Abstract
Cutaneous melanoma originates from pigment producing melanocytes or their precursors and is considered the deadliest form of skin cancer. For the last 40 years, few treatment options were available for patients with late-stage melanoma. However, remarkable advances in the therapy field were made recently, leading to the approval of two new drugs, the mutant BRAF inhibitor vemurafenib and the immunostimulant ipilimumab. Although these drugs prolong patients' lives, neither drug cures the disease completely, emphasizing the need for improvements of current therapies. Our knowledge about the complex genetic and biological mechanisms leading to melanoma development has increased, but there are still gaps in our understanding of the early events of melanocyte transformation and disease progression. In this review, we present a summary of the main contributing factors leading to melanocyte transformation and discuss recent novel findings and technologies that will help answer some of the key biological melanoma questions and lay the groundwork for novel therapies.
Keywords: Molecular biology, oncogenes, tumor biology
Traditional targets and therapies
Melanoma is the most aggressive form of skin cancer, causing 50,000 deaths annually worldwide, and its incidence continues to increase. If the tumor is detected early, before it has invaded into the dermis, surgical excision provides a cure in about 99% of patients. However, the 5-year survival rate falls to 15% and to a median survival of 1 year for those with advanced, disseminated disease (1).
Until recently, there were few approved and successful treatment options for advanced-stage melanoma. Common treatment strategies included conventional chemotherapy with dacarbazine (DTIC) and the cytokines interleukin-2 (IL-2) and interferon-a2b (IFN-a2b). While only 5%–10% of the patients respond to DTIC treatment, IL-2 treatment with the use of IFN-a2b as adjuvant immunotherapy achieves a response in 10%–20% of patients (1). Unfortunately, these responses are generally short-lived and associated with high toxicity. Encouragingly, in recent years the identification of the main genetic aberrations and signaling pathways involved in melanocyte transformation and disease progression has resulted in the development of novel, more effective therapeutic approaches.
The first clues about the molecular basis of melanoma came from studies of families prone to the disease and the identification of two autosomal-dominant high-susceptibility loci: cyclin-dependent kinase inhibitor 2A (CDKN2A) and cyclin-dependent kinase 4 (CDK4). The CDKN2A locus encodes two tumor suppressor proteins, p16INK4a and p14ARF. While p16INK4a is an inhibitor of the cyclin-dependent kinases CDK4 and CDK6 and prevents S-phase entry during cell cycle, p14ARF acts as a positive regulator of p53. Deletions of the CDKN2A locus have been found in up to 50% of melanomas, but inactivation of this locus can also occur through mutations and promoter hypermethylation (2-4). Although undoubtedly important for melanoma development, in terms of therapy, direct targeting and restoring of the function of tumor suppressor proteins have been inherently difficult.
Possibilities for novel therapeutic options came with the realization that the mitogen activated protein kinase (MAPK) pathway is a crucial regulator of melanoma development. In fact, activation of this pathway regulates both proliferation and survival of melanoma cells. The underlying mechanism of MAPK deregulation is attributed to activating mutations in NRAS and BRAF genes, resulting in a constitutive activation of the pathway. In addition, autocrine growth factors, adhesion molecule signaling, and morphogen signaling all contribute to MAPK pathway activation. While NRAS mutations are observed in 15%–25% of melanomas, BRAF is mutated in as many as 50% of the cases (5). In addition, more than 95% of the mutations in BRAF affect a valine residue at the 600 amino acid position (BRAFV600E), while the NRAS mutation is most often a glutamine-to-arginine substitution at position 61 (NRASQ61R).
This increased knowledge about the molecular changes in melanoma led to the application of sorafenib, the first agent targeting the MAPK pathway. Disappointingly, in phase II trials, sorafenib demonstrated poor ability to inhibit MAPK pathway activation and little to no clinical efficacy as a single agent. The real advance came with the development of more potent and selective BRAFV600E inhibitors including vemurafenib (PLX4032). Clinical results from phase I, II, and III trials showed that vemurafenib treatment caused complete or partial tumor regression in 80% of patients carrying BRAFV600E tumors as well as an increased progression-free survival rate (6,7). Given this success, several other pharmacological inhibitors targeting proteins of the MAPK pathway, including MEK and ERK, are being developed (8). Unfortunately, after an initial period of response to vemurafenib, most patients relapse and develop resistance to the drug (7). Resistance can be acquired through several mechanisms including activation of the serine/threonine kinase COT and RAF isoform switching (9). Furthermore, we and others have shown that the elevation of insulin-like growth factor receptor (IGFR) or platelet-derived growth factor receptor (PDGFR) confer resistance to BRAFV600E inhibitors through activation of the PI3K/Akt signaling pathway (10-12), collectively suggesting that targeting of the PI3K/Akt signaling pathway along with the MAPK pathway might provide a new and more effective therapeutic approach for melanoma treatment.
