Abstract
The packaging of DNA with histones to form chromatin represents an important and powerful mechanism to regulate gene expression. Critical aspects of chromatin specific contributions to gene regulation have been revealed by the comparison of the activities from DNA regulatory elements examined both as transiently transfected reporters and stably integrated reporters organized as chromatin. Using the mouse mammary tumor virus promoter as a model, we probed the structural differences between transiently transfected and stably integrated DNA templates. We demonstrated that all four core histones and the linker histone (H1) are associated with the transient template. However, while the core histones were present at a similar stoichiometry between the transient and the stable templates, we found that linker histone H1 molecules are fewer on the transient template. By using supercoiling assay, we show that the transient template shows intermediate levels of nucleosomal assembly. Over expression of H1 resulted in repression of MMTV transcriptional activity and reduced accessibility to restriction endonucleases on the transient MMTV promoter. However, the addition of exogenous H1 failed to impose a normal chromatin structure on the transient template as measured by micrococcal nuclease digestion pattern. Thus, our results suggest that while transiently transfected DNA acquires a full complement of core histones the under representation of H1 on the transient template is indicative of structural differences between the two templates that may underlie the differences in the mechanisms of activation of the two templates.
Transcriptional regulation of eukaryotic promoters is extensively studied by using transient transfection-reporter assays. Although these assays have given abundant information on transcription factor binding sites and the effects of various treatments on transcription, it is undetermined how closely they emulate the activity of endogenous promoters. One of the main concerns about using transient transfection to assay promoter activity is the lack of proper chromatin architecture on this DNA (1). This seems particularly important given the impact of chromatin, histone modifications and chromatin remodeling proteins on transcriptional regulation
Using the glucocorticoid responsive mouse mammary tumor virus (MMTV) promoter as a model promoter, previous work has demonstrated very clear differences between the stably integrated, replicating copies of the promoter and transiently transfected promoter DNA (2). The MMTV promoter is activated by the hormone bound glucocorticoid receptor (GR). Although both the stable and the transient MMTV promoters are dependent on hormone for transcriptional activity, accessibility to restriction endonucleases on the promoter is hormone inducible on the stable template and constitutively accessible on the transient template. This architectural difference was attributed to the lack of proper nucleosomal organization on the transient template as assayed by micrococcal nuclease digestion (2). Recent work using MMTV promoter has demonstrated other such differences between the two templates. BRG1 chromatin remodeling complex was shown to be necessary for activation of the stably integrated promoter but not on the transient promoter (3). Over expression of NFκB relA subunit leads to repression of GR mediated transcriptional activity only from the stable promoter and not from the transiently transfected promoter (4). More recently, we demonstrated that cooperative binding between two site-specific transcription factors, GR and NF1, on the MMTV promoter occurs only on stably transfected and not on the transiently transfected promoter (5). Differences in transcriptional activity between the templates have not been limited to MMTV and have been used to study MyoD activated genes (6).
In this study, we initiated a detailed investigation of the structural differences between these two templates. We demonstrate that the stoichiometry of all the core histones is similar in both these templates but the linker histone H1 is under represented on the transient template. Linker histone H1, acts as a generalized transcriptional repressor on transiently transfected MMTV promoter when over expressed. Under conditions of H1 over expression, there is alteration of the chromatin conformation as measured by reduced accessibility of transient template to restriction endonucleases. These conformational changes on the transient template do not result in regular nucleosomal organization.
Experimental Procedures
Plasmids
pLTRluc has MMTV promoter fused to luciferase reporter gene (7). pCDNA3-H1.3 has the mouse H1.3 cDNA (from 1471.1 cells) cloned in pCDNA3.1 mammalian expression plasmid which over expresses H1.3 with a flag tag at the C-terminal end of the protein.
Cell culture and transfections
C127 is a mouse mammary carcinoma cell line and 1471.1 is its derivative which maintains ~ 300 copies of a bovine papilloma virus with the MMTV LTR fused to chloramphenicol acetyl transferase (CAT) reporter gene (3). Both these cell lines were grown at 37°C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2mM glutamine, 100μg/ml penicillin/streptomycin and 10mM HEPES. Cells were treated with a synthetic glucocorticoid dexamethasone (dex) (10−7M). Transient transfections were performed by the Lipofectamine PLUS method (Invitrogen, Carlsbad, CA) as described by the manufacturer.
Nucleosome assembly assay
Relaxed form of plasmid DNA (Topoisomerase treated) were prepared by treatment with E.coli Topoisomerase I. C127 cells were transfected with supercoiled, topoisomerase treated, linear pLTRluc plasmid for 24hrs. Isolated nuclei and proteinase K treated overnight. Genomic DNA was purified genomic DNA by phenol chloroform extractions and ethanol precipitation. The genomic DNA was digested with KpnI (no KpnI site in pLTRluc) for ease in loading the gel. Two micrograms of DNA was electrophoresed on a 0.8% Agarose TPE gel (8) with 50 μg/ml chloroquine for 48 hrs in the dark and blotted. The blot was probed with vector specific probe (~ 1.0 kb XbaI – AvaI fragment of the luciferase gene).
