Summary
The ring-shaped hetero-oligomeric chaperonin TRiC/CCT uses ATP to fold a diverse subset of eukaryotic proteins. To define the basis of TRiC/CCT substrate recognition, we mapped the chaperonin interactions with the VHL tumor suppressor. VHL has two well-defined TRiC binding determinants. Each determinant contacts a specific subset of chaperonin subunits, indicating that TRiC paralogs exhibit distinct but overlapping specificities. The substrate binding site in these subunits localizes to a helical region in the apical domains that is structurally equivalent to that of bacterial chaperonins. Transferring the distal portion of helix 11 between TRiC subunits suffices to transfer specificity for a given substrate motif. We conclude that the architecture of the substrate binding domain is evolutionarily conserved among eukaryotic and bacterial chaperonins. The unique combination of specificity and plasticity in TRiC substrate binding may diversify the range of motifs recognized by this chaperonin and contribute to its unique ability to fold eukaryotic proteins.
Introduction
Chaperonins are essential components of the cellular machinery that folds newly made and stress-denatured proteins (Bukau and Horwich, 1998; Frydman, 2001; Hartl and Hayer-Hartl, 2002). These large oligomeric assemblies consist of 14–18 subunits arranged in two stacked rings. Each ring contains a central chamber that accommodates the nonnative polypeptide substrates. Chaperonins mediate folding by undergoing a conformational cycle driven by ATP binding and hydrolysis. Although it is clear that the ring-shaped architecture is essential for folding, the exact mechanism by which chaperonins promote folding of a bound substrate remains to be established (Swain and Gierasch, 2005).
Chaperonins are classified into two distinct families. Prokaryotes and the endosymbiotic organelles of eukaryotic cells contain the well-studied group I chaperonins, exemplified by GroEL from E. coli (Bukau and Horwich, 1998). These chaperonins are homo-oligomeric and require the assistance of a small ring-shaped co-chaperone, such as GroES, that acts as a lid for the central cavity. Individual subunits consist of three domains: an equatorial ATP binding domain, an apical domain containing the polypeptide and GroES binding sites, and an intermediate hinge domain that enables communication between the equatorial and apical domains. In the absence of nucleotide, the apical domains expose hydrophobic polypeptide binding sites to the cavity. ATP and GroES binding occlude these hydrophobic binding sites, thus releasing the substrate into the central cavity. The ensuing confinement of the bound polypeptide within this chamber appears to be essential for folding of most GroEL substrates (Brinker et al., 2001).