Melanoma represents a heterogeneous group of neoplasms with variable genotypic and phenotypic traits based on anatomic location, degree of sun exposure, and individual susceptibility. For instance, several genetic studies indicate that acral and mucosal melanomas develop through different etiological pathways than cutaneous melanomas. In these subtypes, the MAPK pathway is not activated through the same mutation as in the cutaneous counterparts. While BRAF mutations are significantly less frequent in these melanoma subtypes, activating mutations in the KIT gene are often seen. KIT encodes a receptor tyrosine kinase (c-Kit) that plays an important role in the development, proliferation, and survival of melanocytes (13). The therapeutic potential of targeting c-KIT in this subgroup of melanomas was validated by two clinical studies where patients with activating mutations in c-KIT showed significant responses to the c-KIT inhibitor imatinib (14,15); however, overall clinical responses to this inhibitor are less pronounced than to BRAF inhibitors.
Numerous other molecular alterations contribute to the complexity of melanoma biology, including mutations of receptor tyrosine kinases ERBB4 and EPH, activation of vascular endothelial growth factor receptors (VEGFR), deregulation of p53, MITF expression, and the developmental signaling pathways Notch and Wnt (16-19). However, attempts to target these pathways therapeutically have not been successful so far.
Improved understanding of tumor immunobiology has also provided novel treatment approaches for melanoma. Ipilimumab is a monoclonal antibody that augments T-cell activation and proliferation by blocking the cytotoxic T-lymphocyte antigen-4, a critical negative regulator of the antitumor T-cell response. In advanced-stage melanoma patients, treatment with ipilimumab resulted in a 20% increased survival up to 4 years after treatment (20). Nevertheless, only a fraction of patients receive durable benefits from ipilimumab therapy.
Thus, even though ipilimumab and vemurafenib have created enthusiasm in the melanoma therapeutic field, it is evident that further improvements are necessary. Likely, the key to further success in therapy lies in combination therapies, in which two or more drugs are combined. The lack of good therapeutic targets outside of the MAPK pathway also underlines the need for further analysis of currently known drivers of melanocyte transformation and melanoma progression as well as identification of new ones.
The path to melanocyte transformation
Melanocytes develop during embryogenesis from melanoblastic precursors that migrate from the neural crest to populate the epidermis, hair follicles, cochlea, and the uveal tract of the eye (21). Once situated in the epidermis, melanocytes remain under tight control by keratinocytes and proliferate only after stimulation by paracrine factors secreted by the keratinocytes (22). Development and progression of melanoma depends on a set of genetic alterations that allow melanocytes to escape regulation by keratinocytes, migrate, invade, and survive in a ‘hostile’ environment. These alterations are termed ‘drivers’ of the disease. In addition to these alterations, the cells also acquire many secondary or ‘passenger’ alterations as a result of general genome instability, which is particularly seen in melanoma cells. The ability to distinguish the essential driver from passenger mutations is critical to the development of effective therapies.
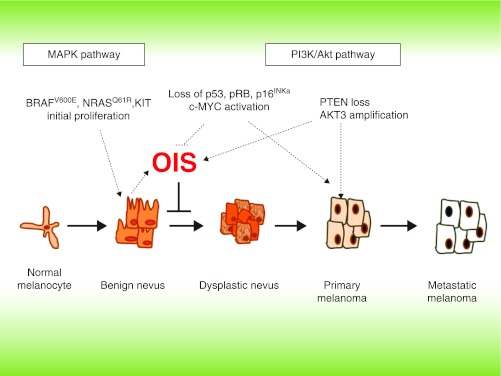
Acquisition of the BRAFV600E mutation is considered an early event in the initiation of melanocytic neoplasia but in itself is not sufficient for full transformation (Figure 1). For example, overexpression of BRAFV600E in melanocytes of transgenic zebrafish and mice induces nevus-like cell growth but does not lead to melanoma development (23,24). In normal human melanocytes (NHM), ectopic introduction of BRAFV600E in vitro leads to cellular senescence, an irreversible form of cell cycle arrest (25). Senescence is often characterized by phenotypic features, such as increased senescence-associated β-galactosidase (SA-β Gal) activity and elevated levels of negative regulators of the cell cycle, such as p16INK4a, p53, and p21CIP/WAF (25,26). Interestingly, both p16INK4a and p53 were reported to be dispensable for BRAFV600E-induced senescence in NHM, and the exact mechanisms of this oncogene-induced senescence (OIS) are still not fully understood (27,28). However, it is assumed that OIS represents a major barrier to oncogenic transformation which makes OIS mechanisms important for further studies. Benign nevi, which are considered non-proliferative melanocyte lesions, represent the best in vivo evidence of OIS. BRAFV600E mutations are found in 60% of benign nevi and are thought to result in the initial increase in proliferation of melanocytic cells followed by induction of cell cycle arrest and senescence (29).
