Abstract
Chaperonins are allosteric double-ring ATPases that mediate cellular protein folding. ATP binding and hydrolysis control opening and closing of the central chaperonin chamber, which transiently provides a protected environment for protein folding. During evolution, two strategies to close the chaperonin chamber have emerged. Archaeal and eukaryotic group II chaperonins contain a built-in lid, whereas bacterial chaperonins use a ring-shaped cofactor as a detachable lid. Here we show that the built-in lid is an allosteric regulator of group II chaperonins, which helps synchronize the subunits within one ring and, to our surprise, also influences inter-ring communication. The lid is dispensable for substrate binding and ATP hydrolysis, but is required for productive substrate folding. These regulatory functions of the lid may serve to allow the symmetrical chaperonins to function as ‘two-stroke’ motors and may also provide a timer for substrate encapsulation within the closed chamber.
Chaperonins are ubiquitous and essential mediators of cellular protein folding that consist of 14–18 subunits arranged in two stacked rings1–4. Each ring encompasses a central chamber that accommodates non-native polypeptides. All chaperonins share a similar subunit architecture, consisting of three distinct domains: an ATP-binding equatorial domain, a distal apical domain harboring the polypeptide-binding sites and an intermediate hinge domain. A key feature of chaperonins is their ability to close their chamber and encapsulate the bound substrate, thus providing a protected environment for protein folding. Opening and closing of the folding chamber is controlled by a conformational cycle driven by ATP binding and hydrolysis. Despite overall structural similarities, there are substantial differences between the eubacterial chaperonins, such as GroEL from E. coli2–5, and the chaperonins from archaea and eukaryotes6,7. Whereas bacterial, so-called group I chaperonins are homo-oligomeric, eukaryotic and archaeal group II chaperonins are generally hetero-oligomeric7. The greatest difference between group I and group II chaperonins resides in their distinct strategies to mediate closure of their central folding chamber. Group I chaperonins use a ring-shaped cochaperone, GroES, as a detachable lid. In the presence of ATP, GroES binds GroEL and closes its central cavity5. In contrast, group II chaperonins have a built-in lid formed by protrusions that extend from the tip of each apical domain8–10. For the eukaryotic chaperonin TRiC (also called CCT), it has been shown that assembly of the iris-like lid is triggered by ATP hydrolysis9. Despite intensive studies on the biochemistry and function of bacterial chaperonins, little is known of the mechanistic and biological significance underlying the unique structural features of eukaryotic and archaeal chaperonins.
The chaperonin folding cycle is crucially dependent on the synchronized action of individual subunits11–13. Accordingly, chaperonins are highly allosteric protein machines11–13. Subunits within each ring are coupled as a functional unit through positive cooperativity in ATP binding11,14–16; this allows them to act in a concerted fashion to create the closed folding chamber. In addition, negative communication between the rings causes ATP binding to one ring to inhibit ATP binding to the adjacent ring14–18. This feature is thought to ensure that only one ring is folding-active at a given time, allowing chaperonins to function as ‘two-stroke’ motors. This unique allosteric behavior is observed in both group I and group II chaperonins11,14,15,19,20 and is described as nested cooperativity, as the positive cooperative transition within each ring is nested into the overall negative cooperativity between them.
The basis of allosteric regulation in group I chaperonins has been extensively studied. Positive and negative cooperativity in GroEL are established independently of the GroES lid19,21, although GroES profoundly influences the conformational changes of GroEL12,21–24. In contrast, little is known about the molecular basis of allostery in eukaryotic and archaeal chaperonins. Their very different mechanisms of lid formation raise the question of how group II chaperonins regulate their conformational cycle. We here investigate the function of the built-in lid in the mammalian chaperonin TRiC and an archaeal chaperonin from Methanococcus maripaludis. Our results show that the built-in lid is important in regulating the conformational cycle of group II chaperonins. We found that the apical protrusions have been incorporated into the allosteric network that communicates ATP-induced conformational changes between subunits. It thus appears that group I and group II chaperonins have evolved very different strategies for intersubunit coordination, despite the overall structural conservation of the chaperonin ring architecture. Our findings help explain how group II chaperonins act like two-stroke motors without the help of an extrinsic cofactor and show how conserved protein scaffolds can achieve allosteric regulation through widely divergent structural features.
RESULTS
The lid couples ATP hydrolysis to substrate folding
To examine the role of the built-in lid in the folding cycle of group II chaperonins, we assessed the effect of eliminating the lid on the eukaryotic chaperonin TRiC (Fig. 1) and the archaeal Cpn from the mesophile M. maripaludis (Fig. 2)25,26. TRiC is central in the folding of many essential structural and regulatory eukaryotic proteins and is perhaps the most complex chaperonin, consisting of eight different subunits7. The simpler M. maripaludis Cpn consists of eight identical subunits per ring and, unlike other archaeal chaperonins from extremophiles6, can be studied at physiologically relevant temperatures and salt concentrations25,26.
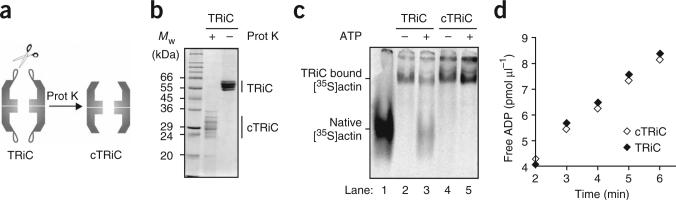
Figure 1.
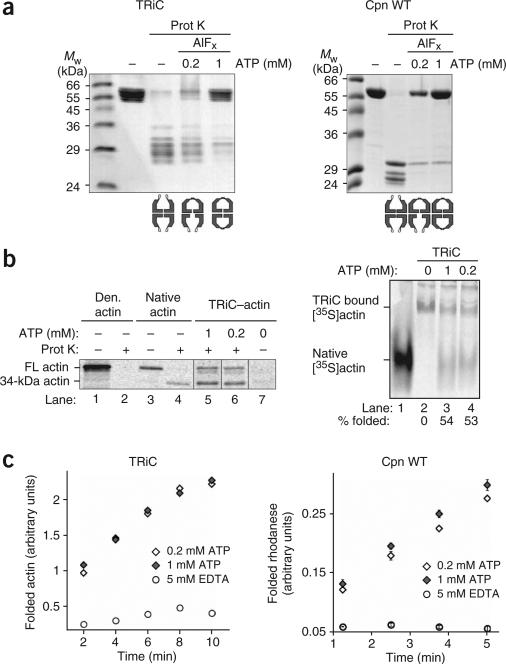
The built-in lid couples ATP hydrolysis to substrate folding in the eukaryotic chaperonin TRiC. (a) cTRiC is a lidless version of TRiC generated by a selective cleavage within the apical protrusions27. Prot K, proteinase K. (b) SDS-PAGE analysis followed by Coomassie staining of TRiC and cTRiC. Cleavage in the apical protrusion of each of the eight different 55- to 60-kDa subunits of TRiC produces sixteen 24- to 36-kDa fragments27. (c) cTRiC is able to bind denatured [35S]actin but cannot promote [35S]actin folding in the presence of ATP. About 60% of bound [35S]actin was refolded by TRiC in the presence of ATP (lane 3). Lane 1, native [35S]actin migration standard. (d) ATP hydrolysis by TRiC and cTRiC measured at 0.6 mM [α-32P]ATP.