DNA replication assay
1471.1 cells were transfected with pLTRLuc for 48 hrs and genomic DNA was isolated by proteinase K digestion followed by phenol chloroform extractions and ethanol precipitation. Genomic DNA was digested to completion using DpnI restriction enzyme. Purified DNA was subjected to PCR using Oligo-344 (5′-TTA AGT AAG TTT TTG GTT ACA AAC T-3′) and Luc-2 (5′-ATT TTA CCA ACA GTA CCG GAA TGC-3′) primer pair (to assay transient template) or Oligo-344 and CAT-2 (5′-GAA CGG TCT GGT TAT AGG TAC ATT GA-3′) (for stable template). The PCR products were electrophoresed on a 1.5% agarose gel.
Chromatin immunoprecipitation assay (ChIP)
Cells were grown on 150mm plates and were fixed with 1% formaldehyde for 10 min at room temperature. Nuclei were isolated as previously described (9). Isolated nuclei were subjected to HaeIII restriction enzyme digestion (4000U/ml) for 15 min at 30°C in 450 μl of restriction enzyme buffer supplied by the manufacturer. Nuclear extracts were prepared by adding 150 μl of lysis buffer (4% Sodium dodecyl sulfate (SDS), 40 mM Ethylenediaminetetraacetic acid (EDTA), 200 mM Tris-HCl, pH 8.0) and incubating on ice for 10 min. The rest of the ChIP procedure was followed as described earlier (10). Antibodies used for ChIP assay are: histone H2A (07-146), H2B (07-371), H3 (05-928) H4 (05-858) and H1 (clone AE-4, 05-457) antibodies (Upstate Biotechnology, Upstate, NY) and normal serum immunoglobulin G (IgG) (Santa Cruz Biotech, Santa Cruz, CA). After immunoprecipitation, the eluted samples were reverse crosslinked for 12 hours at 65°C, treated with proteinase K (Invitrogen) and purified using PCR purification kit (Qiagen, Valencia, CA). Real time PCR was performed on ChIP DNA samples using MMTV template specific (transient and stable) real time PCR primers. For assaying transient template, primer pair (luciferase gene, luc-1 and luc-2) 5′-CCG CTC GTC ACT TAT CCT TCA-3′ and 5′-ATT TTA CCA ACA GTA CCG GAA TGC- 3′ and for stable template, primer set (CAT gene, CAT-1 and CAT-2) 5′-TCT GAG CTT GGC GAG ATT TTC- 3′ and 5′-GAA CGG TCT GGT TAT AGG TAC ATT GA-3′ were used.
Luciferase assay and chloramphenicol acetyl transferase (CAT) assays
Cells were grown in 6-well plates and were washed twice with PBS and lyzed on the plate with 500 μl Reporter Lysis buffer (Promega, Madison, WI) per well. The plates were scraped and lysates were pelleted by centrifugation at 20,800 x g for 1 min at room temperature. Twenty microliters of the lysate was added to 100 μl of luciferase substrate and the activity was monitored. Relative light units were normalized to the total protein measured. For CAT assay, 20μl of the lysate was used following manufacturer’s direction (Promega).
Western blot analysis
Nuclei were isolated as described previously (9). Acid-soluble proteins were isolated from nuclei as previously described (11). Histone proteins (40 μg) were electrophoresed using a 14% SDS-Polyacrylamide gel. The gels were either stained with Coomasie blue or transferred to a nitrocellulose membrane (Amersham Biosciences Corp., Piscataway, N.J.) and probed with H1 antibody (Upstate Biotechnology) or Flag antibody (Sigma).
In vivo restriction enzyme accessibility assay
Nuclei were isolated as previously described (9) and digested with SstI (100U/ml), FokI (400U/ml), DdeI (1000U/ml) and HaeIII (1000U/ml). DNA was purified and then digested to completion with HaeIII or AlwN1. The samples were amplified by reiterative primer extension using 32P labeled luc-2 primer and the products were separated on 6% denaturing polyacrylamide gels. The gels were exposed to PhosphorImager screens for analysis. The accessibility to restriction enzymes was calculated as percent in vivo enzyme cleavage of in vitro enzyme digestion.
Micrococcal nuclease analysis
Isolated nuclei were resuspended in 200 μl wash buffer containing 1mM CaCl2 and digested with 20 units/ml micrococcal nuclease (Worthingtom Biochemicals, Lakewood, NJ) for 5 min at 30°C (12). The reaction was stopped by adding 40 μl of 100 mM EDTA pH 8.0, 10mM EGTA pH 7.5. The samples were purified and digested to completion with PstI prior to southern blot. Ten micrograms of micrococcal nuclease digested DNA was separated on a 2% agarose gel and transferred to Hybond N+ membrane (Amersham-Pharmacia, Piscataway, NJ). The blot was probed with a 32P end labeled 84 bp PCR product corresponding to the nucleosome B region of the MMTV promoter (amplified using nucB-1 and nucB-2: 5′-GGT TAC AAA CTG TTC TTA AAA CGA GGA T-3′ and 5′-CAG AGC TCA GAT CAG AAC CTT TGA-3′ primer pairs). The blot was exposed to PhosphorImager screens for analysis.
