Summary
Characterization of gene expression programs and pathways important for normal and cancer stem cells has become an active area of investigation. Microarray analysis of various cell populations provides an opportunity to assess genomewide expression programs to define cellular identity and to potentially identify pathways activated in various stem cells. Here we describe methods to isolate a leukemia stem cell population, amplify RNA, and perform microarray analyses.
Keywords: Stem cells, Leukemia stem cells, Gene expression profiling, Microarray, MLL-AF9, Leukemia, Gene expression, RNA amplification
1. Introduction
Gene expression is one of the major determinants of the biology of both normal and malignant cells. Gene expression profiling, which has enabled the measurement of expression of thousands of genes in an RNA sample, provides insight into the response of a cell to environmental stimuli, thereby facilitating our understanding of the molecular mechanisms underlying normal and dysfunctional biological processes. Gene expression profiling has also been shown to be very useful in identifying molecular features that distinguish leukemia stem cells (LSCs) from normal stem cells (1). Understanding the gene program of LSCs will be significant for the targeted therapy of leukemia. In this chapter, acute myeloid leukemia (AML) originating from MLL-AF9 transduced granulocyte-macrophage progenitors (GMPs) will be used to illustrate the methods utilized for identification, characterization, and microarray analysis of LSCs.
2. Materials
pMSCV expression system (Clontech).
MLL-AF9 cDNA.
HEK 293T cells (ATCC CRL-1573).
Packaging plasmid: Ψ-Eco (Or any ecotropic retrovirus packaging system).
FuGENE 6 (Roche Molecular Biochemicals).
OptiMEM (Gibco/Invitrogen).
Fetal bovine serum (FBS, heat inactivated, Cellgro).
Penicillin/streptomycin (P/S) (Gibco/Invitrogen).
L-Gluamine (L-Glut) (Gibco/Invitrogen).
2-Mercaptoethanol (Gibco/Invitrogen).
Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco/Invitrogen).
Dulbecco’s Modified Eagle Medium (DMEM) (Gibco/Invitrogen).
293T medium: DMEM supplemented with 10% FBS, 1% P/S, 1% L-Glut, 0.02% plasmocin.
Virus collection medium: IMDM supplemented with 15% FBS and 2-mercaptoethanol.
Polybrene (Sigma-Aldrich).
murine recombinant interleukin-3 (IL-3), IL-6, stem cell factor (SCF) (Peprotech).
Phosphate buffer saline (PBS) (Gibco/Invitrogen).
Antibodies for flow sorting and fluorescence-activated cell sorting (FACS): Nonlabeled CD3 (17A2); CD4 (GK1.5); CD8a (53-6.7); CD19 (6D5); B220 (RA3-6B2); Gr1 (RB-8C5); and TER-119 (TER-119); CD127 (A7R34); CD16/32-PE (Fc-RγII/III, clone 93); Sca1-PE/Cy7 (E13-161.7); Gr1-PE (RB-8C5); B220-PE (RA3-6B2); CD117-APC (c-Kit, 2B8); CD34-Pacific Blue (RAM34); CD3-Alexa647 (17A2); Mac1-APC (M1/70) from eBioscience. Dynal beads coated with goat-anti-rat IgG antibody (cat# 110.35) and goat-anti-rat IgG F(ab)2 – Qdot605 (cat# Q11601MP) from Invitrogen.
Magnetic stand for beads separation (Cat# 120.01; Dynal/Invitrogen).
Viability stain, 7AAD (Molecular Probes/Invitrogen).
U-bottom 96-well plate (BD).
Cell strainer 70 μm (BD/Falcon).
Five milliliters FACS tubes (BD/Falcon REF352054).
C57BL/6 mice (Charles River Laboratories), 6–8 weeks old.
RBC lysis buffer (Puregene).
In vitro transcription (IVT) reagent (Ambion T7 Megascript Kit, Ambion).
Nanoprep RNA Isolation Kit (Stratagene).
RNeasy Mini Kit (Qiagen).
Tryzol (Invitrogen).
Chloroform (Sigma-Aldrich).
Glycogen (Invitrogen).
Ethanol (Sigma-Aldrich).
Phenol:chloroform:isoamyl alcohol (25:24:1, v/v) (Invitrogen).
RETROscript kit (Ambion).
RNase H (Sigma-Aldrich).
RNAse-Free DNAseI (Sigma-Aldrich).
T4 DNA polymerase (NEB).
E. coli DNA polymerase I (NEB).
NH4OAc (Sigma-Aldrich).
Gamma Cell, 137Cs γ-irradiator.
Cell sorter, FACSAria equipped with 407-, 488-, 640-nm layers.