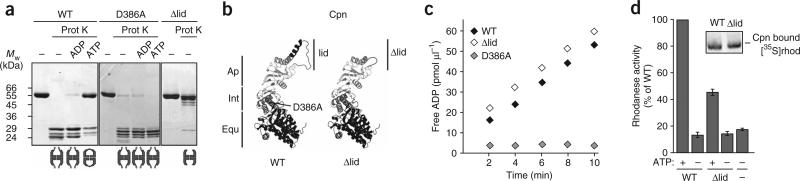
Figure 2.
The built-in lid couples ATP hydrolysis to substrate folding in archaeal group II chaperonins. (a) ATP hydrolysis but not ATP binding induces lid closure in M. maripaludis Cpn. Lid closure in the presence of ATP was monitored for Cpn WT, the ATPase-deficient mutant Cpn D386A and Cpn Δlid, as in Figure 1b9. (b) Homology model of a single subunit of Cpn WT and Cpn Δlid, respectively. The equatorial domain (Equ, black) is linked to the apical domain (Ap, light gray) via the flexible intermediate domain (Int, gray). Position of Asp386 is indicated. In Cpn Δlid, a short linker replaces the apical protrusions (lid, black). (c) ATP hydrolysis by Cpn WT, Cpn Δlid and Cpn D386A measured at 0.5 mM [α-32P]ATP. (d) Cpn Δlid is able to bind denatured [35S]rhodanese ([35S]rhod) but cannot efficiently promote rhodanese folding in the presence of ATP. Error bars represent s.e.m. Inset shows an autoradiogram of [35S]rhodanese–chaperonin complexes analyzed by 4% nondenaturing gel electrophoresis.
The activity of TRiC was compared with that of the previously described9,27 lidless version, called clipped-TRiC or cTRiC (Fig. 1a,b). Cryo-EM analysis revealed that the doublering architecture characteristic of chaperonins is preserved in cTRiC (Supplementary Fig. 1 and Supplementary Methods online), consistent with previous findings9. Furthermore, both the substrate-binding and ATPase domains retain their integrity in cTRiC, as it binds non-native substrates, such as [35S]actin, with an efficiency comparable to that of TRiC (Fig. 1c, compare lanes 2 and 4) and can also effectively hydrolyze ATP (Fig. 1d). Notably, cTRiC is not able to promote actin folding upon incubation with ATP (Fig. 1c, compare lanes 3 and 5). We conclude that an intact lid is required to couple ATP hydrolysis with productive substrate folding in the eukaryotic chaperonin.
To extend the findings for the eukaryotic TRiC complex to other group II chaperonins, we next examined the function of the built-in lid in archaeal chaperonins (Fig. 2). First, the nucleotide requirements for lid closure in the archaeal Cpn were defined. To this end, we exploited the differential protease susceptibility of the lid segments in the open and closed states9 (Fig. 2a). Mild proteolytic treatment of wild-type Cpn (Cpn WT) in the open, nucleotide-free state led to specific cleavage of the lid segments (Fig. 2a, left gel). Notably, cleavage occurred at a position in the Cpn lid equivalent to that cleaved in TRiC subunits27. Incubation with ATP, but not ADP, triggers lid closure, resulting in protection of the lid segments (Fig. 2a, left gel). Analysis of Cpn mutant D386A, which can bind ATP (data not shown) but is unable to hydrolyze it (Fig. 2c), further indicated that ATP binding does not suffice for lid closure, which is mediated by ATP hydrolysis, as observed for TRiC (Fig. 2a, middle gel).
We next compared the properties of M. maripaludis Cpn WT with a lidless variant, Cpn Δlid, in which the apical protrusions (Ile241–Lys267) are replaced with a short flexible linker (Fig. 2b). Deletion of the lid segments rendered the chaperonin resistant to protease digestion even in the absence of nucleotide, supporting the idea that the highly dynamic nature of the lid segments is responsible for providing the proteolytic cleavage site (Fig. 2a, left gel). The built-in lid of Cpn was dispensable for both ATP hydrolysis (Fig. 2c) and substrate binding (Fig. 2d, inset). However, deletion of the lid impaired the ability of Cpn to fold rhodanese in the presence of ATP (Fig. 2d).
Together, our data suggest that, despite the evolutionary distance spanned by group II chaperonins, the mechanism of closure and the function of the lid are conserved among them all. In both archaeal and eukaryotic chaperonins, lid closure is triggered by ATP hydrolysis9 (Fig. 2a). Furthermore, in both cases the lid is required to fold a bound substrate upon ATP hydrolysis (Fig. 1c and Fig. 2d). The characterization of lidless variants of TRiC and Cpn provided an opportunity to define how the built-in lid regulates the ATP-driven conformational cycle of group II chaperonins.
The lid establishes allosteric coupling within one ring
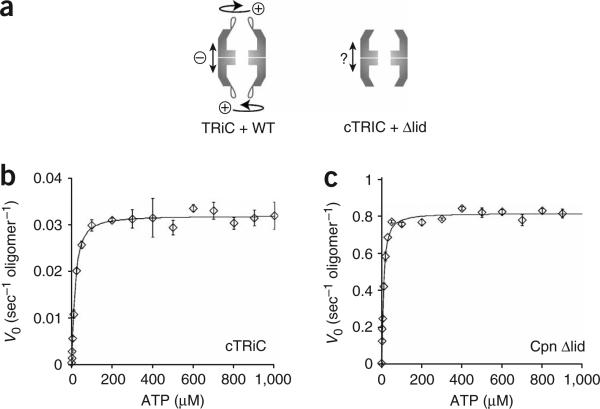
In group II chaperonins, all eight apical protrusions within a ring must interact tightly to form the iris-like lid8. Accordingly, ATP should produce a concerted conformational change in all subunits of one ring. Given the overall conservation of the chaperonin structure, communication between subunits could be independent of the lid segments, as observed in GroEL, where positive intra-ring cooperativity is independent of GroES19 (Fig. 3a, model 1). Alternatively, the apical protrusions themselves could synchronize the subunits within one ring, thus coordinating lid formation (Fig. 3a, model 2). These models make distinct predictions for the contribution of the built-in lid to positive intra-ring cooperativity. Whereas coupling between subunits would be retained in lidless chaperonin variants according to the first model, it would be lost according to the second one. To distinguish between these possibilities, we compared the intra-ring cooperativity in wild-type and lidless variants of both TRiC and M. maripaludis Cpn (Fig. 3). The allosteric properties of these chaperonins were determined by measuring the initial rates of ATP hydrolysis as a function of ATP concentration (Fig. 3).