RESULTS
Nucleosome assembly on transiently transfected DNA
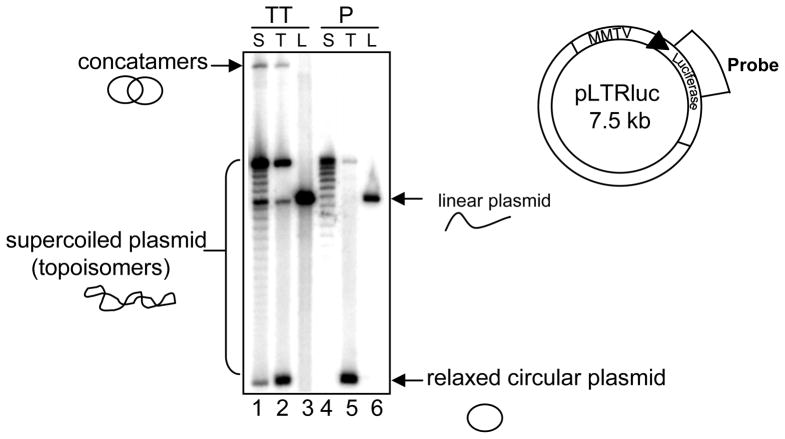
To understand the chromatin architecture of a transiently transfected DNA, we first wanted to assess if nucleosomes associate with plasmid DNA introduced into mammalian cell. Previous work demonstrated that transiently transfected DNA does not assume proper nucleosomal organization as assayed by micrococcal nuclease digestion (2,13). It was also shown that using DEAE dextran method of transfection for 24 hours, relaxed DNA was converted to supercoiled DNA (14) which is indicative of nucleosome deposition. Nucleosome deposition on a circular DNA creates changes in superhelical densities. We wanted to assess changes in linking number of the plasmid DNA after transfection under our experimental conditions. To analyze nucleosome assembly on transient template, we performed supercoiling assay with genomic DNA from C127 cells transfected with topoisomerase treated (relaxed) and untreated pLTRluc (supercoiled). As a control we also transfected linear DNA (pLTRluc digested with a single cutter SstI restriction enzyme) for 24 hours. The various species of plasmid DNA with different linking numbers were separated on chloroquine-agarose gels to analyze their superhelical distribution. In the lane with supercoiled DNA transfection (Fig 1, lane 1), there appears to be some faint bands in between the highly supercoiled and the relaxed form of the plasmid DNA that is not in the original introduced plasmid (Fig 1, lane 4). This suggests some intermediate levels of nucleosomal assembly on the plasmid when introduced in the cell. However, when we introduce relaxed pLTRLuc into cells, we do not observe significant changes from the relaxed DNA originally transfected (Fig 1, compare lanes 2 and 5). We know that relaxed replicating plasmids assume supercoiling using cell free extracts indicating nucleosome deposition (15,16). So the lack of supercoiling on transiently introduced relaxed DNA implies that assembly on this DNA is not similar to what is observed on the stably integrated DNA.
FIG. 1.
Transient template has intermediate levels of nucleosomal assembly. (A) Schematic of pLTRluc plasmid to show the region of the probe. (B) C127 cells were transfected with supercoiled (S), topoisomerase treated (T) or Sst1 restriction enzyme treated (Linear-L) pLTRluc for 24 hrs. Genomic DNA was isolated from nuclei and the DNA was digested with KpnI (lanes 1–3; TT-transiently transfected DNA) (There is no KpnI site in pLTRluc). The DNA was electrophoresed on 0.8% agarose TPE gel with chloroquine 50 μg/ml for 48 hrs in the dark. As controls, supercoiled, topoisomerase treated and linear (digested with Sst1) pLTRluc (lanes 4–6; P- plasmid control) were also loaded on the gel. The gel was blotted and probed with 32P labeled with luciferase specific probe.
Association of histones with the transiently transfected DNA
The results from the supercoiling assay suggests that there is some nucleosomal organization on a transiently transfected DNA. We next wanted to investigate the association of histones with the transiently transfected DNA and to understand if there are differences in the stoichiometry between the histones associated with this template when compared with the stable template. We utilized an experimental set up as previously described (2) followed by chromatin immunoprecipitation assay. In this setup, 1471.1 cells which have ~300 copies of the MMTV promoter fused to the CAT reporter gene on stably replicating episome were transfected with pLTRluc construct which has the MMTV promoter fused to the luciferase reporter gene. This allows us to assay transcription and chromatin remodeling of both templates in the same cell. Under these conditions, earlier work demonstrated that while both the templates were hormone inducible for transcriptional activity, accessibility to restriction endonucleases is hormone inducible on the stable template and constitutive on the transient template. This architectural difference was attributed to the lack of proper nucleosomal organization on the transient template as assayed by micrococcal nuclease digestion.