Figure 1.
Genetic alterations leading to bypass of oncogene-induced senescence (OIS) and transformation of melanocytes.
In addition to BRAFV600E, overexpression of constitutively active mutants of RAS, most commonly HRASG12V and NRASQ61R, also leads to OIS in NHM. However, there are differences between mechanisms of OIS induction in each case. For example, HRASG12V-specific senescence can be mediated by the endoplasmatic reticulum (ER)-associated unfolded protein response (UPR) (27). Moreover, p16INK4a, p14ARF, and p53 do not appear to play a prominent co-operating role in the HRASG12V-induced senescence. In the case of NRASQ61R, contribution of both p53 and pRb is evident due to the fact that depletion of either factor makes NHM refractory to senescence induction (30). Collectively, these observations underline mutation-specific differences in senescence induction. Interestingly, mutation specificity is also seen in melanoma cells and has to be taken into consideration when evaluating therapeutic approaches. In melanoma cells RAS mutations are not equivalent to BRAF mutations since they activate the MAPK pathway in different ways. While BRAF mutations result in direct phosphorylation and activation of MEK, RAS mutations induce a switch in signaling from BRAF to CRAF, allowing CRAF to signal to MEK (31,32).
Since approximately 30% of melanomas arise from nevi, these cells must have acquired additional alterations during the transformation process allowing them to overcome OIS. Recently it has been shown that the ectopic expression of the oncogenic transcription factor c-MYC can significantly suppress OIS in NHM (28). A similar effect is obtained by depletion of the B56α subunit of the PP2A tumor suppressor complex, which also leads to the up-regulation of endogenous c-MYC (33). Hömig-Hölzel et al. demonstrated that suppression of the transcription factor and putative tumor suppressor gene TGFβ-stimulated clone 22 (TSC22D1) allows NHM to bypass BRAFV600E-induced senescence (34). Another protein recently reported to modulate OIS in NHM is pirin, a highly conserved nuclear protein belonging to the cupin superfamily14. Down-regulation and overexpression experiments showed that pirin is involved in the negative control of OIS and that its expression is necessary to overcome the senescence barrier (35). In addition, pirin controls melanoma cell migration through the transcriptional regulation of snail homolog 2 (SNAI2) (36).
It is interesting to speculate whether acquisition of the alterations allowing melanocytes to bypass OIS is sufficient for the development of the malignant phenotype. In animal models, BRAFV600E induces melanoma when combined with p16INK4a/p14ARF or p53 deficiency, as well as when combined with the activation of β-catenin that leads to silencing of p16INK4a (24,37-40). However, Dhomen et al. recently reported that BRAFV600E can induce melanoma in mice without p16INK4a inactivation. Still, these lesions appeared after a long latency period, and it is possible that additional unidentified genetic alterations have occurred during this time (23).
Previously it has been shown that NHM immortalized with the oncogenic simian virus 40 (SV40) large-T, human telomerase reverse transcriptase, and transduced with either HRASG12V or c-Met, gain the ability to form tumors in vivo (41). In addition, combined expression of MITF and BRAFV600E also led to transformation of NHM (42). In our in vitro models, transformation of NHM without immortalization by the SV40 can be achieved by combining BRAFV600E and inactivating both the p53 and Rb pathways (43).