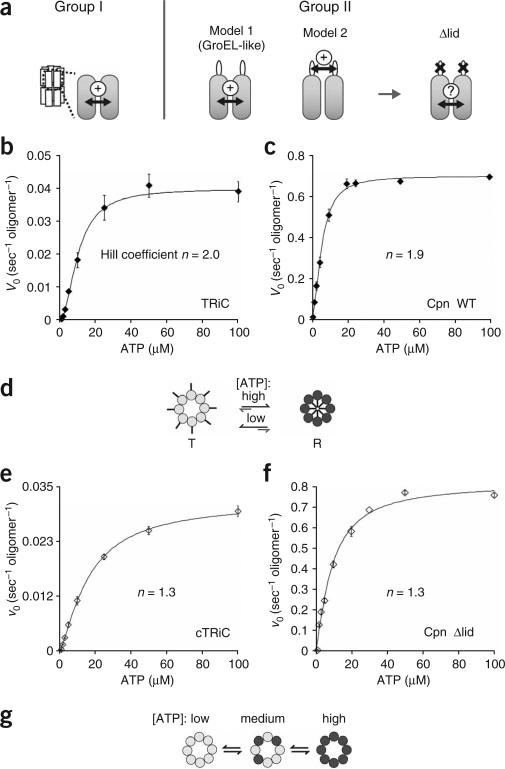
Figure 3.
The built-in lid is required for positive cooperativity between the subunits of one ring. (a) Two possible models for allosteric coupling of subunits in group II chaperonins. In the group I chaperonin GroEL, intra-ring allosteric coupling is achieved by interactions between the apical and intermediate domains of neighboring subunits. In group II chaperonins, two models are possible. Model 1: intra-ring coupling is independent of the lid, similar to GroEL. Model 2: the lid segments are required to orchestrate synchronized action between subunits in one ring. (b,c) Wild-type eukaryotic and archaeal chaperonins show positive cooperativity among the subunits of one ring. Initial velocities of ATP hydrolysis by TRiC (b) and Cpn WT (c) were plotted against ATP concentration and fit to the Hill equation (equation (2), see Methods). Each data point shows the average of at least three independent experiments. Error bars represent s.e.m. (d) Model of the ATP-induced conformational change of one ring of TRiC and Cpn WT. Positive cooperativity causes subunits in one ring to undergo a concerted conformational change leading to lid closure. (e,f) Allosteric coupling of subunits in one ring is impaired in a lidless chaperonin. Initial velocities of ATP hydrolysis by cTRiC (e) and Cpn Δlid (f) were plotted against ATP concentration and fit to equation (2) as above. Each data point shows the average of at least three independent experiments. Error bars represent s.e.m. (g) Model of the ATP-induced conformational changes in lidless cTRiC and Cpn Δlid. No allosteric coupling between the subunits can be observed, indicating that the subunits bind ATP independently of one another.
The first allosteric transition of TRiC and Cpn results from intra-ring communication20,26. Accordingly, we assessed coupling between subunits in one ring by comparing the kinetics of these chaperonins at ATP concentrations below 100 μM20,26. The kinetics obtained for both intact TRiC and Cpn WT were sigmoidal (Fig. 3b,c and Supplementary Fig. 2 online), indicating positive cooperativity between the subunits of one ring (P < 0.001 for both chaperonins). The apparent ATP-binding constant (K1) and the Hill coefficient (n) for this first allosteric transition, as well as the maximal turnover rate (vmax), were calculated by fitting the data points to the Hill equation (equation (1) and Table 1). For TRiC, the values for K1 and n are 10.1 ± 0.5 μM and 2.0 ± 0.2, respectively, in very good agreement with previous observations20. Comparison of the respective values for the catalytic rate (kcat, calculated to be 0.04 s–1 for TRiC and 0.7 s–1 for Cpn WT) reveals that the archaeal chaperonin is a much more efficient ATPase than TRiC, a property Cpn shares with its bacterial counterpart, GroEL19. Notably, although Cpn hydrolyzes ATP much more rapidly than TRiC, the affinity of the archaeal chaperonin for ATP (K1 = 5.8 ± 0.3 μM) as well as its degree of allosteric coupling (n = 1.9 ± 0.1) were very similar to those of TRiC (Table 1). Thus, for both chaperonins, positive intra-ring cooperativity drives a concerted conformational change in all subunits of one ring that results in lid closure (Fig. 3d). This allosteric switch converts the subunits of one ring from a ‘tense’ T state with low affinity for ATP to a ‘relaxed’ R state with high affinity for ATP (Fig. 3d)11.
Table 1.
Parameters defining kinetic properties of group II chaperonins
| TRiC | cTRiC | Cpn WT | Cpn Δlid | |
|---|---|---|---|---|
| K1 (μM) | 10.1 ± 0.5 | 16.5 ± 1.0 | 5.8 ± 0.3 | 9.0 ± 0.6 |
| K2 (μM) | 593 ± 23 | NA | 562 ± 175 | NA |
| n | 2.0 ± 0.2 | 1.3 ± 0.2 | 1.9 ± 0.1 | 1.3 ± 0.1 |
| m | 10.8 ± 4.2 | NA | 2.9 ± 1.8 | NA |
| kcat R/T′ (oligomer–1 sec–1) | 0.04 | 0.03 | 0.7 | 0.8 |
| kcat R′/R′ (oligomer–1 sec–1) | 0.028 | NA | 0.54 | NA |
Errors are s.e.m. NA, not applicable. See Methods for definitions of parameters.
We next examined the allosteric coupling of lidless variants of TRiC and Cpn (Fig. 3e,f). Loss of the lid segments did not affect the overall affinity for ATP, K1, or the maximal hydrolysis rate, vmax, of either chaperonin (Table 1). However, positive cooperative coupling between subunits was weaker (Fig. 3e,f and Supplementary Fig. 2), as indicated by significantly reduced values for the Hill coefficients (n; Table 1; P = 0.01 for nTRiC compared with ncTRiC, P = 0.005 for nCpn WT compared with nCpn Δlid). Of note, deletion of the entire apical protrusion in Cpn (Fig. 3f) has a similar effect on the positive cooperativity as cleaving the lid-forming segments in TRiC (Fig. 3e). These lidless chaperonins hydrolyze ATP with Michaelis-Menten kinetics, which is typical of enzymes without allosteric regulation, indicating that the subunits in one ring bind ATP independently of one another (Fig. 3g).
These experiments suggest that the presence of intact lid segments is important to establish positive cooperativity among the subunits of one ring. Thus, the built-in lid is required to synchronize the ATP-induced conformational change of subunits within one ring.
Analysis of negative inter-ring communication
TRiC and M. maripaludis Cpn show a second allosteric transition above 400 μM ATP that results from inter-ring communication (Fig. 4a,b)20,26. These higher ATP concentrations overcome the negative cooperativity between the rings and, as a result, the trans-ring also starts binding and hydrolyzing ATP (Fig. 4c)20,26.
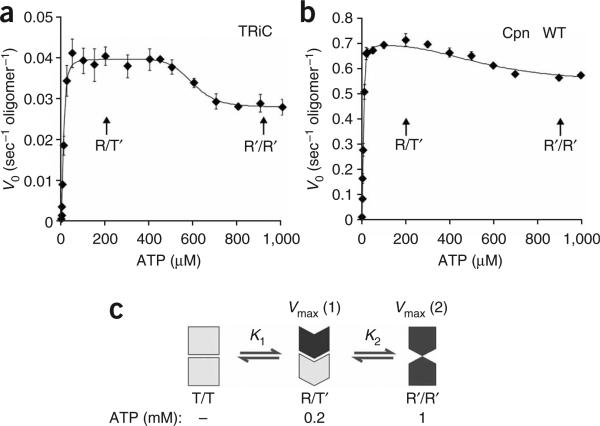
Figure 4.