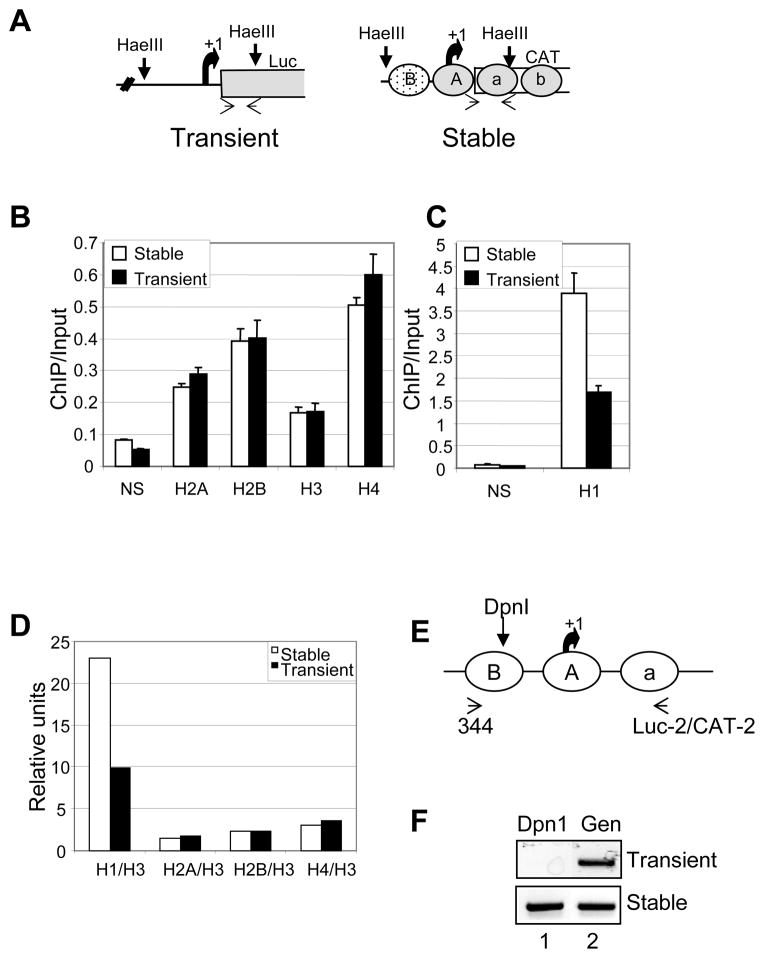
Using the above mentioned experimental set up, we performed ChIP assay with cells transfected with pLTRLuc for 24 hrs. In order to avoid cytoplasmic portion of the transfected DNA, we performed ChIP assay with isolated nuclei. To assess the association of histones on both templates from the same immunoprecipitated samples, we fragmented the DNA with HaeIII restriction endonuclease. HaeIII enzyme digests DNA in the linker regions on the stably integrated MMTV-CAT gene releasing approximately 3 nucleosome length of DNA which encompasses the nuclesome A and B region and part of the coding region of both these reporter genes (17) (Fig 2A). By using reporter specific primers, we can analyze the differences between these two templates. We immunoprecipitated nuclear lysate with all the core histones and linker histone H1 antibodies. We found all the histones associated with the transient template (Fig 2B). Interestingly, the stoichiometries of core histones as assayed by ChIP/input ratio were similar on both the transient and stable template (Fig 2B). This is intriguing given the lack of micrococcal ladder with the transiently transfected DNA (2) and intermediate levels of nucleosome assembly. The association of histones with the transient template was detected as early as 4 hrs after transfection (data not shown). Surprisingly, when we compared ChIP/input ratio of histone H1 on these two templates, we found that there were half the number of histone H1 associated with the transient template when compared with the stable template (Fig 2C and 2D). Since the comparison of histone binding to the two templates is done in the same cell, the effect of transfection on histone binding is not a concern. Taken together, these results suggest that there are similar amounts of core histones associated with the transient template when compared with the stable template, but the linker histone is underrepresented on the transient template.
FIG. 2.
Differences in histone-DNA interactions between transient and stable templates. (A) Schematic to show HaeIII restriction enzyme sites on both pLTRluc (transient) and BPV plasmid containing MMTV-CAT (stable). (B–D) Association of H1 is less on the transient template as compared with the stable template. 1471.1 cells were transfected with pLTRluc for 24 hrs and formaldehyde crosslinked for 10 min at room temperature. Isolated nuclei were digested with HaeIII and the chromatin was immunoprecipitated with the (B) core histone and (C) H1 antibodies. Real time PCR was performed on ChIP samples using template specific primers (as shown in 1A). (D) Relative ratios of ChIP/input values of all histones in comparison with histone H3 values in both stable and transiently transfected DNA templates. (E) Schematic of the MMTV promoter depicting the DpnI restriction endonuclease site and PCR primer sites. (F) 1471.1 cells were transfected with pLTRluc for 48 hrs. Purified DNA was digested to completion using DpnI enzyme. Using the primers shown in the schematic, the region was amplified by PCR and the product was electrophoresed on 1.5% agarose gel. As a control total genomic DNA (uncut with DpnI) was amplified with the same primer sets.
Since the transiently transfected behaves like the stable template with respect to association of core histones, we wanted rule out the possibility of DNA replication on this template. Although pLTRluc does not have a eukaryotic origin of replication, we still wanted to exclude any repair induced DNA replication. To analyze DNA replication on transient template, we isolated genomic DNA from 1471.1 cells transfected with pLTRLuc DNA for 48 hrs. Genomic DNA was subjected to digestion by restriction enzyme DpnI which is methylation dependent (GmATC). The transfected plasmid DNA which is bacterial in origin is digestible but if this DNA undergoes replication in a eukaryotic cell, then the DNA becomes resistant to DpnI digestion due to the loss of methylation. MMTV promoter sequence has DpnI site in the nucleosome B region (Fig 2E). When PCR was performed using template specific primers encompassing the DpnI site, we fail to observe resistance to DpnI digestion upon transfection (Fig 2F). In contrast and as expected the stable template showed complete resistance to DpnI (Fig 2F). This suggests that transiently transfected DNA did not undergo replication even after 48 hrs of transfection.