3. Methods
The methods described below outline (1) identification of leukemia stem cells, (2) characterization of leukemia stem cells, (3) RNA isolation and amplification, and (4) microarray analysis.
3.1. Identification of Leukemia Stem Cells
Identification of LSCs is described in Subheadings 3.1.1–3.1.4. This includes (a) isolation of GMPs from 6- to 8-week-old C57BL/6 mice using FACS (see Notes 1 and 2) (we chose to initiate leukemia from GMPs since this population of committed myeloid progenitors cannot self-renew (2), and introduction of MLL-ENL into committed myeloid progenitors (CMPs and GMPs) has been shown to induce leukemia (3)), (b) retroviral transduction of MLL-AF9 into GMPs (see Note 3), (c) transplantation of MLL-AF9 transduced GMPs into sublethally irradiated C57BL/6 mice, and (d) flow-sorting leukemia stem cells.
3.1.1. Flow-Sorting GMPs (4)
Harvest bone marrow (BM) cells from the tibia, femur, and humerus of 3–5 mice and filter through the cell strainer to prepare a single cell suspension (5).
Pelletize the cells at 500g for 5 min at 4°C.
Lyse red blood cells (RBCs) in 3–5 mL of Puregene RBC lysis buffer on ice for 10 min.
Wash BM mononuclear cells with PBS to remove debris, and filter through a cell strainer.
Resuspend 5 × 106 cells in 300 μL of PBS supplemented with 0.1% of FBS (PBS–FBS) in a conical 15-mL tube.
Set aside approximately 1 × 106 cells to prepare “single color” controls for FACS.
Incubate BM cells for 30 min on ice with unlabeled lineage antibodies 2 μg/mL each: CD3, CD4, CD8, CD19, CD127, CD45R, Ter119, and Gr1.
Wash the cells with PBS, pelletize at 500g for 5 min at 4°C, and resuspend the cells in 300 μL of PBS–FBS.
While washing cells, wash Dynal magnetic beads with PBS-FBS twice.
Add the cell suspension to the magnetic beads and incubate with slow rotations at 4C for 40 min.
Collect the cell in suspension not bound to magnetic beads using a magnetic stand.
Pelletize the cells form the suspension at 500g for 5 min at 4°C, and resuspend in 300 μL of PBS–FBS.
Add 2 μL of Qdot605-labeled Goat F(ab′)2anti-rat IgG (H + L) secondary antibody, and incubate on ice for 30 min.
Wash the cells with PBS, resuspend in 250 μL of PBS with 10 μL of rat IgG (20 mg/mL), and incubate on ice for 10 min.
Add 2 μL each of CD34-Pacific Blue, Sca1-PE/Cy7, Fc-RγII/III-PE, and c-Kit-APC, and incubate on ice for 30 min.
Wash cells with PBS, spin down cells, and resuspend in 300 μL of PBS.
Filter the cell suspension through a 70-μm cell strainer cap attached to a 5-mL FACS tube to remove cell clumps, and add 20 μL of 7-AAD (10 μg/mL) solution.
Adjust forward scatter and side scatter to bring the BM cells to the center of the axes; gate on the lymphocyte population.
Set up compensation using unstained and single-color stained cells (Qdot605, Pacific Blue, PE, 7AAD, PE/Cy7, and APC alone) (see Note 2).
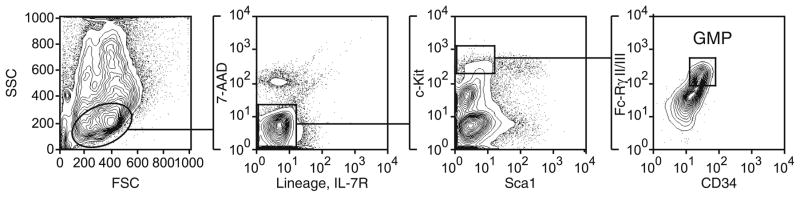
Collect GMPs as 7AAD− Lin−, Sca-1−, c-Kithigh, CD34+, Fc-RγII/IIIhigh cells by flow-sorting (see Fig. 1) in PBS supplemented with 20% horse serum or FBS. If the cell sorted is properly aligned and controls are properly compensated, the double sorting does not increase purity.
Fig. 1.
Gating strategy for isolation of GMPs from murine bone marrow. GMPs are isolated as Lin− Sca1− c-Kit+ Fc-RγII/III+ CD34+ cells.
3.1.2. Retroviral Transduction
The steps described in the following two subheadings outline the procedure for the production of retroviral supernatants and retroviral transduction of GMP (see Note 3).