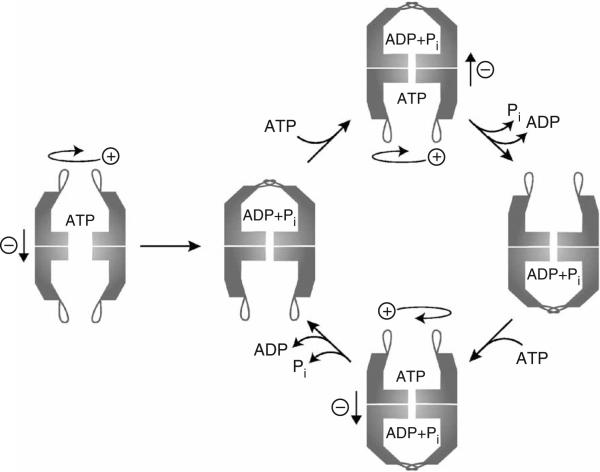
Negative allosteric coupling between rings affects ATP binding and hydrolysis. (a,b) Negative allosteric coupling between rings results in a second allosteric transition occurring at high ATP concentrations in wild-type chaperonins. Initial velocities of ATP hydrolysis by TRiC (a) and Cpn WT (b) were plotted against ATP concentration and fit to equation (2) (see Methods). Each data point shows the average of at least three independent experiments. Error bars represent s.e.m. (c) Proposed conformational states for group II chaperonins at different ATP concentrations. T/T state: in the absence of nucleotide, both rings reside in the T state, and the lid structure is not formed. This T/T complex has a low affinity for ATP. R/T′ state: at intermediate ATP concentrations (0.2 mM ATP), the cis-ring reaches saturation for ATP binding and hydrolysis and therefore assumes the R state. The trans-ring adopts a conformational state T′ with low affinity for ATP as a consequence of negative cooperativity between the rings. This asymmetric state has optimal ATPase activity (vmax(1)). R′/R′ state: at high ATP concentrations (1 mM ATP), the negative cooperativity is overcome and both rings bind and hydrolyze ATP simultaneously. The two rings hinder each other, leading to the observed drop in the hydrolysis rate vmax.
To examine the nature of inter-ring communication in group II chaperonins, we extended our analysis to a broader range of ATP concentrations20. The second allosteric transition observed for both TRiC and Cpn was reflected by a decreased hydrolysis rate at higher ATP concentrations (Fig. 4a,b), in contrast with previous measurements for TRiC20. Notably, similar results were obtained for TRiC and Cpn WT; thus, our results probably reflect a general property of group II chaperonins. As both chaperonins used here were fully competent for substrate folding (Fig. 1c and Fig. 2d), the discrepancy with previous TRiC measurements could be explained by weakened interring contacts in previous protein preparations, which may impair negative cooperativity.
The observation that the second allosteric transition produces a marked decrease in the rate of ATP hydrolysis suggests that negative inter-ring communication prevents both ATP-bound rings from hydrolyzing ATP simultaneously at an optimal rate. Our findings suggest a model for how nested allosteric interactions in group II chaperonins lead them to function as two-stroke motors (Fig. 4c). In the absence of nucleotide, the two rings are virtually identical and in the symmetrically open T state (Fig. 4c). At intermediate ATP concentrations (Fig. 4c, 0.2 mM ATP), the subunits in one ring (the cis-ring) undergo an allosteric transition to the R state and bind ATP with positive cooperativity14,15. As a result of the negative inter-ring cooperativity, ATP binding to the cis-ring induces a conformational change in the subunits of the trans-ring to a T′ state with lower affinity for ATP. This asymmetric state is characterized by an optimal ATPase activity (vmax(1)). At higher ATP concentrations (Fig. 4c; 1 mM ATP), the trans-ring also binds ATP, but overall ATP hydrolysis becomes less efficient. We propose that this change corresponds to a different state of the enzyme, the R′/R′ state, where the negative cooperativity for ATP binding has been overcome and both rings are forced to adopt a conformation with a suboptimal ATPase rate (vmax(2)). Notably, similar results were observed for both TRiC and Cpn, despite their widely different overall catalytic rates and subunit composition, suggesting that this type of allosteric regulation is conserved in all group II chaperonins.
Allosteric coupling drives a two-stroke folding cycle
The model suggested above predicts the formation of an asymmetric R/T′ state at intermediate ATP concentrations and a symmetric R′/R′ state at high ATP concentrations. This prediction was tested by exploiting the differential protease sensitivity of the lid in the open T state and the closed R state. TRiC (Fig. 5a, left gel) and M. maripaludis Cpn (Fig. 5a, right gel) were incubated in the absence of nucleotide or in the presence of either 0.2 or 1 mM ATP to generate the three states proposed by our model (Fig. 4c). The γ-phosphate mimic AlFx was included in the reactions to stabilize the closed state9,28. As expected, both rings were in the open conformation in the nucleotide-free T/T state (Fig. 5a). In contrast, at a high ATP concentration (Fig. 5a, 1 mM ATP), virtually all the apical lid segments were protected in both TRiC and Cpn WT, yielding full-length chaperonin, consistent with a R′/R′ state, in which both rings are closed. Notably, incubation with an intermediate ATP concentration (Fig. 5a, 0.2 mM) reduced the level of protection of the lid segments of either mammalian or archaeal chaperonin to about half of the value obtained at the high ATP concentration, consistent with the idea that negative inter-ring cooperativity prevents the trans-ring from binding ATP. Moreover, this lower ATP concentration yields the maximal ATP hydrolysis rates, suggesting that the asymmetric conformation of the chaperonin induced by this ATP concentration is optimized for ATP cycling.
Figure 5.
Group II chaperonins sample two different conformational states at intermediate and high ATP concentrations. (a) In the absence of nucleotide, chaperonins predominantly sample the T/T state, in which both rings are in the open conformation and therefore susceptible to protease digestion. TRiC and Cpn WT are fully protected from protease cleavage at 1 mM ATP-AlFx and therefore reside in a symmetrically closed complex, the R′/R′ state. The partial protection pattern at 0.2 mM ATP-AlFx indicates the existence of an asymmetrically closed R/T′ state at intermediate ATP concentrations. (b,c) The asymmetric R/T′– state of group II chaperonins supports optimal substrate folding. b shows TRiC-mediated [35S]actin folding at 1 and 0.2 mM ATP, examined by protease digestion (left) and nondenaturing PAGE (right), followed by autoradiography. FL, full-length. c shows substrate-folding rates of TRiC and Cpn at 0.2 and 1 mM ATP. Left, time course of actin folding by TRiC in the absence of nucleotide (5 mM EDTA) and in the presence of 0.2 and 1 mM ATP. Right, time course of rhodanese folding by Cpn WT in the absence of nucleotide (5 mM EDTA) and in the presence of 0.2 and 1 mM ATP. Error bars represent s.e.m.