Another point we wanted to ensure in our ChIP analysis is that it is not limited to a pool of transiently transfected DNA that is associated with octamer proteins. So we compared the ratio of transient template to stable template in ChIP input samples and total genomic DNA and found the ratio to be similar in both these cases (data not shown).
Over expression of H1 leads to repression of transcriptional activity
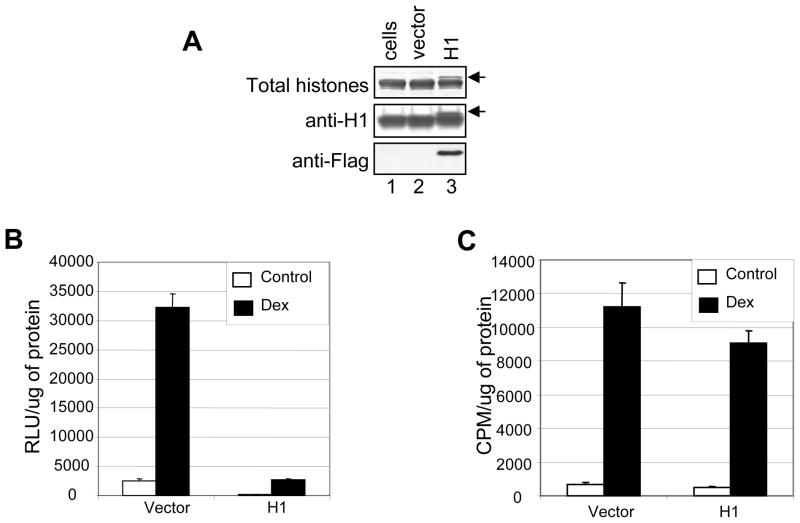
Our data so far shows that histones are associated with the transient template, however there is less histone H1 represented on the transient template when compared with the stable template. Since H1 is implicated in nucleosomal organization and higher order chromatin structure (18–20), we wanted to determine if insufficient levels of H1 in the cells was the cause for the structural differences between these two templates. We over expressed H1 in 1471.1 cells, co-transfected with pLTRLuc DNA (Fig 3A). We observed repression of both basal transcription and glucocorticoid mediated transactivation of the MMTV promoter (Fig 3B) consistent with the idea that H1 is a generalized transcriptional repressor. We also observed repression of transcription from pGL3-control vector with SV-40 promoter fused to luciferase reporter gene (data not shown). When we analyzed transcription from the stable template by performing CAT assay, over expression of H1 had very minimal effect on GR mediated transcriptional activation of the promoter (Fig. 3C). Therefore, H1 over expression results in generalized repression of transcription from a transiently transfected DNA.
FIG. 3.
Over expression of H1.3 construct in 1471 cells. (A) Western blot analysis. 1471.1 cells were transfected with H1 or vector construct and histones were isolated by acid extraction. Acid extracted proteins were electrophoresed on 14% SDS-PAGE, transferred and probed with anti-histone H1 and anti-flag antibodies. Arrow indicates over expressed histone H1 with a decreased mobility due to the C-terminal flag tag. Total histone H1 was obtained by electrophoresing acid extracted histones on a gel and staining with coomasie blue. (B) H1 over expression represses transcription from transiently transfected MMTV promoter. 1471.1 cells were transfected with H1.3 or vector alone constructs and co-transfected with pLTRluc for 24 hrs. The cells were treated with dexamethasone (10−7 M) for 16 hrs. Cell lysates were assayed for luciferase activity. (C) Cell lysates were also assayed for CAT activity.