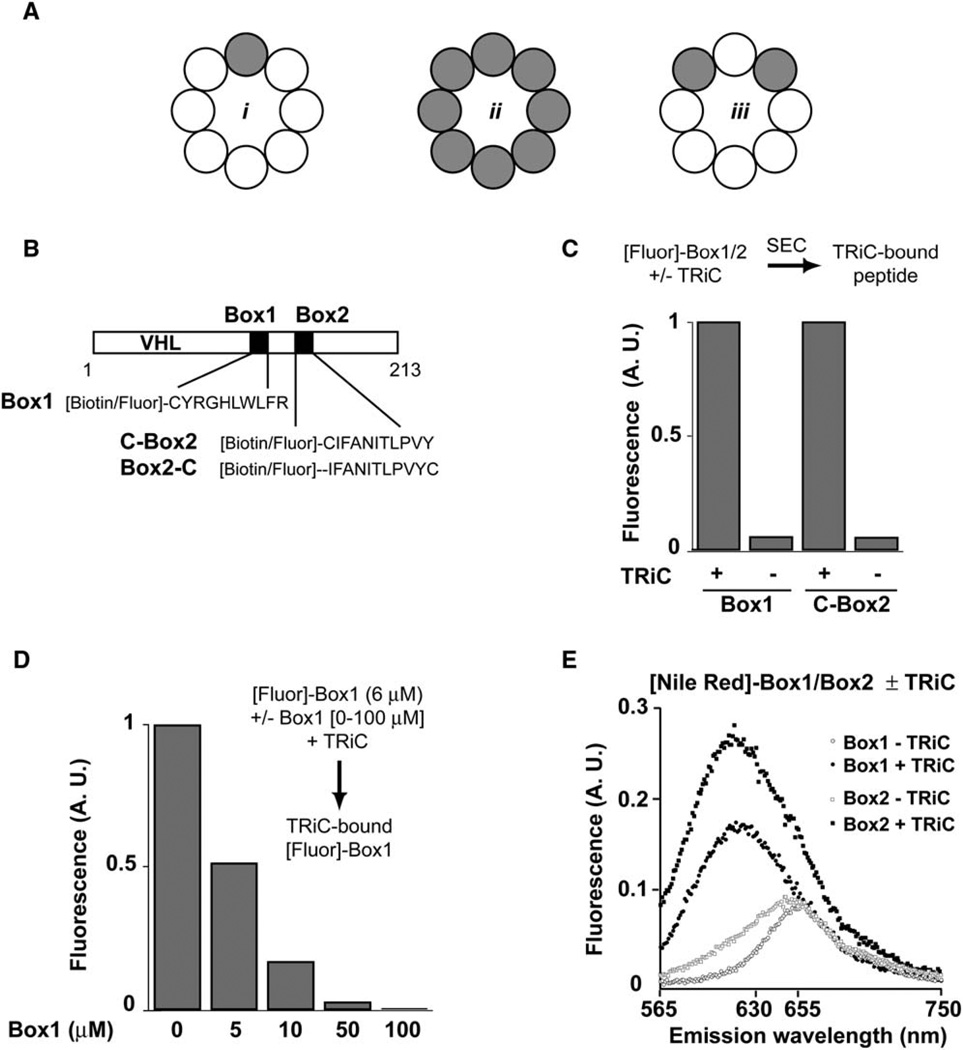
Group II chaperonins, found in eukaryotic cells and in archaea, have a similar overall architecture and subunit domain arrangement as group I chaperonins but differ from their bacterial counterparts in two major aspects (Gutsche et al., 1999; Spiess et al., 2004). First, they lack a cochaperone that acts as a lid. Instead they have an entirely different mechanism to confine the substrate in the central cavity, which relies on the ATP-dependent formation of a built-in lid from protrusions found at the tip of the apical domains (Meyer et al., 2003). A second major departure from bacterial chaperonins is that group II complexes are hetero-oligomeric. For example, the eukaryotic chaperonin TCP-1 ring complex (TRiC, also called CCT) consists of eight paralogous subunits (Spiess et al., 2004). The functional relevance of subunit diversity in group II chaperonins is not well understood. Because most of the sequence divergence within the paralogous subunits is in the apical domains (Kim et al., 1994; Archibald et al., 2001), the simplest hypothesis is that the subunits of TRiC differ in their substrate specificity. However, there is little experimental evidence on this issue because the substrate binding sites of TRiC/CCT have not been defined. Although cryo-electron microscopy (cryo-EM) reconstructions led to the suggestion that TRiC subunits are highly specific for defined sequence motifs in the substrates actin and tubulin (Gomez-Puertas et al., 2004; Llorca et al., 1999, 2000) (Figure 1Ai), photocrosslinking analysis of actin-TRiC contacts during cotranslational folding demonstrated significant overlap in the substrate specificity of the subunits (Etchells et al., 2005) (Figure 1Aiii).
Figure 1. The TRiC-Interaction Motifs of VHL Bind the Chaperonin in a Hydrophobic Environment.
(A) Possible roles of TRiC subunits in binding substrate determinants: recognition of the binding sequence may be (i) subunit specific or (ii) nonspecific. Scenario iii shows a binding event based on overlapping specificity of a subset of subunits.
(B) Schematic representation of the TRiC binding determinants Box1 and Box2 within VHL highlighting the sequences of corresponding peptides used in this study.
(C) Box1 and Box2 peptides bind directly to TRiC. Peptides (6 µM) were fluorescently labeled with Alexa Fluor 546. After 30 min incubation with TRiC (0.1 µM), the sample was analyzed by size-exclusion chromatography (SEC) to separate the TRiC-bound from unbound peptide and the fluorescence of the TRiC fraction analyzed.
(D) Concentration-dependent competition of Alexa Fluor 546-Box1 binding to TRiC by unlabeled Box1 peptide. Experiment was done as in (C) but in the presence of indicated concentrations of unlabeled Box1 peptide.