The observation of an asymmetric R/T′ state at intermediate ATP concentrations raised the possibility that negative allosteric coupling between rings allows the inherently symmetrical group II chaperonins to function as two-stroke motors. To relate the regulation of ATPase activity to chaperonin function, we compared the protein-folding activity of TRiC and Cpn at either 0.2 or 1 mM ATP (Fig. 5b,c). TRiC-mediated actin folding was assessed using two independent folding assays, nondenaturing PAGE (Fig. 5b, right gel) and protease susceptibility (Fig. 5b, left gel)9. Comparable folding yields and rates were observed at 0.2 and 1 mM ATP (Fig. 5b,c). Of note, the protease-susceptibility assay can distinguish between released folded actin, which yields a protease-resistant actin fragment of 34 kDa, and TRiC-encapsulated actin, as lid closure protects the full-length polypeptide (Fig. 5b, left gel)9. The levels of [35S]actin protection observed at 0.2 and 1 mM ATP were similar, suggesting that only one ring is functional for substrate folding and encapsulation, even at the higher ATP concentrations. The same conclusions were reached for Cpn, as comparable yields and rates of rhodanese folding were observed at 0.2 and 1 mM ATP (Fig. 5c, right chart). Thus, the asymmetric R/T′ state supports optimal substrate-folding rates.
Together, these results support the idea that at intermediate ATP concentrations, negative inter-ring cooperativity establishes an asymmetric R/T′ state, with only one ring having a closed lid. Consistent with our model, structural evidence for an asymmetric cycle of lid closure has been obtained for TRiC29 and for archaeal thermosomes30,31. Our analysis provides a rationale as to how allosteric regulation of group II chaperonins allows these symmetrical complexes to function as a two-stroke machine without the assistance of an external GroES-like cofactor.
The lid influences inter-ring communication
To examine the role of the apical lid segments in establishing the negative inter-ring cooperativity, we next extended our analysis to cTRiC and Cpn Δlid (Fig. 6). Notably, there were substantial differences in the kinetics of the lidless and wild-type chaperonins. For both the archaeal and the eukaryotic chaperonins, absence of a functional lid completely abolished the second allosteric transition at higher ATP concentrations (Fig. 6b,c and Table 1; compare with Fig. 4a,b for wild-type). We conclude that the lid segments not only synchronize subunits within one ring but also are important in communication between the rings.
Figure 6.
The built-in lid affects inter-ring communication of group II chaperonins. (a) Nested allosteric coupling in group II chaperonins. Positive cooperativity among the subunits of one ring is nested into negative cooperativity between the two rings. (b,c) The second allosteric transition, occurring at high ATP concentrations, is absent in the lidless chaperonin variants. Initial velocities of ATP hydrolysis by cTRiC (b) and Cpn Δlid (c) were plotted against ATP concentration and fit to equation (2) (see Methods). Each data point shows the average of at least three independent experiments. Error bars represent s.e.m.
DISCUSSION
Opening Andino closing of the built-in lid in group II chaperonins requires the coordinated action of all subunits in a ring, controlled by ATP binding and hydrolysis in the distant equatorial domains. This study uncovers a remarkable degree of mechanistic and functional conservation between group II chaperonins from eukaryotic and archaeal origin, despite their evolutionary distance. In both cases, the built-in lid is in the open state upon ATP binding and closes during ATP hydrolysis. The lid segments are not required for maintenance of the double-ring chaperonin architecture but help integrate the subunits within a ring into an allosteric unit (Fig. 7). Although the lid is far away from the inter-ring contacts, the lid structure also functions in modulating inter-ring communication (Fig. 7). This negative inter-ring coupling enables group II chaperonins to function as two-stroke motors, where the two rings alternately sample the folding-active state during the reaction cycle.
Figure 7.
The built-in lid controls the ATPase cycle of group II chaperonins. ATP binding to subunits in one ring is a cooperative event communicated by the apical protrusions. Subsequent ATP hydrolysis within the cis-ring is accompanied by a synchronized conformational change that results in lid closure9. The lid probably remains closed in the post-hydrolysis ADP + Pi state. Reopening of the stable iris-like lid structure seems to be the rate-limiting step in the conformational cycle18. Owing to negative inter-ring cooperativity14,17,18, ATP hydrolysis in the trans-ring can occur only once the products of ATP hydrolysis have dissociated from the cis-ring. The precise mechanism that unravels the stable lid structure remains undefined.
Strategies of allosteric coupling in chaperonins
The phenomenon of nested cooperativity has been observed in all chaperonins. We found that group I and group II chaperonins use different strategies to establish the same type of allostery. Allosteric coupling of subunits within one ring is intrinsic to GroEL and is modulated only by the GroES cofactor19,21–24. In contrast, we found that group II chaperonins depend on their built-in lids to coordinate intra-ring communication.
The role of the built-in lid in establishing positive cooperativity in group II chaperonins is surprising, given their overall similarities with their bacterial counterparts. The distinct allosteric strategies of group II chaperonins may originate from the unique mechanistic requirements of having a built-in lid. In GroEL, the lid is already preformed, and lid closure requires merely an increased affinity of GroEL for GroES. In contrast, group II chaperonins must create the lid in a coordinated manner during ATP hydrolysis within a ring. The structural challenges posed by the built-in lid may be incompatible with the allosteric regulation of bacterial chaperonins, forcing the emergence of novel allosteric networks in group II chaperonins.
Understanding how the built-in lid contributes to allosteric coupling in group II chaperonins must await a better characterization of the open state. Lid formation could involve a concerted interaction between all apical protrusions or, alternatively, the conformational change could propagate sequentially through domino-like interactions between apical protrusions. Notably, cryo-EM analysis of TRiC has revealed the presence of asymmetry in one ring at low ATP concentrations32, suggesting a sequential type of allosteric transition. Given that substrate proteins might be bound to more than one subunit within a ring33, these different models have different implications for the mechanism of substrate release from the binding sites, which in turn will influence the folding mechanism.
The built-in lid regulates inter-ring communication
Our analysis indicates that the built-in lid also affects inter-ring communication (Fig. 6). We envision two possible models that could account for these observations. First, removing the lid could abolish both positive and negative allosteric coupling, so that all 16 subunits bind and hydrolyze ATP independently. As all subunits in both rings would be hydrolyzing ATP simultaneously under saturating conditions, this scenario leads to the prediction that the lidless variants should reach a higher maximal hydrolysis rate, vmax, than intact chaperonins. Indeed, such behavior has been observed for GroEL mutants with distorted inter-ring communication34,35. Our findings, however, are not consistent with this possibility (compare Fig. 4a,b and Fig. 6b,c). Accordingly, we favor a second model, wherein the formation of a functional lid structure is not required for negative cooperativity but serves to slow down the ATPase cycle by stabilizing the closed state. This model is consistent with various lines of evidence obtained for TRiC and the archaeal thermosome, which indicate that lid reopening is the slowest, rate-limiting step in the ATPase cycle9,18. First, steady-state measurements at high ATP concentrations indicate that for TRiC, the closed post-hydrolysis state dominates the kinetic ATPase cycle9. In agreement with this idea, kinetic analysis of the thermosome has revealed that ADP + Pi release is rate limiting18, also indicating a long-lived post-hydrolysis state. On the basis of these results, it has been proposed that the trans-ring of the thermosome is prevented from hydrolyzing ATP until Pi and ADP are released from the cis-ring18. Upon consideration of this together with our results, a picture emerges in which formation of the closed lid delays the release of the hydrolysis products, ADP and Pi, thus extending the duration of the ATPase cycle in the cis-ring and delaying ATP hydrolysis in the trans-ring. At saturating ATP concentrations, this would effectively slow down the steady-state ATPase rate, as observed experimentally for both TRiC (Fig. 4a) and M. maripaludis Cpn (Fig. 4b). As the lidless chaperonins do not have a closed lid to slow down release of ADP and Pi in the cis-ring, the inhibition of hydrolysis in the trans-ring should not be observed; this is consistent with the faster turnover rate observed at high ATP concentrations for the lidless chaperonins (Fig. 6b,c). Moreover, in addition to explaining all available data, this model reveals that the lid acts as a timing device that regulates the duration of the folding-active state.