Over expression of H1 alters chromatin architecture of the transient template
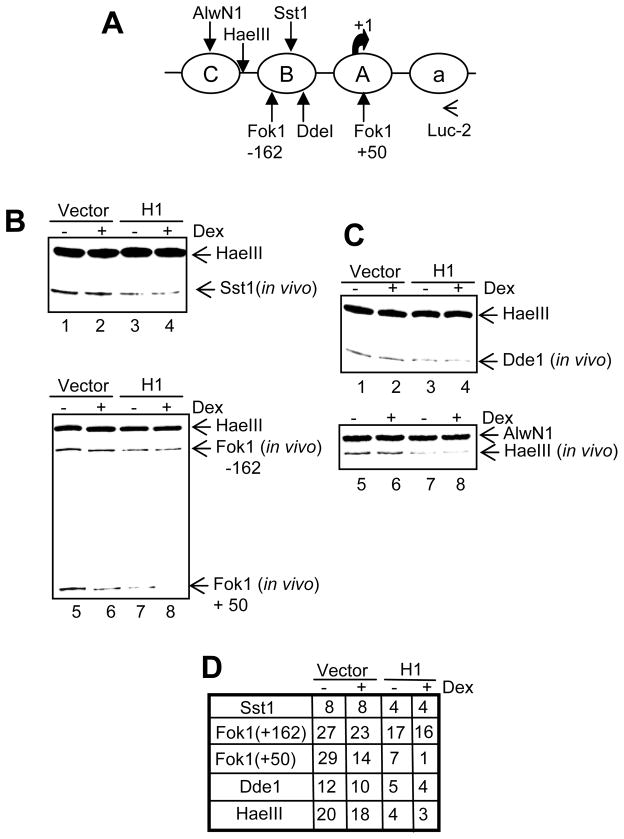
Next we wanted to understand the structural effect of histone H1 over expression on the transiently transfected DNA. Using MMTV promoter, it has been previously shown that while the stable template undergoes chromatin remodeling upon hormone induction as evidenced by increased accessibility to restriction enzymes, the transient template is constitutively accessible to restriction enzymes (2). Given our results thus far, it seems that the constitutive accessibility to restriction enzymes may be due to fluidity in nucleosomal positioning on the transient template. We wanted to examine if over expression of H1 has any effect on the constitutively open chromatin organization of the transiently transfected DNA. To achieve this, we assayed the accessibility of SstI restriction enzyme which has a cleavage site in the nucleosome B region (Fig 4A). As expected, in the absence of H1 over expression, we do not observe changes in accessibility to SstI upon hormone treatment (Fig 4B, lanes 1 and 2). Interestingly, when H1 was over expressed, SstI accessibility was reduced indicating a conformational change (Fig 4B, lanes 3 and 4). This reduced SstI cutting was the same under both control and hormone treatment conditions. This is consistent with the transcriptional activity of the MMTV promoter in which we observed reduction in both basal and hormone induced transcription on the transient template (Fig 3B). Although the basal cutting in the presence of H1 over expression mimicked what is observed on the stable template, the hormone induced hypersensitivity associated with the stable template was not evidenced on the transient template. Similar to the SstI enzyme, accessibility to other restriction enzymes such as the Fok1 (−162) (Fig 4B, lanes 5–8, 4D) and the DdeI (Fig 4C, lanes 1–4, 4D) in the nucleosome B region on the transient template was reduced upon H1 over expression. To understand if this change in conformation is restricted to the nucleosome B region of the MMTV promoter, we analyzed accessibility to enzymes with sites outside of nucleosome B region (Fig 4A) such as HaeIII (Fig 4C, lanes 5–8, 4D) and Fok1 (+50) (Fig 4B, lanes 5–8, 4D) and we find reduced cutting upon H1 over expression. Thus over expression of H1 results in conformational change on the transient template that is evidenced all along the promoter and possibly the entire plasmid. Our results thus far suggests that the transiently transfected DNA has higher nucleosomal mobility which may be restricted by binding of H1 under conditions of H1 over expression.
FIG. 4.
Over expression of H1.3 construct leads to alteration in chromatin architecture on the transient template. (A) Schematic of MMTV promoter region showing restriction enzyme sites and primer for linear PCR. (B and C) 1471.1 cells were transfected with pLTRluc and vector or H1 construct for 24 hrs and treated with dex for 1 hour. Isolated nuclei were subjected to limited digestion with various restriction enzymes. Purified DNA was digested to completion with HaeIII or AlwN1. Five micrograms of DNA was amplified by linear PCR using Taq DNA polymerase and 32P labeled primer as depicted in Fig. 4A. The PCR products were then separated on 6% denaturing polyacrylamide gels. (D) The results in 4B and 4C is quantitated as percent cutting.
H1 over expression does not alter nucleosomal organization on the transiently transfected DNA
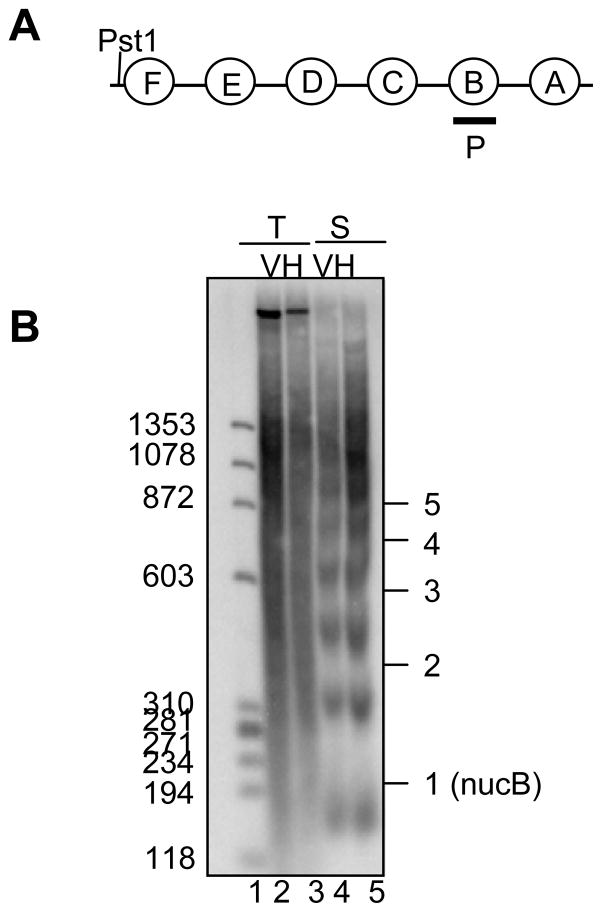
To determine if over expression of H1, results in alteration of nucleosomal organization on the transient template, we performed micrococcal nuclease assay. For this assay we used C127 cells transfected with pLTRluc cotransfected with vector or H1 construct to analyze transient template and we used 1471.1 cells transfected with vector or H1 construct to examine the stable template. As expected, we observed ordered chromatin organization on the stable template (Fig 5A and Fig 5B, lane 4) as evidenced by the regularly spaced ladder generated by the micrococcal nuclease enzyme. Unlike the stable template and consistent with the previous results (2), we fail to detect the nucleosomal ladder (Fig 5B, lane 2) on the transient template. However interestingly, we observed no change in the pattern of micrococcal nuclease digestion on the transient template upon H1 over expression (Fig 5B, lanes 1 and 2). This suggests that over expression of H1 does not result in the ordered nucleosomal organization on the transient template. Therefore, we conclude that under conditions of H1 over expression, excess H1 probably binds to all accessible sites on the plasmid in a very random way and thus reducing the nucleosome fluidity on the DNA but not organizing the nucleosomes.