Transfection of 293T Cells to Produce Retroviral Supernatants
Prepare “Fugene mix” by mixing 1.9 mL of OptiMEM and 95 μL of FuGENE.
Mix 10 μg of Ψ-Eco (or other packaging plasmid) and 10 μg of pMSCV-MLLAF9-MIG plasmids in a 15-mL conical tube.
Add 1.8 mL of FuGENE mix to the DNA mix and incubate at room temperature for 15 min.
Replace media in 30–50% confluent 293T plates with 4.2 mL of fresh 293T media and then gently apply 1.8 mL of the DNA/FuGENE mix to the plate to ensure uniform mixing.
Sixteen hours later (approximately next morning) remove the media from the plates and add 5 mL of the virus collection media to the cells.
After 8 h, harvest the first retroviral supernatant and add 6 mL of the virus collection media.
Filter the retroviral supernatant through a 0.45-μm filter, aliquot, and store at −70°C.
Repeat steps 6 and 7 two more times at 8-h intervals.
Retroviral Infection of GMP
Pelletize 1–5 × 104 freshly sorted GMPs from step 18 of Subheading 3.1.1 by centrifugation (500g; 5 min), resuspend in 250 μL of retroviral supernatant, and transfer into a well in a 96-well plate.
Add 25 μL of 10× cytokine media (IMDM supplemented with 50% FBS, 200 ng/mL of SCF, 100 ng/mL of each IL-3, and IL-6) and 7 μg/mL of polybrene.
Centrifuge the plate for 1 h at 37°C and 600g and transfer the plate to a 37°C incubator for 16 h.
Next morning, remove 200 μL of the supernatant and add 200 μL of fresh IMDM media supplemented with 15% FBS, 20 ng/mL of SCF, 10 ng/mL of IL-3, and 10 ng/mL of IL-6. (Optional: repeat steps 2–3 twice.)
Forty-two to 48 hours after the initiation of retroviral transduction, resort GFP+ 7AAD− cells.
3.1.3. Transplantation of MLL-AF9 Transduced GMP
Irradiate syngeneic recipients (C57BL/6) with a single dose of 600 rad using a γ 137Cs irradiator.
Transplant 1–10 × 103 GFP+ 7AAD− cells resuspended in 250 μL PBS via the tail vein or retro-orbitally into the sublethally irradiated mice.
Monitor the mice daily for development of sickness; tail-bleed the mice weekly for peripheral white blood cell counts.
3.1.4. Isolation of Leukemia Stem Cells Using Flow Cytometry
This consists of the same steps as described in Subheading 3.1.1 except that (1) normal bone marrow cells (BMCs) from C57BL/6 mice are used to set up the gates for flow-sorting LSCs (see Figs. 1 and 2), and (2) seven-color rather than six-color compensation is performed since leukemia cells are GFP+.
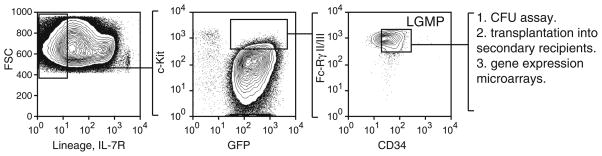
Fig. 2.
LGMPs are expanded in bone marrow of mice with MLL-AF9 AML. Normal bone marrow cells are used to set up gates, and l-GMPs are identified in bone marrow cells from leukemic mice.
3.2. Characterization of Leukemia Stem Cells – Transplantations into Secondary Recipient Mice
Using FACS we isolate several cell populations from bone marrow of mice with AML: GFP+ Lin+; GFP+ Lin− Kit−; GFP+ Lin− Kit+ Fc-Rγ+ CD34+. These cell populations are assayed for colony forming units (CFUs) in vitro and for the frequency of leukemia initiating cells in vivo by limiting dilution transplantation assay. The cell population with the most primitive immunophenotype, GFP+ Lin− Kit+ Fc-Rγ+ CD34+, similar to normal GMP, was found to contain the greatest percentage of CFUs and leukemia initiating cells (frequency of 1 in 6 ± 2 cells). This cell population is designated as leukemic GMPs (LGMPs). The immunophenotype of primary AML should be compared with that of secondary AML to determine whether the leukemia stem cell population recapitulates the primary AML.
3.2.1. Transplantation of Leukemia Stem Cells
Irradiate syngeneic recipients (C57BL/6) with a single dose of 600 rad using a γ 137Cs irradiator.
Transplant the purified LGMPs via tail vein or retro-orbitally into sublethally irradiated mice.
Monitor the mice daily for development of sickness; tail-bleed the mice weekly for peripheral white blood cell counts.