(E) TRiC binds the VHL determinants in a hydrophobic environment. Box1 and Box2 peptides (18 nM), labeled with the polarity-sensitive dye Nile Red, were incubated in bufferwith or without TRiC (280 nM) and the fluorescence emission spectrum recorded after excitation at 550 nm. Binding to the chaperonin causes spectral changes characteristic of a transfer of Box1/Box2 peptides to a hydrophobic environment.
The structural and mechanistic differences between group I and group II chaperonins have profound functional consequences on their ability to fold proteins (Tian et al., 1995; Kerner et al., 2005). Several eukaryotic proteins, such as actin, can only be folded by TRiC (Tian et al., 1995; Frydman et al., 1992). Conversely, bacterial proteins that require GroEL assistance are unable to fold in the eukaryotic cytosol (Kerner et al., 2005). Thus, despite their shared overall architecture, group I and group II chaperonins influence the folding landscape of their substrates in different ways.
An important factor underlying the distinct specificity of group I and group II chaperonins is thought to be the chemical nature of their interaction with substrates. Substrate binding to GroEL involves recognition of exposed hydrophobic surfaces characteristic of unfolded polypeptides (Ashcroft et al., 2002; Chen and Sigler, 1999; Kobayashi et al., 1999). In contrast, cryo-EM and evolutionary analyses led to the suggestion that different TRiC subunits harbor highly specific binding sites for unique polar motifs found in selected cellular proteins (Hynes and Willison, 2000; Llorca et al., 1999, 2000; McCormack et al., 2001). On the other hand, biochemical analysis of the determinants that direct association of TRiC with several substrates, including actin (Rommelaere et al., 1999), the von Hippel-Lindau (VHL) tumor suppressor (Feldman et al., 2003), and the Gβ WD-40 protein (Kubota et al., 2006), implicate hydrophobic interactions in TRiC binding.
We reasoned that identifying the substrate binding sites in TRiC should clarify the principles governing substrate recognition and their possible links to subunit heterogeneity. To this end, we took advantage of previous observations that the VHL tumor suppressor, an obligate substrate of TRiC (Feldman et al., 1999; Melville et al., 2003), binds to the chaperonin through two short linear motifs (Feldman et al., 2003). These determinants, called Box1 and Box2, are necessary and sufficient for chaperonin binding. Of note, tumor-causing mutations within these sites, as well as alanine replacements, destabilize binding to TRiC and lead to severe misfolding of VHL in mammalian cells, even though the polypeptide retains the ability to fold to the native state in vitro upon stepwise dialysis from denaturant (Feldman et al., 2003). Because the discrete chaperonin binding determinants are required for both TRiC association and folding in vivo (Feldman et al., 2003), it constitutes a unique system to dissect how the chaperonin engages its substrates. Using photocrosslinking and fluorescence spectroscopy, we establish here that different subunits in the complex are specialized to recognize defined substrate motifs; however, there is also redundancy between the substrate specificity of different subunits. Substrate binding and specificity reside within a helical region of the apical domain of each subunit that is structurally equivalent to the GroEL substrate binding site (Chen and Sigler, 1999; Kobayashi et al., 1999), thus revealing a fundamental conservation between bacterial and eukaryotic chaperonins. However, the diversification of substrate binding sites in TRiC/CCT results in a unique combination of structural plasticity and specificity that may underlie its ability to facilitate folding of a subset of eukaryotic proteins.
Results
The TRiC-Interaction Motifs of VHL Bind the Chaperonin in a Hydrophobic Environment
VHL contains two short linear motifs, Box1 and Box2, that are necessary and sufficient for binding to the chaperonin in vivo, suggesting that these sequences bind TRiC directly (Feldman et al., 2003). Therefore, we used peptides corresponding to Box1 and Box2 (herein Box1 and Box2, see Figure 1B) carrying either fluorescent probes or photoactivatable crosslinkers to probe the vicinity of the substrate recognition surface in TRiC and more precisely define the chemical nature and the location of the chaperonin binding site(s). Binding of fluorescently labeled Box1 and Box2 peptides to TRiC was first examined by size-exclusion chromatography (SEC) (Figure 1C for Alexa Fluor 546-labeled peptides). The labeled peptides coeluted with TRiC, and no fluorescence was observed in the high molecular weight fractions if TRiC was either omitted or incubated with the unreacted (quenched or free) fluorescent dye (Figures 1C and 1D and data not shown). TRiC-associated fluorescence was reduced by competition with unlabeled peptide in a concentration-dependent manner, indicating that the peptide moiety is responsible for the observed binding (Figure 1D). Similar results were obtained when labeling the peptides with other fluorescent dyes such as fluorescein or Texas Red (data not shown).