FIG. 5.
Over expression of H1 does not alter chromatin organization on the transiently transfected MMTV promoter (A) Schematic of MMTV promoter region to show the probe region for B. (B) For transient template, C127 cells were transfected with pLTRluc with pCDNA3 (vector) or H1.3 construct. For stable template, 1471.1 cells were tranfected with salmon sperm DNA with pCDNA3 or H1.3 construct. Transfections were carried out for 24 hrs. Isolated nuclei were digested with micrococcal nuclease (20U/ml). Purified DNA samples were digested with PstI enzyme. Ten micrograms DNA from 1471.1 cells and 1 μg DNA from transient transfection samples plus 9 μg of sheared salmon sperm DNA were electrophoresed on a 2% agarose gel. V denotes pCDNA3, H denotes H1.3 construct. T and S denote transient and stable respectively. The numbers on the side of the gel indicates the number of nucleosome length of the DNA.
DISCUSSION
Functional differences in transcriptional activity between MMTV promoters embedded in chromosomes and within transiently transfected plasmids help to illuminate contributions that chromatin make to regulate gene expression. In this study we wanted to understand the structural differences between transiently transfected DNA and DNA within chromosomes. Using a Lipofectamine method of transient transfection, we demonstrated intermediate levels of nucleosome deposition on the transiently transfected DNA. In an attempt to address the structural disparities between these two templates, we evaluated the differences in stoichiometry in association of histones using chromatin immunoprecipitation assay followed by realtime PCR. We demonstrate association of all four core histones and the linker histone with the transiently transfected DNA. Interestingly, the stoichiometry of the core histones on the transient template is very similar to what is observed on the stable template. This suggests that the histones associated with the transient template have similar nucleosomal composition as that of the stably replicating DNA. Since the introduction of supercoils on a plasmid DNA in the supercoiling assay can also be indicative of association of H3–H4 tetramer and non-histone proteins, our result suggesting histone octamer association with the transient template is a significant one. Since transiently transfected template does not undergo replication, this octamer deposition on the DNA is replication independent deposition. However, this association of octamers with the transient template does not lead to a proper micrococcal nuclease ladder pattern. Therefore the absence of a micrococcal nuclease ladder pattern may suggest a random placement of octamers on the transient template. This is consistent with structural data available from earlier work suggesting that nucleosomes are deposited on transiently transfected template but the organization of nucleosomes is not similar to what is found on a replicating endogenous DNA (2,13,14).
Intriguingly, in contrast to the core histones, the linker histone association was found to be less on the transient template when compared with the stable template. This under-representation of histone H1 on the transient template can perhaps explain the constitutive accessibility of transient template to restriction endonucleases. This also implies that the arrangement of histone octamers on the transiently transfected DNA may be very dynamic and fluid due to fewer H1 molecules. Consistent with this idea and in contrast with the stably integrated promoter, the transiently transfected MMTV promoter does not require BRG1 chromatin remodeling complex for GR mediated transcriptional activation (4). In vitro studies have shown similar role of linker histone H1 in modulating nucleosomal remodeling by human SWI/SNF (21). However, the lack of a micrococcal nuclease ladder on a transient template cannot be associated with underrepresentation of H1 on this template given the fact that changes in stoichiometry of H1 in vivo does not result in a loss of micrococcal nuclease ladder. Changes in H1 levels in the cells leads to alteration in the spacing between nucleosomes (22). There are cell types specific differences in the levels of H1 per nucleosome in higher eukaryotes (20). Interestingly, the embryonic stem cells have 0.5 H1 per nucleosome with a nucleosome repeat length of 189 bp (23). Moreover, recent imaging studies have demonstrated that H1 molecules in the cells are very mobile with a residence time of 3 minutes compared to several hours for the core histones (24).