Lid formation and the catalytic cycle of group II chaperonins
Our results uncover several functions for the built-in lid during the conformational cycle of group II chaperonins (Fig. 7). In addition to physically encapsulating the substrate, the built-in lid is also key in regulating chaperonin function and ensuring its activity as a two-stroke molecular machine. Because the closed conformation is presumably the folding-active state, lid opening upon release of ADP and Pi serves as a timer mechanism that regulates the length of substrate encapsulation in the folding chamber. Notably, despite having similar mechanisms, TRiC and M. maripaludis Cpn have very different ATP turnover rates, suggesting that the substrate residence time within the chamber may be fine-tuned to suit the folding requirements of different cellular environments.
What drove the evolution of such distinct mechanisms of substrate encapsulation within the chaperonin chamber? Given that several eukaryotic proteins, such as actin, can be folded only by TRiC and not by group I chaperonins36, the distinct structural features of group II chaperonins probably evolved to suit the specific folding requirements of their substrates. In turn, the built-in lid drove a remodeling of the allosteric network in group II chaperonins such that the apical protrusion communicates conformational changes between subunits. As ATP hydrolysis probably hides the substrate-binding sites from the cavity33, it is possible that incorporating the lid into the allosteric network may help ensure that the lid is formed before substrate release to prevent premature escape from the cavity.
It is noteworthy that the incorporation of a slight structural variation, namely the built-in lid, into the conserved chaperonin architecture forced the archaeal and eukaryotic complexes to evolve a different strategy in order to maintain the same type of regulation through nested cooperativity. Defining the structural basis of inter-subunit communication in eukaryotic and archaeal chaperonins may provide insights into the plasticity of allosteric networks13.
METHODS
Materials
[α-32P]ATP was obtained from PerkinElmer Life and Analytical Sciences and all other chemicals were purchased from Sigma-Aldrich, unless mentioned otherwise.
Plasmids
Genomic DNA from M. maripaludis strain LL (see Acknowledgments) was used as a template to amplify the M. maripaludis cpn gene by PCR. The PCR fragment was inserted into the vector pET21a+ (EMD Chemicals) using the NdeI and BamHI restriction sites, resulting in the vector pET21MmCpnWT. Replacement of the helical protrusion region (Ile241–Lys267) by a short linker (ETASE) as well as exchange of Asp386 to alanine was done by site-directed mutagenesis using the QuikChange kit (Stratagene). Information about the oligonucleotides used can be obtained from the authors upon request.
Protein purifications
TRiC was purified from bovine testis essentially as described37. M. maripaludis Cpn WT, Cpn Δlid and Cpn D386A were purified as follows. Chaperonin proteins were overproduced in Escherichia coli strain Rosetta (DE3) pLysS (EMD Biosciences) harboring plasmid pET21MmCpnWT, pET21MmcpnΔlid or pET21aMmCpnD386A, respectively. The cells were harvested by centrifugation, resuspended in MQ-A buffer (20 mM HEPES-KOH (pH 7.4), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% (v/v) glycerol, 1 mM DTT, 0.1 mM PMSF) and disrupted using a French Press at a pressure of 16,000 pounds per square inch. The lysate was centrifuged at 15,000g for 30 min to pellet cell debris. The supernatant of an ammonium sulfate cut using ammonium sulfate to 55% saturation was dialyzed against MQ-A buffer and loaded on a Q Sepharose FF column (60 ml, GE Healthcare) equilibrated in MQ-A buffer. Bound proteins were eluted by a NaCl gradient ranging from 0 to 1 M NaCl. Fractions containing Cpn were pooled and diluted 1:1 in MQ-A buffer before they were loaded on a Hi-Trap Heparin column (20 ml, GE Healthcare) equilibrated in MQ-A buffer containing 0.2 M NaCl. Bound proteins were eluted by a NaCl gradient ranging from 0.2 to 1 M NaCl. Fractions containing Cpn were pooled, concentrated using an Amicon Ultra-15 10K concentrator (Millipore) and loaded on a Superose 6 10/300 GL column (GE Healthcare). Fractions containing the oligomeric chaperonin were pooled, and aliquots were flash-frozen in liquid nitrogen. All Cpn variants were well expressed and correctly assembled, and all had similar stability to the wild-type Cpn. The protein concentration was determined using the BCA-Assay (Pierce) with BSA as a standard.
cTRiC
cTRiC was generated essentially as described9. Briefly, 0.25 μM purified TRiC protein was preincubated in TRiC ATPase buffer (50 mM Tris-HCl (pH 7.4), 50 mM KCl, 5 mM MgCl2, 1 mM EGTA) for 5 min at 25 °C. After addition of 20 μmg ml–1 proteinase K and further incubation for 10 min at 25 °C, protease activity was inhibited by adding PMSF to the reaction at a final concentration of 5 mM. After incubation on ice for approximately 10 min, the quantitative conversion of TRiC to cTRiC was confirmed by SDS-PAGE analysis. Reactions containing cTRiC were kept on ice for no more than 2 h before they were used for further biochemical analysis.
Proteinase K protection assay
Purified TRiC (0.25 μM) and purified Cpn WT or Cpn D386A (0.25 μM) were respectively preincubated in TRiC ATPase buffer and Cpn ATPase buffer (20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 10% (v/v) glycerol), which was supplemented with EDTA (5 mM), ADP (1 mM) or ATP (0.2 or 1 mM). To generate AlFx, Al(NO3)3 (5 mM) and NaF (30 mM) were included in the reaction. The reactions were incubated for 10 min (30 min for reactions containing AlFx) at 30 °C for Cpn and 25 °C for TRiC. After addition of 20 μg ml–1 proteinase K and further incubation for 10 min at 25 °C, PMSF was added at a final concentration of 5 mM to inhibit protease activity. The reaction was then incubated on ice for 10 min and analyzed by SDS-PAGE. For N-terminal sequencing, the protein fragments were transferred to a PVDF membrane and visualized by amido-black staining. The protein bands were cut from the membrane and submitted to the Stanford PAN facility. The open Cpn lid is cleaved at position Met259, equivalent to the site cleaved in the TRiC lids, giving rise to two fragments, of 26 and 30 kDa (Fig. 2a). The smaller fragment (24 kDa, Fig. 2a) results from the additional cleavage of the first 15 N-terminal amino acid residues. Protease treatment of Cpn Δlid produces only N- and C-terminal cleavage events. All three fragments share the same N terminus, starting with Tyr15.