3.2.2. FACS Analysis
This step is performed to immunophenotypically compare AML from primary recipient mice with AML from secondary recipients.
Collect the BMCs from tibia, femur, and humerus of sick mice.
Lyse RBCs by incubating BMCs in Puregene RBC lysis on ice for 5–10 min.
Filter the cell suspension through a 70-μm cell strainer.
Count the viable cells and add 2 × 105 cells to each FACS tube.
Incubate the cells in each tube with one or two antibodies (e.g., Gr1-PE/Mac1-APC, CD3-APC/B220-PE).
Set appropriate compensations using single color controls.
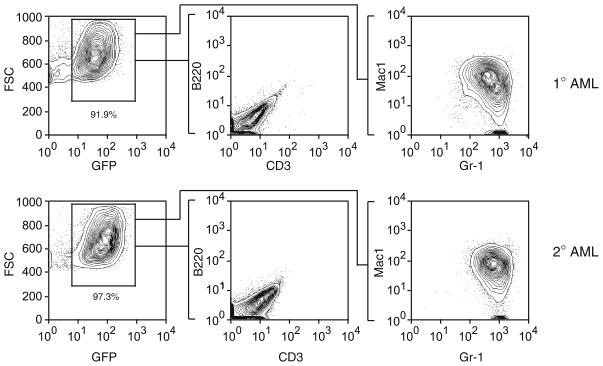
FACS-analyze the stained cells using normal BMCs as a negative control (see Fig. 3).
Fig. 3.
The leukemias from the secondary (2°) recipient mice are immunophenotypically indistinguishable from the primary (1°) disease, demonstrating that LGMPs are enriched for leukemia stem cells.
3.3. RNA Amplification
Because of experimental limitation, number of cells is frequently limited for gene expression analysis. In our case, the sorted populations often contains 1–10 × 104 cells, which yield 1–10 × 10−9 g of total RNA (Subheading 3.3.1). Generally, for hybridization with microarrays it is optimal to have 5 μg of biotinylated anti-sense RNA (aRNA). Therefore, to obtain enough labeled aRNA for hybridization it is necessary to perform RNA amplification. To increase the yield, two consecutive rounds of amplification are performed. The output RNA from round 1 is used as input RNA for round 2. Following is the description of RNA amplification by in vitro transcription (IVT) (see Note 4). We note that there are a number of recently developed commercial methods for RNA amplification that may work as well as what is described here.
3.3.1. Trizol-Based Isolation of RNA
Pelletize the cells of interest in a 1.5-mL Eppendorf-type tube at 500g, and aspirate the supernatant.
Add 1 mL of Tryzol reagent.
Mix by vortexing.
Incubate for 5 min at room temperature.
Add 200 μL of chloroform.
Vortex for 1 min.
Centrifuge at 13,000 g for 10 min at room temperature.
Gently aspirate the upper (clear) phase, transfer to a new 1.5-mL tube, and add 1 μL of glycogene (20 mg/mL).
Add 500 μL of isopropanol.
Precipitate RNA overnight at −20°C.
Pelletize RNA at 13,000g for 15 min at +4°C.
Wash the RNA pellet with 70% ethanol twice; air-dry the pellet.
Resuspend in 50 μL of diethylpyrocarbonate (DEPC) treated deionized water.
Spectrophotometrically measure the RNA concentration.
3.3.2. First Round of RNA Amplification
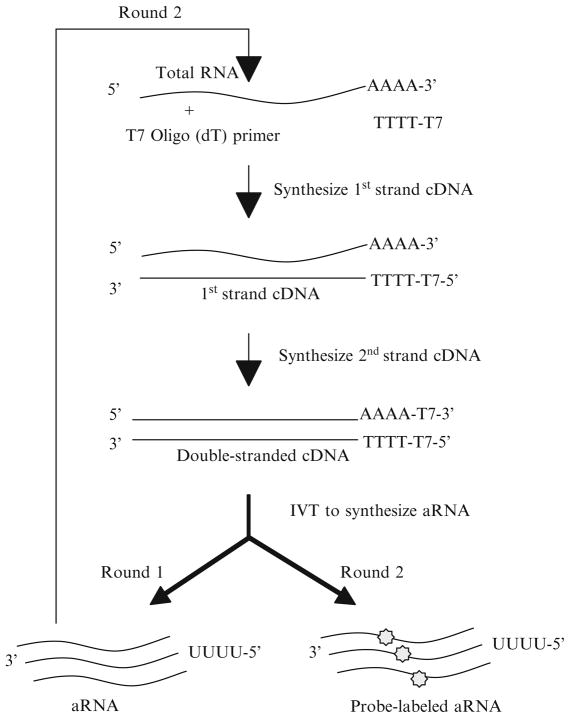
Round 1 of RNA amplification is described in the following subheadings (see Fig. 4), including (a) first-strand cDNA synthesis, (b) second-strand cDNA synthesis, (c) double-stranded cDNA (dscDNA) purification, (d) in vitro transcription (IVT) to synthesize antisense RNA (aRNA or cRNA), and (e) aRNA purification.