Increasing evidence supports the hypothesis that activation of the PI3K/Akt pathway, which acts upstream of both β-catenin and c-MYC, may also be involved in melanocyte senescence suppression and transformation (28,44). Chudnovsky et al. showed that invasive human melanocytic neoplasia can be induced by activation of PI3K when combined with Rb and p53 inhibition as well as hTERT expression (45). Interestingly, in the same study, BRAFV600E did not show the same oncogenic potential as PI3K, suggesting that the effect of different genes on transformation depends on specific combinations and the timing of mutational events as well as the experimental model used. In contrast, expression of BRAFV600E combined with PTEN gene silencing induced melanomas with 100% penetrance, short latency, and metastatic disease in a mouse model (44). In NHM, however, PTEN deficiency combined with BRAFV600E activation induced a melanoma in situ-like phenotype without dermal invasion in organotypic human skin culture. Only after further addition of cell autonomous TGFβ did these lesions develop an invasive phenotype (46).
In addition to the MAPK and PI3K/Akt pathways, other signaling networks are likely important for OIS escape and melanocyte transformation. The Notch signaling pathway, for example, plays a critical role in the proper development of melanocytes from neural crest precursors (reviewed in (47)). Upon ligand activation, Notch receptors are proteolytically cleaved, resulting in the liberation of the intracellular domain (NIC) that translocates into the nucleus and initiates transcription of target genes. Transduction of primary melanocytes with this intracellular domain leads to activation of the Notch signaling pathway and induces a transformed phenotype in vitro (48). However, NIC-transfected cells do not form tumors in NOD-SCID mice, suggesting that Notch overexpression in itself is not sufficient for neoplastic transformation.
Future perspectives; new knowledge through new models
The identification of novel drivers of OIS escape and melanocyte transformation and the further characterization of melanoma based on genetic subgroups are critical for improving current therapies. In the last few years, complex melanoma genetics have been addressed by extensive genome-wide association studies (GWAS). By comparing frequencies of genetic polymorphisms, between melanoma patients and healthy individuals, these studies aim to identify novel melanoma susceptibility genes. Most of the studies have analyzed single nucleotide polymorphisms (SNPs).The main advantage of this approach is the capability for highly detailed and robust data collection. In melanoma, GWAS validated the importance of the well-known, high-penetrance alleles CDKN2A and CDK4, while identifying several novel low-risk alleles associated with increased melanoma risk, including ATM, MX2, and CASP8 (49,50). Furthermore, SNPs in the TERT and TRF1 genes, that play important roles in the regulation of telomerase activity and telomere length, are significantly associated with an increased melanoma risk (51). Interestingly, GWAS also found a novel TERT locus (TERT CLPTM1) that is positively associated with shorter telomere length and is inversely associated with the melanoma risk. Previously, it has been shown that there is a positive association between the number and size of nevi and telomere length (52,53). Few studies have addressed the role of telomerase in evasion of OIS and transformation of melanocytes, although recently Soo et al. reported that deregulation of telomerase function occurs late in the disease progression and not in early stages (54). Another susceptibility locus was identified at 1q21.3 that spans a region with ten genes, of which several are good candidate genes for melanoma susceptibility, including ARNT (Hif1β) and SETDB1 (49,55). In fact, it was shown that SETDB1, which methylates histone H3 on lysine 9 (H3K9), significantly accelerates melanoma formation in zebrafish (56).
Additional advances in melanoma genetics have also been made with improved next generation sequencing technology. This allowed the comparison of the complete DNA sequence of cancer cells to their normal counterparts. Pleasance et al. catalogued the complete spectrum of somatic mutations and rearrangements in the metastatic cell line COLO-829 using a lymphoblastoid cell line from the same patient as a reference (57). This resulted in the identification of an astonishing total of 33,345 base substitutions of which 292 were somatic mutations (187 non-synonymous and 105 synonymous) in protein coding sequences. Since the ratio between non-synonymous and synonymous mutations did not significantly exceed random expectation, the majority of these mutations were catalogued as ‘passengers’. Interestingly, 70% of these mutations could be traced back to DNA damage induced by ultraviolet radiation, emphasizing its dominating role in melano magenesis. Previously identified driver mutations in CDKN2A, BRAF, and PTEN genes were confirmed in this study, while the oncogenes GLI1, ETV5, and tumor suppressor genes DCC and TP63 were suggested as novel potential drivers. In addition, mutations were seen in the matrix metalloproteinase MMP28 gene. Other novel but currently poorly characterized potential ‘driver’ genes included the transcription factor encoding SPDEF, the serine/threonine kinase 19 encoding STK19 and XIRP2, which encodes xin actin-binding repeat containing 2.