Based on our observations, we wanted to understand the effect of over expression of histone H1 on a transiently transfected DNA. We found that over expression of H1 had a generalized repressive effect on transcription from a transient template (both basal and induced). Over expression of histone H1 has very little effect on transcription from stably integrated MMTV promoter. This selective transcriptional repression from the transient template may be due to the fact that the transient template architecture is more accessible to binding of factors. This allows H1 to bind at random sites and inhibit transcription. Consistent with this idea, transcription profiling of H1 knockout cells indicates effects on very few genes when compared with the wild type cells in mice (25). Along with the transcriptional repression on transiently transfected DNA, H1 over expression resulted in structural changes of the template as evidenced by reduced restriction enzyme accessibility. These data are consistent with a mechanism by which H1 stabilizes histone octamer-DNA interactions by reducing its mobility on the DNA (26), thus reducing the accessibility of restriction enzymes to DNA. In spite of the structural changes on the transient template upon H1 over expression, the nucleosome arrangement was not regularized. This result is consistent with the idea that limiting amounts of H1 in the cells is not a reason for improper nucleosome positioning on the transient template. Similarly the random binding of H1 on the transiently transfected DNA, which reduces the histone octamer fluidity, is not sufficient to organize a phased array of nucleosomes as observed on the stable template.
Taken together, our results suggest that transiently transfected DNA is associated with histone octamers which are randomly positioned on the DNA. Apart from this random positioning, the nucleosome arrangement is dynamic and fluid possibly due to the underrepresentation of histone H1 on this template. Over expression of H1 diminishes accessibility of the DNA to transcription machinery as assessed by reduced transcription and restriction endonuclease access. Our results imply a more basic role of H1 in DNA replication mediated chromatin organization. Histone H1 is known to deposit on newly replicated DNA immediately after the core histone deposition (27). Since the nucleosome assembly proteins are so closely linked with the DNA replication machinery, there is immediate deposition of nucleosomes on both the copies of DNA. We propose that in the case of transiently transfected DNA, the large circular DNA molecule is presented to the nuclear pools of histones all at once which results in random deposition and positioning of octamers on the template. Coupled with the reduced levels of H1, the newly deposited octamers may have few restrictions on their mobility relative to the octamers within the chromosomes. Therefore random organization of octamers on transient template depicts a scenario in which DNA replication and nucleosome deposition steps are uncoupled in vivo.
Acknowledgments
We would like to thank Sayura Aoyagi and Kevin Trotter for carefully reviewing the paper. We thank Dr David Clark (NIH) for helpful suggestions with nucleosome assembly assay. We would like to thank Dr Maureen Bunger for the pCDNA3-H1.3 expression construct. This research was supported by the Intramural Research Program of the NIH, and NIEHS.
References
- 1.Smith CL, Hager GL. J Biol Chem. 1997;272(44):27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 2.Archer TK, Lefebvre P, Wolford RG, Hager GL. Science. 1992;255(5051):1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 3.Archer TK, Cordingley MG, Wolford RG, Hager GL. Mol Cell Biol. 1991;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trotter KW, Archer TK. Mol Cell Biol. 2004;24(8):3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebbar PB, Archer TK. J Biol Chem. 2007;282(11):8284–8291. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Genes Dev. 1997;11(4):436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 7.Bresnick EH, John S, Berard DS, LeFebvre P, Hager GL. Proc Natl Acad Sci U S A. 1990;87 (10):3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garner MM, Felsenfeld G, O’Dea MH, Gellert M. Proc Natl Acad Sci U S A. 1987;84(9):2620–2623. doi: 10.1073/pnas.84.9.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archer TK, Lee HL. Methods. 1997;11(2):235–245. doi: 10.1006/meth.1996.0410. [DOI] [PubMed] [Google Scholar]
- 10.Hebbar PB, Archer TK. Mol Cell Biol. 2003;23(3):887–898. doi: 10.1128/MCB.23.3.887-898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson MA, Ricci AR, Deroo BJ, Archer TK. J Biol Chem. 2002;277(17):15171–15181. doi: 10.1074/jbc.M200349200. [DOI] [PubMed] [Google Scholar]
- 12.Deroo BJ, Archer TK. Mol Biol Cell. 2001;12(11):3365–3374. doi: 10.1091/mbc.12.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong S, Stein A. Nucleic Acids Res. 1994;22(3):370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cereghini S, Yaniv M. Embo J. 1984;3(6):1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith S, Stillman B. Cell. 1989;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 16.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 17.Lee HL, Archer TK. Embo J. 1998;17(5):1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoma F, Koller T, Klug A. J Cell Biol. 1979;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Proc Natl Acad Sci U S A. 1998;95(24):14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodcock CL, Skoultchi AI, Fan Y. Chromosome Res. 2006;14(1):17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran A, Omar M, Cheslock P, Schnitzler GR. J Biol Chem. 2003;278(49):48590–48601. doi: 10.1074/jbc.M309033200. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. Mol Cell Biol. 2003;23(13):4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Cell. 2005;123(7):1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Nature. 2000;408(6814):877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Inselman A, Han X, Xu H, Zhang W, Handel MA, Skoultchi AI. J Biol Chem. 2004;279(22):23525–23535. doi: 10.1074/jbc.M400925200. [DOI] [PubMed] [Google Scholar]
- 26.Pennings S, Meersseman G, Bradbury EM. Proc Natl Acad Sci U S A. 1994;91(22):10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bavykin S, Srebreva L, Banchev T, Tsanev R, Zlatanova J, Mirzabekov A. Proc Natl Acad Sci U S A. 1993;90(9):3918–3922. doi: 10.1073/pnas.90.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]