Fig. 4.
Graphical representation of RNA amplification for microarray hybridization. dsDNA from the first round is used as input material for the second round of amplification. After the second round of amplification, the aRNA is used for labeling.
Reverse Transcription to Synthesize First-Strand cDNA Using Retroscript Kit
Add 1 μL of 100 μM T7 oligo (dT) primer in 9 μL of total RNA in DEPC-treated water.
Denature RNA by heating at 70°C for 10 min and anneal T-primer for 5 min at 42°C; then chill on ice.
Add on ice to RNA/primer mixture (9 μL per tube): 4 μL of 5× first strand buffer, 2 μL of 0.1 M DTT (dithiothreitol), 1 μL of 10 mM dNTPs, 1 μL (200 U) of SuperScript III Reverse Transcriptase (RT enzyme), and 1 μL (40 U) of RNaseOUT.
Gently mix and incubate at 51°C for 1 h.
Heat-inactivate the RT enzyme at 70°C for 15 min, chill on ice, and proceed to dscDNA synthesis.
Second-Strand cDNA Synthesis (Optional Stopping Point Used in These Experiments)
Add the following reagents to a tube containing freshly synthesized first-strand DNA (20 μL): 30 μL of 5× second-strand buffer, 3 μL of 10 mM dNTPs, 4 μL (40 U) of DNA polymerase I, 1 μL (10 U) of E. coli DNA ligase, 1 μL (2 U) of RNaseH, and 91 μL of DEPC water (130 μL total).
Incubate at 16°C for 2 h.
Add 10 U of T4 DNA polymerase and incubate at 16°C for 15 min.
Purification of Double-Stranded cDNA
Add equal volume buffer saturated phenol:chloroform:isoamyl alcohol (25:24:1, v/v) to dscDNA (approximately 154 μL).
Vortex and spin at 16,000g for 5 min.
Transfer the upper phase to a new 1.5-mL tube.
Add 0.5 volume (approximately 75 μL) of 7.5 M NH4OAc, 1 μL (20 μg) of glycogen, and 2.5 volumes (375 μL) of 100% pre-chilled ethanol.
Vortex and chill at −80°C for 20 min.
Precipitate DNA by centrifuging for 20 min 16,000g at 4°C.
Wash the pellet with 80% ethanol, and spin at 16,000g for 2 min (optional: repeat this step twice).
Quick-spin and aspirate the remaining ethanol to allow the pellet to dry completely, and then resuspend it in IVT reagent mix.
In Vitro Transcription to Synthesize Antisense RNA Using Ambion T7 Megascript Kit
Thaw all reagents and keep them at room temperature to avoid precipitate formation (see Note 5).
Make an IVT mix (18 μL per reaction): 2 μL of 75 mM ATP, 2 μL of 75 mM GTP, 2 μL of 75 mM CTP, 2 μL of 75 mM UTP, 2 μL of 10× buffer, and 8 μL of DEPC water (all reagents are from the kit).
Resuspend purified dscDNA pellet in 18 μL of IVT master mix.
Add 2 μL of enzyme mixture and incubate at 37°C for 6 h.
aRNA Purification Using Nanoprep RNA Isolation Kit
Add an equal volume (approximately 100 μL) of 70% ethanol to the aRNA solution and vortex.
Transfer this mixture to an RNA-binding nano-spin cup and spin for 1 min 16,000g.
Remove the spin cup, discard the filtrate, and re-seat the spin cup in the same 2-mL collection tube.
DNase Treatment: Add 300 μL of 1× Low-Salt Wash Buffer to the spin cup, spin the sample for 1 min at 13,000g, discard the filtrate, re-seat the spin cup in the collection tube, transfer the cup to a new collection tube, and spin for 2 min to dry the RNA-binding matrix.
Prepare the DNase solution by combining 2.5 μL of reconstituted RNase-Free DNaseI with 12.5 μL of DNase Digestion Buffer for each sample.
Add the 15 μL of DNaseI solution directly onto the fiber matrix inside the spin cup and incubate the sample at 37°C for 15 min.
