a tumor suppressor. J Biol Chem. 2011;286:12450–12460. doi: 10.1074/jbc.M110.170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlen A. Lendeckel U. Dralle H. Langner J. Hoang-Vu C. Biological significance of aminopeptidase N/CD13 in thyroid carcinomas. Cancer Res. 2003;63:8500–8506. [PubMed] [Google Scholar]
- Kim Y.J. Yoon S.Y. Kim J.T. Song E.Y. Lee H.G. Son H.J., et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis. 2009;30:598–605. doi: 10.1093/carcin/bgp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic Z. Fu D. Richardson D.R. The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochim Biophys Acta. 2008;1783:1981–1992. doi: 10.1016/j.bbamcr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Lin B. Madan A. Yoon J.G. Fang X. Yan X. Kim T.K., et al. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One. 2010;5:e10210. doi: 10.1371/journal.pone.0010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.L. Bai W.T. Luo W. Zhang D.X. Yan Y. Xu Z.K., et al. Downregulation of NDRG1 promotes invasion of human gastric cancer AGS cells through MMP-2. Tumour Biol. 2011;32:99–105. doi: 10.1007/s13277-010-0103-z. [DOI] [PubMed] [Google Scholar]
- Ma J. Jin H. Wang H. Yuan J. Bao T. Jiang X., et al. Expression of NDRG2 in clear cell renal cell carcinoma. Biol Pharm Bull. 2008;31:1316–1320. doi: 10.1248/bpb.31.1316. [DOI] [PubMed] [Google Scholar]
- Malette B. Cherry E. Lagace M. Bernard M. Gosselin D. Hugo P., et al. Large scale validation of human N-myc downstream-regulated gene (NDRG)-1 expression in endometrium during the menstrual cycle. Mol Hum Reprod. 2003;9:671–679. doi: 10.1093/molehr/gag084. [DOI] [PubMed] [Google Scholar]
- Melotte V. Lentjes M.H. Van den Bosch S.M. Hellebrekers D.M. De Hoon J.P. Wouters K.A. Daenen K.L., et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- Mordalska A. Latek J. Ferenc T. Pomorski L. Galecka E. Zygmunt A., et al. Evaluation of NDRG2 gene expression in primary papillary thyroid carcinoma and in metastases of this neoplasm to regional lymph nodes. Thyroid Res. 2010;3:6. doi: 10.1186/1756-6614-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling S.H. Hjelmeland A.B. Radiloff D.R. Liu I.M. Wakeman T.P. Fielhauer J.R., et al. NDRG4 is required for cell cycle progression and survival in glioblastoma cells. J Biol Chem. 2009;284:25160–25169. doi: 10.1074/jbc.M109.012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. Zhao Z.Y. Wang Y.Z. Ji S.P. Liu X.P. Liu X.W., et al. Immunohistochemical detection of Ndrg2 in the mouse nervous system. Neuroreport. 2008;19:927–931. doi: 10.1097/WNR.0b013e32830163d0. [DOI] [PubMed] [Google Scholar]
- Shi H. Jin H. Chu D. Wang W. Zhang J. Chen C., et al. Suppression of N-myc downstream-regulated gene 2 is associated with induction of Myc in colorectal cancer and correlates closely with differentiation. Biol Pharm Bull. 2009;32:968–975. doi: 10.1248/bpb.32.968. [DOI] [PubMed] [Google Scholar]
- Tepel M. Roerig P. Wolter M. Gutmann D.H. Perry A. Reifenberger G., et al. Frequent promoter hypermethylation and transcriptional downregulation of the NDRG2 gene at 14q11.2 in primary glioblastoma. Int J Cancer. 2008;123:2080–2086. doi: 10.1002/ijc.23705. [DOI] [PubMed] [Google Scholar]
- Tschan M.P. Shan D. Laedrach J. Eyholzer M. Leibundgut E.O. Baerlocher G.M., et al. NDRG1/2 expression is inhibited in primary acute myeloid leukemia. Leuk Res. 2010;34:393–398. doi: 10.1016/j.leukres.2009.08.037. [DOI] [PubMed] [Google Scholar]
- Wang W. Li Y. Hong A. Wang J. Lin B. Li R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124:521–530. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
- Zhou R.H. Kokame K. Tsukamoto Y. Yutani C. Kato H. Miyata T. Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics. 2001;73:86–97. doi: 10.1006/geno.2000.6496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.