Substrate-folding assays
Rhodanese folding by Cpn WT and Cpn Δlid was assayed as described38. In brief, 0.25 μM protein was incubated in Cpn ATPase buffer supplemented with 20 mM sodium thiosulfate. Purified rhodanese was denatured in 6 M guanidinium-HCl containing 5 mM DTT and rapidly diluted 1:100 into the reaction mix to a final concentration of 30 μM. After incubation for 5 min at 37 °C, the reaction was started by addition of 2 mM ATP and allowed to proceed for 50 min at 37 °C. To detect the presence of refolded rhodanese, 10 μl of the reaction was withdrawn and subjected to a rhodanese activity assay as described38.
The actin-folding assay was done as described39. In brief, 0.25 μM TRiC or cTRiC was incubated in buffer A (20 mM HEPES-KOH (pH 7.5), 100 mM potassium acetate, 5 mM MgCl2, 1 mM DTT, 10% (v/v) glycerol and 1% (w/v) PEG 8,000). [35S]actin denatured in 6 M guanidine-HCl39 was then rapidly diluted 1:100 into the reaction mix to a final concentration of 30 μM. After incubation for 10 min at 4 °C, the reaction was supplemented with 1 mM ATP and incubated for 40 min at 30 °C. Generation of native [35S]actin was determined by nondenaturing gel electrophoresis using folded [35S]actin as a control, as described39. The gel was exposed on a phosphorimaging screen (Kodak), which was scanned in a Typhoon 9410 imager (GE Healthcare). The radioactive signal was quantified using ImageQuant 5.2 (Molecular Dynamics). The amount of actin migrating with native mobility is expressed as the percentage of total radioactivity per lane.
To determine actin-folding rates at various ATP concentrations, the appearance of native [35S]actin was measured by a protease protection assay as described9. To this end, TRiC and actin were incubated as described above to form a binary complex. After addition of 5 mM EDTA and 0.2 or 1 mM ATP, samples were withdrawn at the indicated time points and the folding reaction was stopped by incubation on ice and by supplementing the reactions with 10 mM trans-1,2-cyclo-hexanediaminetetraacetate (CDTA). The samples were then incubated with 20 μg ml–1 proteinase K for exactly 5 min at 25 °C before protease activity was inhibited by addition of PMSF at a final concentration of 5 mM. After incubation on ice for 10 min, the samples were analyzed by SDS-PAGE. The gel was exposed on a phosphorimaging screen (Kodak), which was scanned in a Typhoon 9410 imager. Protease treatment of native actin results a previously described 34-kDa fragment9 (Fig. 5b, left gel, lanes 3 and 4). Denatured actin, however, is not protected against protease digestion (lanes 1 and 2) and can thereby be distinguished from native actin. Lanes 5–7 show populations of actin present in a reaction mix after 15 min of incubation in the presence of 1 mM ATP (lane 5) or 0.2 mM ATP (lane 6), or in the absence of nucleotide (lane 7). The full-length actin band in lanes 5 and 6 corresponds to encapsulated substrate protein that is protease protected, as shown previously9. The radioactive signal corresponding to released native actin9 was quantified using ImageQuant 5.2.
Rhodanese binding assay
Cpn WT or Cpn Δlid (0.25 μM) was incubated in Cpn ATPase buffer. Purified [35S]rhodanese denatured in 6 M urea40 was rapidly diluted 1:100 into the reaction mix to a final concentration of 26 μM. After incubation for 15 min at 4 °C, chaperonin-bound rhodanese was detected by nondenaturing gel electrophoresis, as described for TRiC39. The migration behavior of the chaperonin proteins was analyzed by Coomassie staining. To visualize comigration of [35S]rhodanese, the gel was exposed on a phosphor-imaging screen (Kodak), which was scanned in a Typhoon 9410 imager.
ATPase assay
ATP hydrolysis by wild-type chaperonins and chaperonin variants was measured at 25 °C for TRiC and cTRiC and 37 °C for Cpn WT, Cpn Δlid and Cpn D386A, in their respective ATPase buffers, in the presence of 1–1,000 μM [α-32P]ATP. After 5 min of preincubation, the reaction was started by mixing 5 μl [α-32P]ATP solution (0.01 μCi μl–1) with 20 μl 1.25-fold concentrated reaction mix. At the indicated time points, 2-μl samples were taken and transferred onto PEI-cellulose F thin-layer chromatography plastic sheets (EMD Chemicals). The plates were developed in a solvent system containing 1 M LiCl and 0.5 M formic acid in H2O, air-dried and exposed to a phosphorimager (Kodak). After the screen was scanned in a Typhoon 9410 imager, the amount of [α-32P]ATP was quantified using ImageQuant 5.2.
Data fitting
We analyzed allosteric properties with respect to ATP by directly fitting the data to equation (1) and (2), as indicated in the figure legends, using KaleidaGraph Version 4.0 (Synergy Software).
| (1) |
v0 is the observed initial rate of ATP hydrolysis; [S] is the ATP concentration; vmax(1) and vmax(2) are the maximal initial rates of ATP hydrolysis by a single ring and by both rings of group II chaperonins, respectively; n and m are the Hill coefficients for the first and second allosteric transition, respectively; and K1 and K2 are the respective apparent binding constants of ATP to the first and second ring20.
| (2) |
v0 is the observed initial rate of ATP hydrolysis; [S] is the ATP concentration; vmax is the maximal initial rate of ATP hydrolysis; n is the Hill coefficient; and K1 is the respective apparent binding constant for ATP.
To determine whether the allosteric parameters derived above using the Hill equation explain the experimental data more accurately than a simple nonallosteric Michaelis-Menten model, the statistical significance of the difference between the two fits was calculated by an F-test, using the Prism package (GraphPad Software). Briefly, for each chaperonin, the fit obtained using equation (2) with a free Hill coefficient (n) value was compared with a fit done using equation (2) but with the Hill coefficient (n) value arbitrarily fixed at 1 (modeling no allosteric regulation, according to the Michaelis-Menten equation). This analysis indicated that the parameters derived using the Hill equation shown in Table 1 were significantly better at explaining the experimental data than a Michaelis-Menten model (P = 0.0004 for TRiC and P < 0.0001 for Cpn). A similar approach was used to assess the significance of the loss of cooperativity observed upon removal of the lid segments in TRiC or Cpn. Briefly, for both TRiC and Cpn, an F-test was used to compare the global fits obtained for the intact and lidless versions of the chaperonin, using either two independently fit n values or an identical n value for both curves. This test indicated that the loss of positive cooperativity measured in the fits is statistically significant, as the experimental data are best explained by two different n values for intact and lidless versions (P = 0.01 and for TRiC versus cTRiC P = 0.004 for Cpn WT versus Cpn Δlid).
Molecular modeling
The homology model for Cpn was obtained by submitting the protein sequence to the SWISS-Model server41 (http://swissmodel.expasy.org//SWISS-MODEL.html). The figures were prepared by using MacPyMOL (http://pymol.sourceforge.net).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Thanbichler, R. Andino and members of the Frydman laboratory for discussions and comments on the manuscript. S.R. thanks A. Böck for advice and discussions. J. Leigh (University of Washington) kindly provided M. maripaludis DNA. This research was supported by the US National Institutes of Health, the US National Institutes of Health Roadmap Initiative on Nanomedicine, the Studienstiftung des Deutschen Volkes and the Robert Welch Foundation.