Pre-warm elution Buffer at 37°C, and elute the RNA in 50 μL of the elution buffer from the spin cups. (Optional: use the collected eluate for second elution of the remaining RNA from the spin cups.)
3.3.3. Second Round of RNA Amplification
The second round of RNA amplification is described in the following subheadings (see Fig. 4), including: (a) first-strand cDNA synthesis, (b) second-strand cDNA synthesis, (c) double-stranded cDNA (dscDNA) purification, (d) in vitro transcription (IVT) incorporating RNA labeling, and (e) the purification of probe-labeled aRNA.
Reverse Transcription to Synthesize First-Strand cDNA Using Retroscript Kit
Add 1 μg of random hexamer primers to 600 ng of aRNA (from step 7 in Subheading “aRNA Purification Using Nanoprep RNA Isolation Kit”) in DEPC water and bring the total volume to 10 μL.
Heat at 70°C for 5 min, chill on ice for 3 min, spin, and warm up to room temperature.
Add (9 μL per tube) 4 μL of 5× first strand buffer, 2 μL of 0.1 M DTT, 1 μL of 10 mM dNTPs, 1 μL (200 U) of SuperScript III RT, and 1 μL (40 U) of RNaseOUT.
Mix, spin, heat at 25°C for 10 min increasing the temperature by 1°C every 12 s to 51°C, and incubate at 51°C for 1 h.
Add 1 μL (2 U) of RNAse H and incubate at 37°C for 20 min.
Inactivate RNAse H at 95°C for 2 min, chill on ice for 2 min, spin, and proceed to second strand cDNA synthesis.
Second-Strand cDNA Synthesis
Add 1 μL of 100 μM T7 oligo (dT) primer on ice.
Incubate at 70°C for 5 min, then at 42°C for 10 min, and chill on ice.
Make the master mixture (128 μL per tube): 30 μL of 5× second-strand buffer, 3 μL of 10 mM dNTPs, 4 μL (40 U) of DNA polymerase I, 1 μL (2 U) of RNase H, and 90 μL of DEPC water. Incubate at 16°C for 2 h.
Add 10 U of T4 DNA polymerase, incubate at 16°C for 15 min, and chill on ice.
Purification of Double-Stranded cDNA
Place all dscDNA solution and an equal volume (approximately 155 μL) of buffer saturated phenol:chloroform:isoamyl alcohol (25:24:1, v/v) into a Phase-Lock tube, vortex, and spin for 5 min (16,000g).
Transfer the upper phase to a new 1.5-mL tube.
Add 0.5 volume (75 μL) of 7.5 M NH4OAc, 1 μL (20 μg) of glycogen, and 2.5 volumes (375 μL) of 100% cold ethanol mix and place at −80°C for 20 min.
Spin at 16,000g for 20 min at 4°C and discard the supernatant without disturbing the pellet.
Wash the pellet with 1 mL of 80% ethanol, spin at 16,000g for 2 min, discard the supernatant, and repeat the wash again.
Remove all ethanol, spin for 1 min, aspirate the remaining ethanol, and allow the pellet to dry completely; then resuspend in IVT reagent mix.
In Vitro Transcription to Synthesize Antisense RNA Incorporating RNA Labeling
Thaw and warm the reagents to room temperature (see Note 5).
Make IVT master mixture (18 μL per tube): 2 μL of 75 mM ATP, 2 μL of 75 mM GTP, 1.5 μL of 75 mM CTP, 1.5 μL of 75 mM UTP, 3.75 μL of 10 mM Bio-11-CTP, 3.75 μL of 10 mM Bio-16-UTP, 2 μL of 10× buffer (Ambion), and 1.5 μL of DEPC water.
Dissolve the dscDNA pellet in 18 μL of IVT master mix.
Add 2 μL of enzyme mix (Ambion), shake, and quick-spin.
Incubate at 37°C for 6 h.
Purification of Probe-Labeled aRNA Using Rneasy Mini Kit
Add 80 μL of DEPC water and 350 μL of RLT buffer to a tube with IVT reaction (20 μL) from the previous step.
Add 250 μL of 100% EtOH, mix well, and transfer the sample to RNeasy spin column.
Spin at 16,000g for 1 min, empty the collection tube, and add 500 μL of RPE buffer.
Spin at 16,000g for 1 min and empty the collection tube.
Add 500 μL of RPE buffer and spin at 16,000g for 2 min.
Transfer the spin column to a new capped collection tube.
Add 50 μL of DEPC water to the membrane and let it soak for 4 min; spin at max speed for 1 min.
Repeat steps 1–7 using the first eluate for the second elution (optional).