We next examined the proximate environment in the chaperonin binding site by labeling the peptides with the polarity-sensitive dye Nile Red (NR) (Kim et al., 2005). As previously shown for GroEL-substrate interactions, the emission spectrum of NR has a pronounced blue shift and increases its quantum yield as its environment becomes more hydrophobic (Figures S1 A and S1B in the Supplemental Data available with this article online) (Kim et al., 2005). NR-labeled Box1 and Box2 in buffer had emission spectra characteristic of the dye in a polar environment. However, incubation of the peptides with TRiC was accompanied by the blue shift in the emission spectrum and increase in fluorescence intensity characteristic of binding in a hydrophobic environment (Figure 1E). Importantly, these spectral changes were abolished if binding of the NR peptide to TRiC was competed with unlabeled peptide (data not shown). These experiments indicate that, upon binding, VHL Box1 occupies a hydrophobic environment, in agreement with previous observations that TRiC binding is mediated by hydrophobic residues within VHL (Feldman et al., 2003).
Box1 and Box2 Contact a Subset of TRiC/CCT Subunits
We next used photocrosslinking to map the contacts between the Box1 and Box2 binding motifs and individual chaperonin subunits. We used the photoactivatable bifunctional crosslinker BPIA (Figure 2A), because its highly reactive benzophenone moiety can form covalent crosslinks independent of the chemical nature of the binding environment (Kramer et al., 2002). In addition, the short length of BPIA (10 Å, [Buskiewicz et al., 2004]), equivalent to an extended polypeptide chain of three amino acids, would increase the likelihood that the crosslinks only occur to the TRiC subunit that directly contacts the binding motif and not to other subunits within the complex. BPIA was attached to the N-(or C-) terminal cysteine of Box1 and Box2. The peptides also contained an N-terminal biotin tag (or fluorescent probe, see below, Figure S4) to facilitate detection and analysis of the photoadducts. After incubation of the peptides with TRiC and photolysis, the chaperonin subunits were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The TRiC subunits, migrating as closely spaced bands between 50 and 60 kDa, were visualized by protein staining (Figures 2B–2D), whereas covalent adducts with the biotinylated peptide were detected by using streptavidin-HRP (Figure 2).
Figure 2. VHL Box1 and Box2 Contact a Subset of TRiC Subunits.
(A) Experimental Strategy and structure of the crosslinker Benzophenone-4-iodoacetamide (BPIA).
(B) Photocrosslinks of Box1 to TRiC are strictly dependent on the presence of the crosslinker BPIA. After transfer to nitrocellulose, the crosslinked adducts were detected by probing for the N-terminal biotin moiety of the peptide and the chaperonin subunits observed by staining with Ponceau S.
(C) Dependence of Box2-TRiC crosslinks on the relative position of the crosslinker. TRiC crosslinks were obtained with Box2 peptides carrying the crosslinker at the C terminus (Box2-C), but not at the N terminus (C-Box2), indicating that the peptide binds TRiC in a defined orientation. Of note, both Box2-C and C-Box2 bind to TRiC. Box1 is included as a positive control.
(D) Formation of Box1-TRiC adducts requires access to the central chaperonin cavity. To block access to the substrate binding sites within the central chamber, TRiC was preincubated with ATP and AlFx.
Initially we examined the crosslinks of Box1 with TRiC. Upon photolysis, several crosslinks with TRiC subunits were detected by streptavidin reactivity (Figures 2B– 2D). Importantly, the crosslinks were strictly dependent on the presence of BPIA (Figure 2B) and increased with the concentration of Box1 peptide (Figure S2). Box2 was also crosslinked to several subunits of TRiC (Figure 2C). Surprisingly, the ability of this peptide to crosslink to the chaperonin was highly dependent on the position of the crosslinker relative to the binding motif (Figure 2C). Whereas Box2 peptides carrying an N- or C-terminal cysteine both associated with TRiC with comparable efficiency and in a similar environment (Figure S3), only peptides carrying BPIA on the C-terminal cysteine were able to form crosslinks with the chaperonin subunits (Figure 2C). The all-or-nothing reaction for Box2 depending on the placement of the crosslinker suggests that the peptide binds to its cognate TRiC subunits in a defined orientation that may, for instance, place the BPIA moiety facing away from the chaperonin. Of note, the N-terminally labeled Box2 serves as a negative control, indicating the crosslinks are not mediated by BPIA itself.