Collectively, GWAS and the next generation sequencing studies have identified several novel genes that might play a role in melanoma development and progression. However, extensive validation of these genes will be necessary to elucidate the mechanisms by which they contribute to melanocyte transformation. In order to perform such validations we must use and develop biologically adequate experimental models. Most of our current knowledge is based on in vitro studies, experimental grafting, and transgenic animal melanoma models. Difficulties in accessing relevant biological materials and recapitulating genetic complexity have hampered the direct analysis of disease progression in humans. This could possibly be overcome in the near future by the use of human induced pluripotent stem cells (hIPSCs). These pluripotent cells acquire most of the features of embryonic stem cells (ESCs) and can be differentiated into any cell type, including melanocytes (58).
Recently, Nissan et al. demonstrated that use of the cytokine bone morphogenic protein 4 allows generation of fully functional melanocytes from pluripotent stem cell lines of either induced or of embryonic origin (59). These melanocytes exhibit all the characteristic features of their adult counterparts, including the enzymatic machinery required for the production and functional delivery of melanin to keratinocytes. Furthermore, these cells integrate appropriately into an organotypic epidermis reconstructed in vitro. By applying such hIPSC technology, we will be able to obtain melanocytes from patients with genotypes of particular interest via a simple skin biopsy using easily accessible fibroblasts; hIPSC-derived melanocytes will retain the patients' complex genetic background, making it possible to study the impact of the genotype on disease development under a variety of experimentally controlled conditions, which would lead to a substantially better understanding of melanoma biology. Furthermore, using melanocytes in the correct genetic background would allow for testing targeted therapies within the context of proper host cells and thus provide new opportunities for target identification and validation.
Acknowledgements
Parts of these studies were funded by NIH grants CA 25874, CA 114046, CA 076674, and CA 047159 and the Norwegian Cancer Society.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 2.Monahan KB, Rozenberg GI, Krishnamurthy J, Johnson SM, Liu W, Bradford MK, et al. Somatic p16(INK4a) loss accelerates melanomagenesis. Oncogene. 2010;29:5809–17. doi: 10.1038/onc.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 4.Meyle KD, Guldberg P. Genetic risk factors for melanoma. Hum Genet. 2009;126:499–510. doi: 10.1007/s00439-009-0715-9. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Fisher DE. New strategies in metastatic melanoma: oncogene-defined taxonomy leads to therapeutic advances. Clin Cancer Res. 2011;17:4922–8. doi: 10.1158/1078-0432.CCR-10-2612. [DOI] [PubMed] [Google Scholar]
- 6.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inamdar GS, Madhunapantula SV, Robertson GP. Targeting the MAPK pathway in melanoma: Why some approaches succeed and other fail. Biochem Pharmacol. 2010;80:624–37. doi: 10.1016/j.bcp.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Herlyn M. Resistance to BRAF Inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 2011;71:7137–40. doi: 10.1158/0008-5472.CAN-11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulikakos PI, Rosen N. Mutant BRAF melanomas—dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 14.Hodi FS, Friedlander P, Corless CL, Heinrich MC, Mac Rae S, Kruse A, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008;26:2046–51. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 15.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21:492–3. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 16.Box NF, Terzian T. The role of p53 in pigmentation, tanning and melanoma. Pigment Cell Melanoma Res. 2008;21:525–33. doi: 10.1111/j.1755-148X.2008.00495.x. [DOI] [PubMed] [Google Scholar]
- 17.Easty DJ, Gray SG, O'Byrne KJ, O'Donnell D, Bennett DC. Receptor tyrosine kinases and their activation in melanoma. Pigment Cell Melanoma Res. 2011;24:446–61. doi: 10.1111/j.1755-148X.2011.00836.x. [DOI] [PubMed] [Google Scholar]
- 18.O'Connell MP, Weeraratna AT. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment Cell Melanoma Res. 2009;22:724–39. doi: 10.1111/j.1755-148X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strizzi L, Hardy KM, Kirsammer GT, Gerami P, Hendrix MJ. Embryonic signaling in melanoma: potential for diagnosis and therapy. Lab Invest. 2011;91:819–24. doi: 10.1038/labinvest.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernfors P. Cellular origin and developmental mechanisms during the formation of skin melanocytes. Exp Cell Res. 2010;316:1397–407. doi: 10.1016/j.yexcr.2010.02.042. [DOI] [PubMed] [Google Scholar]
- 22.Haass NK, Herlyn M. Normal human melanocyte homeostasis as a paradigm for understanding melanoma. J Investig Dermatol Symp Proc. 2005;10:153–63. doi: 10.1111/j.1087-0024.2005.200407.x. [DOI] [PubMed] [Google Scholar]