Measure the optical density (OD) using a 1:50 dilution, and biotin-labeled aRNA is now ready for microarray hybridization.
3.3.4. Hybridization of Labeled aRNA to Affymetrix Microarrays
Usually, specially trained personnel at institutional core facilities perform microarray hybridizations and scanning since the equipment is rather expensive to purchase for individual laboratories. For our studies we used mouse 430 A 2.0 microarrays which includes approximately 22,000 probe sets. Affymetrix has released a newer version of mouse microarrays 430 2.0, which includes all probe sets from mouse 430 A 2.0 microarrays in addition to approximately new 17,000 probe sets, making it possible to analyze the expression of a total of approximately 39,000 transcripts. We would encourage using the newer version of the microarrays since the data generated with the new arrays is compatible with the old arrays.
3.4. Microarray Analysis
3.4.1. Data Normalization
After hybridization, the raw expression data generally has to be normalized to account for differences in chip intensities. We use dChip, a freely available software for data normalization and analysis that can be found at http://biosun1.harvard.edu/complab/dchip/. While we routinely use the Li and Wong method for data normalization found in dChip, there are a number of other approaches that may be useful for specific applications. However, we have found this method to work well for our analyses.
3.4.2. Data Analysis
For gene expression analysis, we routinely use either GenePattern, available from MIT (http://www.broad.mit.edu/tools/software.html), or cluster obtained from http://rana.lbl.gov/EisenSoftware.htm. First, we filter the data to remove probe sets that do not vary across the dataset or that have very low or high expression values. The preprocessing values we use are to set a minimum and maximum expression value, a max/min filter, and max – min filter. This filtering is performed in GenePattern, and the filtered data exported for analysis by other software. The filtering for the analyses shown in Fig. 5 is max/min >2 and max – min >80.
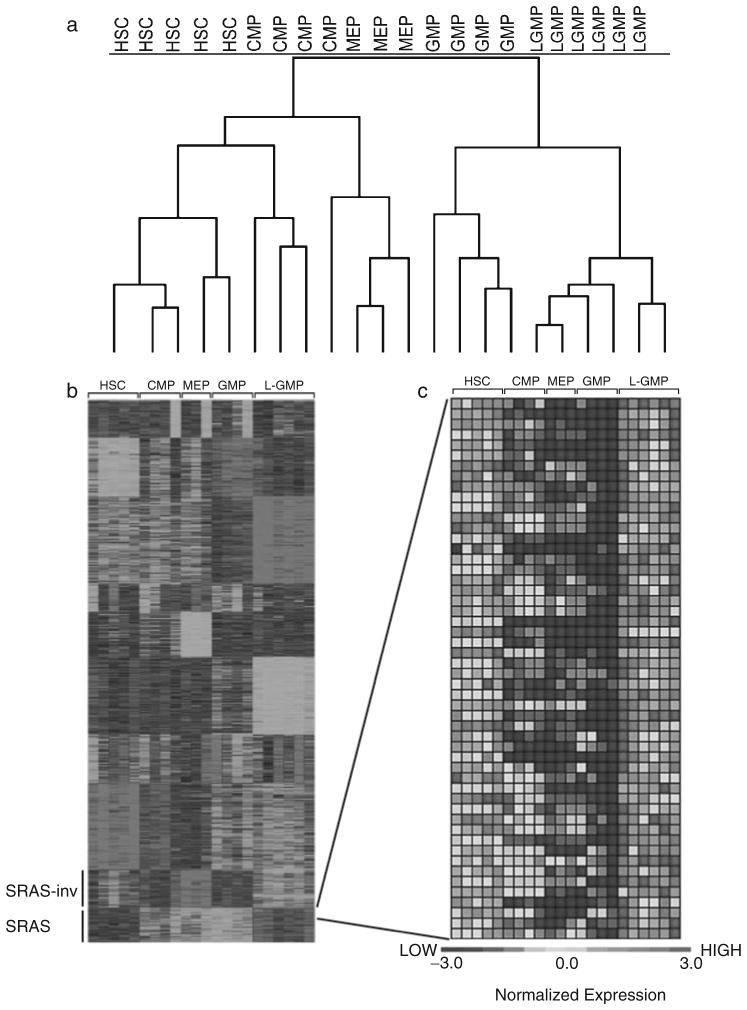
Fig. 5.
Gene expression analysis of normal stem and progenitor cells and LGMPs. (a) Hierarchical clustering of samples identifies the gene expression profile of LGMPs as more similar to GMPs than any other population. (b) K-means analysis identifies a number of clusters of genes that have a similar gene expression pattern labeled 1–3. (c) Supervised marker gene analysis identifies the genes whose expression is most highly correlated with the distinction of interest, in this case cells with self-renewal potential. This is a black-and-white version of a figure taken from (1).