To corroborate that the VHL-derived peptides interact with the chaperonin in a substrate-like manner, we examined the effect of nucleotide-induced lid closure on the crosslinking reaction. We previously demonstrated that incubating TRiC with mimics of the trigonal-bipyramidal transition state of ATP hydrolysis induces symmetric lid closure in both rings, thereby blocking substrate binding to TRiC (Meyer et al., 2003). Strikingly, inducing lid closure prior to addition of Box1 strongly reduced the appearance of crosslinks upon photolysis (Figure 2D). We conclude that the VHL-derived peptide indeed behaves as a TRiC substrate.
The one-dimensional (1D) gel analysis of the crosslinking experiments suggests that both Box1 and Box2 interact with some, but not all, subunits within the complex. These results did not correspond unambiguously with either model of subunit specificity. Thus, if the different subunits in TRiC are highly specific for distinct binding motifs within substrates (Figure 1Ai), then we would predict that either peptide would crosslink to one subunit. On the other hand, if, similar to GroEL, all subunits have a broad specificity, then we would expect that all subunits would crosslink with Box1 and Box2 (Figure 1Aiii). In contrast, our results suggest that these peptides interact with a subset of subunits within the complex (Figure 1Aiii). To gain further insight into the specificity of TRiC binding, we next identified the subunits that interact with Box1.
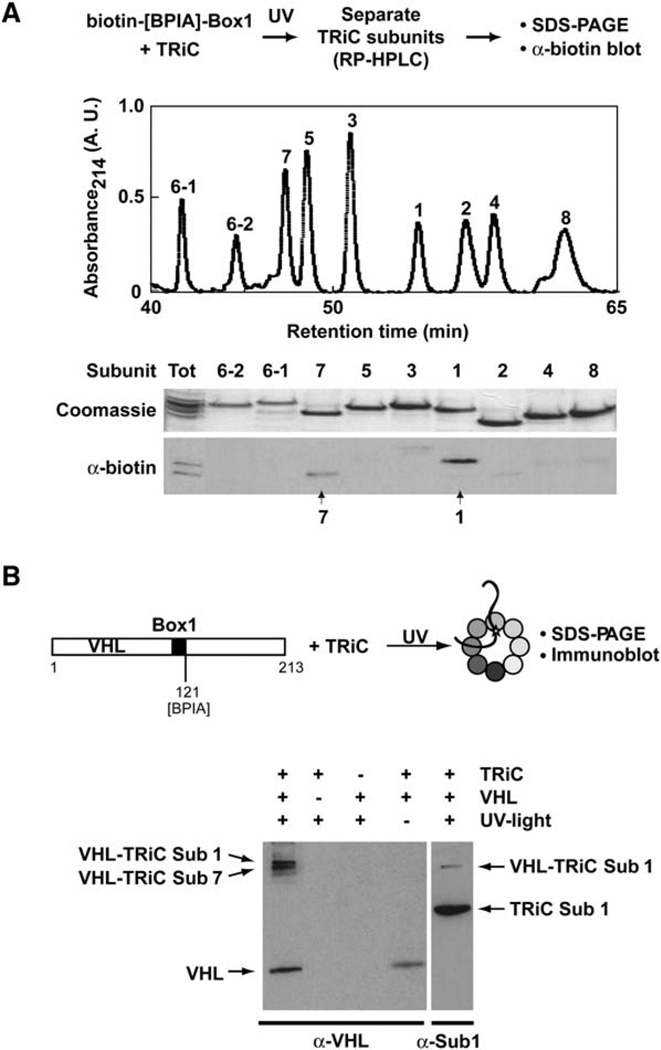
Box1 Crosslinks to TRiC/CCT Subunits 1 and 7
The size similarity of paralogous subunits of TRiC, together with the low efficiency of photocrosslinking, precluded identification of the particular subunits that crosslink to the VHL-derived peptides by simple 1D SDS-PAGE analysis (see Figures 2B–2D) or mass spectrometry (data not shown). We therefore developed an approach based on reverse-phase chromatography (RP-HPLC) separation of individual TRiC subunits followed by subunit identification by mass spectrometry (Figure 3, Figure S4, and data not shown). The elution profile of TRiC subunits was not affected by crosslinking to the Box1 peptide (data not shown). After separation, fractions containing individual TRiC subunits were analyzed by SDS-PAGE and visualized by Coomassie staining. Probing for the biotin tag identified the Box1-crosslinked subunits as TRiC subunits 1 and 7 (Figure 3A).