- 23.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 24.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–54. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Bansal R, Nikiforov MA. Pathways of oncogene-induced senescence in human melanocytic cells. Cell Cycle. 2010;9:2782–8. doi: 10.4161/cc.9.14.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death—apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 27.Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–63. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–34. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 30.Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging (Albany NY) 2009;1:542–56. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–91. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 32.Pavey S, Johansson P, Packer L, Taylor J, Stark M, Pollock PM, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23:4060–7. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 33.Mannava S, Omilian AR, Wawrzyniak JA, Fink EE, Zhuang D, Miecznikowski JC, et al. PP2A-B56alpha controls oncogene-induced senescence in normal and tumor human melanocytic cells. Oncogene. 2011 Aug 8; doi: 10.1038/onc.2011.339. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homig-Holzel C, van Doorn R, Vogel C, Germann M, Cecchini MG, Verdegaal E, et al. Antagonistic TSC22D1 variants control BRAF(E600)-induced senescence. EMBO J. 2011;30:1753–65. doi: 10.1038/emboj.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licciulli S, Luise C, Scafetta G, Capra M, Giardina G, Nuciforo P, et al. Pirin inhibits cellular senescence in melanocytic cells. Am J Pathol. 2011;178:2397–406. doi: 10.1016/j.ajpath.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyazaki I, Simizu S, Okumura H, Takagi S, Osada H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat Chem Biol. 2010;6:667–73. doi: 10.1038/nchembio.423. [DOI] [PubMed] [Google Scholar]
- 37.Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci USA. 2007;104:10968–73. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 2007;21:2923–35. doi: 10.1101/gad.450107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann J, Beermann F. The fibroblast growth factor-2 is not essential for melanoma formation in a transgenic mouse model. Pigment Cell Res. 2005;18:315–19. doi: 10.1111/j.1600-0749.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 40.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–34. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 43.Yu H, McDaid R, Lee J, Possik P, Li L, Kumar SM, et al. The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am J Pathol. 2009;174:2367–77. doi: 10.2353/ajpath.2009.081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet. 2005;37:745–9. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo RS, Witte ON. Transforming growth factor-beta activation promotes genetic context-dependent invasion of immortalized melanocytes. Cancer Res. 2008;68:4248–57. doi: 10.1158/0008-5472.CAN-07-5671. [DOI] [PubMed] [Google Scholar]
- 47.Schouwey K, Beermann F. The Notch pathway: hair graying and pigment cell homeostasis. Histol Histopathol. 2008;23:609–19. doi: 10.14670/HH-23.609. [DOI] [PubMed] [Google Scholar]
- 48.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–20. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20:5012–23. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43:1108–13. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nan H, Qureshi AA, Prescott J, De Vivo I, Han J. Genetic variants in telomere-maintaining genes and skin cancer risk. Hum Genet. 2011;129:247–53. doi: 10.1007/s00439-010-0921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bataille V, Kato BS, Falchi M, Gardner J, Kimura M, Lens M, et al. Nevus size and number are associated with telomere length and represent potential markers of a decreased senescence in vivo. Cancer Epidemiol Biomarkers Prev. 2007;16:1499–502. doi: 10.1158/1055-9965.EPI-07-0152. [DOI] [PubMed] [Google Scholar]
- 53.Han J, Qureshi AA, Prescott J, Guo Q, Ye L, Hunter DJ, et al. A prospective study of telomere length and the risk of skin cancer. J Invest Dermatol. 2009;129:415–21. doi: 10.1038/jid.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soo JK, Mackenzie Ross AD, Kallenberg DM, Milagre C, Heung Chong W, Chow J, et al. Malignancy without immortality? Cellular immortalization as a possible late event in melanoma progression. Pigment Cell Melanoma Res. 2011;24:490–503. doi: 10.1111/j.1755-148X.2011.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet. 2011;43:1114–18. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–17. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang R, Jiang M, Kumar SM, Xu T, Wang F, Xiang L, et al. Generation of melanocytes from induced pluripotent stem cells. J Invest Dermatol. 2011;131:2458–66. doi: 10.1038/jid.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nissan X, Larribere L, Saidani M, Hurbain I, Delevoye C, Feteira J, et al. Functional melanocytes derived from human pluripotent stem cells engraft into pluristratified epidermis. Proc Natl Acad Sci USA. 2011;108:14861–6. doi: 10.1073/pnas.1019070108. [DOI] [PMC free article] [PubMed] [Google Scholar]