The specific type of analysis is dependent on the question being addressed, and details of the decision process for computational analysis are beyond the scope of this chapter. However, the analysis described here will allow a first-pass assessment of gene expression in various populations of cells. One approach to determine global gene expression relationships between various samples is hierarchical clustering. When the question is solely the relationship between samples, one-dimensional clustering of the samples is performed (Fig. 5a). A number of unsupervised methods are available for clustering genes. We generally use K-means clustering (Fig. 5b). In this case, the number of clusters (K) was set at 10. Determining the “ideal” number for K in any experiment is a topic of considerable debate. The K-means and hierarchical clustering are performed with http://rana.lbl.gov/EisenSoftware.htm. Finally, in order to identify marker genes that distinguish two groups, we use the GenePattern software. We first filter the data as described above, and then compare the means of gene in the two groups using a signal-to-noise statistic (Fig. 5c). A first-pass assessment of significance is easily performed in gene pattern using the permutation analysis available in the software. For more details of these analyses the reader is referred to a number of sources (6–9).
Acknowledgments
We thank all the members of the Armstrong lab for their help in developing these protocols. Also, we would like to thank members of the Broad Institute cancer program and Todd R. Golub for help in developing these approaches and for ongoing collaborative efforts.
Footnotes
Dynabeads M-450 Sheep anti-rat IgG can be used for lineage depletion after the cells are labeled with the lineage-specific antibodies. The lineage-positive cells will bind to the magnetic beads, which can be removed in a magnetic field. The cells not expressing lineage markers will remain in the suspension. This procedure removes approximately 80–95% of the lineage-positive cells, which will dramatically reduce the time required for cell sorting.
Compensation is the process by which the fluorescence “spill-over” originating from a fluorochrome other than the one specified for a particular channel detector is subtracted as a percentage of the signal from another channel detector. To set the fluorescence compensation, the percentage (%) compensation/subtraction is adjusted while observing a display of data being run on a two-color histogram for every channel. Compensation is correct when an imaginary line drawn across the top of the negatives and positives is level. Compensation may not be correctly set up with dimly stained cells. If the voltage is subsequently changed on any of the four detectors, or the detection filters are changed, the compensation procedure must be repeated.
Twenty-four hours prior to transfection, plate approximately 3–4 × 106 293 cells in 8 mL of 293T media in a 60-mm tissue culture dish. The culture should be approximately 40% confluent for transfection. Less or more confluence will reduce the transfection efficiency. It is necessary to wait for approximately 36 h post transfection to obtain high-titer retroviral supernatants. It may be possible to harvest and replenish the supernatant every 12 h up to 72 h post transfection without a significant drop in retroviral titer. Retroviral half-life is 3–6 h at 37°C (10). To maintain high titers, supernatants should be aliquoted and frozen (−80°C) following harvest. Freezing does not appear to cause more than twofold drop in titer. To thaw frozen retroviral supernatants, warm for a few minutes in a 37°C water bath and immediately apply onto the cells.
aRNA quality is important to obtaining reproducible microar-ray data. The reverse transcription step of RNA amplification is the key for generating high yields of aRNA with minimal change to the initial representation of message abundance in total RNA sample (11). RNA with trace contaminants are poorly reverse-transcribed and yield less aRNA than pure samples. Therefore it is critical to use a purification method that yields RNA free of contaminants. The protocols used in this report have been optimized and the two rounds of reactions are carried through, facilitating potent amplification of total RNA from samples containing as few as 5 × 103 cells. Moreover, it is important to ensure that each reaction sample contains the same mass amount of total RNA. Differences in the amount of total RNA will affect aRNA yields and amplification efficiency. Furthermore, there are commercial kits such as Ribo-SPIA RNA amplification kit (Nugen) and MessageAmp II aRNA Amplification Kit (Ambion) that also work well for RNA amplification.
Heat 10× buffer (Ambion) at 55°C to dissolve precipitates, and then cool to room temperature.
References
- 1.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 2.Na Nakorn T, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest. 2002;109(12):1579–85. doi: 10.1172/JCI15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–7. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 5.Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strobber W. Current protocols in immunology, isolation of murine macrophages. New York: Wiley; 1994. [Google Scholar]
- 6.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 8.Tamayo P, Scanfeld D, Ebert BL, Gillette MA, Roberts CW, Mesirov JP. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci U S A. 2007;104(14):5959–64. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 10.Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5(12):3133–42. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87(5):1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]