Footnotes
AUTHOR CONTRIBUTIONS J.F. and S.R. designed the project and experiments; J.F. was the project leader and S.R. carried out all experiments; C.P. and S.R. carried out the data fitting; C.R.B. and W.C. carried out EM analysis of the complexes; S.R. and J.F. wrote the manuscript. All authors made contributions to the final manuscript.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 2.Horwich AL, Farr GW, Fenton WA. GroEL-GroES-mediated protein folding. Chem. Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 3.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Sigler PB, et al. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 6.Gutsche I, Essen LO, Baumeister W. Group II chaperonins: new TRiC(k)s and turns of a protein folding machine. J. Mol. Biol. 1999;293:295–312. doi: 10.1006/jmbi.1999.3008. [DOI] [PubMed] [Google Scholar]
- 7.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ditzel L, et al. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 9.Meyer AS, et al. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- 10.Shomura Y, et al. Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J. Mol. Biol. 2004;335:1265–1278. doi: 10.1016/j.jmb.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Horovitz A, Fridmann Y, Kafri G, Yifrach O. Review: allostery in chaperonins. J. Struct. Biol. 2001;135:104–114. doi: 10.1006/jsbi.2001.4377. [DOI] [PubMed] [Google Scholar]
- 12.Saibil HR, Horwich AL, Fenton WA. Allostery and protein substrate conformational change during GroEL/GroES-mediated protein folding. Adv. Protein Chem. 2001;59:45–72. doi: 10.1016/s0065-3233(01)59002-6. [DOI] [PubMed] [Google Scholar]
- 13.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Kafri G, Horovitz A. Transient kinetic analysis of ATP-induced allosteric transitions in the eukaryotic chaperonin containing TCP-1. J. Mol. Biol. 2003;326:981–987. doi: 10.1016/s0022-2836(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 15.Yifrach O, Horovitz A. Transient kinetic analysis of adenosine 5′-triphosphate binding-induced conformational changes in the allosteric chaperonin GroEL. Biochemistry. 1998;37:7083–7088. doi: 10.1021/bi980370o. [DOI] [PubMed] [Google Scholar]
- 16.Cliff MJ, Limpkin C, Cameron A, Burston SG, Clarke AR. Elucidation of steps in the capture of a protein substrate for efficient encapsulation by GroE. J. Biol. Chem. 2006;281:21266–21275. doi: 10.1074/jbc.M601605200. [DOI] [PubMed] [Google Scholar]
- 17.Bigotti MG, Clarke AR. Cooperativity in the thermosome. J. Mol. Biol. 2005;348:13–26. doi: 10.1016/j.jmb.2005.01.066. [DOI] [PubMed] [Google Scholar]
- 18.Bigotti MG, Bellamy SR, Clarke AR. The asymmetric ATPase cycle of the thermosome: elucidation of the binding, hydrolysis and product-release steps. J. Mol. Biol. 2006;362:835–843. doi: 10.1016/j.jmb.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 19.Yifrach O, Horovitz A. Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry. 1995;34:5303–5308. doi: 10.1021/bi00016a001. [DOI] [PubMed] [Google Scholar]
- 20.Kafri G, Willison KR, Horovitz A. Nested allosteric interactions in the cytoplasmic chaperonin containing TCP-1. Protein Sci. 2001;10:445–449. doi: 10.1110/ps.44401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranson NA, et al. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- 22.Ranson NA, et al. Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nat. Struct. Mol. Biol. 2006;13:147–152. doi: 10.1038/nsmb1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roseman AM, Chen S, White H, Braig K, Saibil HR. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 24.Inbar E, Horovitz A. GroES promotes the T to R transition of the GroEL ring distal to GroES in the GroEL-GroES complex. Biochemistry. 1997;36:12276–12281. doi: 10.1021/bi9714870. [DOI] [PubMed] [Google Scholar]
- 25.Kusmierczyk AR, Martin J. Nucleotide-dependent protein folding in the type II chaperonin from the mesophilic archaeon Methanococcus maripaludis. Biochem. J. 2003;371:669–673. doi: 10.1042/BJ20030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusmierczyk AR, Martin J. Nested cooperativity and salt dependence of the ATPase activity of the archaeal chaperonin Mm-cpn. FEBS Lett. 2003;547:201–204. doi: 10.1016/s0014-5793(03)00722-1. [DOI] [PubMed] [Google Scholar]
- 27.Szpikowska BK, Swiderek KM, Sherman MA, Mas MT. MgATP binding to the nucleotide-binding domains of the eukaryotic cytoplasmic chaperonin induces conformational changes in the putative substrate-binding domains. Protein Sci. 1998;7:1524–1530. doi: 10.1002/pro.5560070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iizuka R, et al. Characterization of archaeal group II chaperonin-ADP-metal fluoride complexes: implications that group II chaperonins operate as a “two-stroke engine”. J. Biol. Chem. 2005;280:40375–40383. doi: 10.1074/jbc.M506785200. [DOI] [PubMed] [Google Scholar]
- 29.Llorca O, et al. 3D reconstruction of the ATP-bound form of CCT reveals the asymmetric folding conformation of a type II chaperonin. Nat. Struct. Biol. 1999;6:639–642. doi: 10.1038/10689. [DOI] [PubMed] [Google Scholar]
- 30.Schoehn G, Hayes M, Cliff M, Clarke AR, Saibil HR. Domain rotations between open, closed and bullet-shaped forms of the thermosome, an archaeal chaperonin. J. Mol. Biol. 2000;301:323–332. doi: 10.1006/jmbi.2000.3952. [DOI] [PubMed] [Google Scholar]
- 31.Schoehn G, Quaite-Randall E, Jimenez JL, Joachimiak A, Saibil HR. Three conformations of an archaeal chaperonin, TF55 from Sulfolobus shibatae. J. Mol. Biol. 2000;296:813–819. doi: 10.1006/jmbi.2000.3505. [DOI] [PubMed] [Google Scholar]
- 32.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat. Struct. Mol. Biol. 2005;12:233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 33.Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aharoni A, Horovitz A. Inter-ring communication is disrupted in the GroEL mutant Arg13 → Gly; Ala126 → Val with known crystal structure. J. Mol. Biol. 1996;258:732–735. doi: 10.1006/jmbi.1996.0282. [DOI] [PubMed] [Google Scholar]
- 35.Sewell BT, et al. A mutant chaperonin with rearranged inter-ring electrostatic contacts and temperature-sensitive dissociation. Nat. Struct. Mol. Biol. 2004;11:1128–1133. doi: 10.1038/nsmb844. [DOI] [PubMed] [Google Scholar]
- 36.Tian G, Vainberg IE, Tap WD, Lewis SA, Cowan NJ. Specificity in chaperonin-mediated protein folding. Nature. 1995;375:250–253. doi: 10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
- 37.Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol. Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 38.Weber F, Hayer-Hartl M. Refolding of bovine mitochondrial rhodanese by chaperonins GroEL and GroES. Methods Mol. Biol. 2000;140:117–126. doi: 10.1385/1-59259-061-6:117. [DOI] [PubMed] [Google Scholar]
- 39.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 40.Weissman JS, Kashi Y, Fenton WA, Horwich AL. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 41.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.