Figure 3. Box1 Crosslinks to TRiC/CCT Subunits 1 and 7.
(A) The isolated VHL-Box1 motif contacts TRiC/CCT subunits 1 and 7. Box1 carrying an N-terminal biotin moiety was photocrosslinked to TRiC as in Figure 2. Individual chaperonin subunits were then separated by C4-RP-HPLC. Individual fractions were resolved by SDS-PAGE (top) and subunit identity determined by mass spectrometry. The subunits forming adducts with Box1 were identified by probing for the biotin tag.
(B) The Box1 motif within full-length VHL contacts TRiC/CCT subunits 1 and 7. Full-length VHL carrying BPIA on a unique cysteine at position 121, at the C terminus of Box1, was bound to TRiC. The TRiC-VHL complex was isolated by SEC, and VHL was crosslinked to TRiC by UV irradiation. Proteins were resolved by 12% SDS-PAGE and probed with VHL and anti-subunit 1-specific antibodies. Two TRiC- and UV-dependent VHL immunoreactive adducts were observed: one corresponding to a VHL crosslink to subunit 1 (lane 5) and one corresponding to a crosslink to subunit 7 (see Figure S5).
To examine whether changing the tag on the Box1 peptide affected the crosslinks to TRiC subunits, we carried out a similar experiment using a Box1 peptide with an N-terminal fluorescein tag. After photolysis, the TRiC subunits were separated by RP-HPLC and the fluorescence signal determined. The fluorescence signal coeluted with fractions containing subunit 1 or 7 (Figure S4). We conclude that Box1 crosslinks to subunits 1 and 7 of TRiC regardless of the tag used to detect the crosslinked adduct.
The Subunit Specificity of Box1-TRiC Contacts Is Preserved in Intact VHL
Although analysis of TRiC interactions with the isolated Box1 motif revealed an interaction with subunits 1 and 7, it is in principle possible that the chaperonin contacts of Box1 within intact VHL are affected by other sequences within the substrate protein. For instance, formation of compact folding intermediates or the presence of additional TRiC binding motifs in VHL could modify the subunit interactions of the Box1 motif within VHL. To examine this question, we introduced a single cysteine residue at the C terminus of the Box1 motif (amino acid position 121; VHL-121C) (Figure 3B). The amino acid substitutions in VHL-121C did not affect its ability to bind to TRiC nor to fold in vitro (E.J.M., unpublished data). Irradiation of a BPIA-labeled VHL-121C complex with TRiC yielded two VHL adducts that were readily detectable by anti-VHL immunoblot analysis (Figure 3B). The VHL adducts were strictly dependent on the presence of TRiC and on irradiation with UV light, suggesting that they corresponded to specific crosslinks between VHL and TRiC (Figure 3B). Furthermore, their apparent molecular mass corresponded to that of a VHL adduct with a single TRiC subunit. Immunoblot analysis with an antibody directed against subunit 1 of TRiC (TCP-1, [Etchells et al., 2005]) revealed that one of the crosslinks consisted of an adduct between VHL and subunit 1 (Figure 3B, lane 5). The identity of the TRiC subunits that crosslink to VHL-121C was further defined by mass spectrometry (Figure S5). After irradiation of the VHLTRiC complex, VHL-associated proteins were immunoprecipitated and analyzed by SDS-PAGE followed by silver staining (Figure S5). Two bands, corresponding in molecular weight to VHL-TRiC adducts, were exclusively observed in the UV-irradiated, but not in the control, sample. Excision and mass-spectrometry analysis of these bands revealed they were crosslinks of VHL with TRiC subunit 1 and subunit 7 (Figure S5). Thus, the Box1 motif in intact VHL contacts the same TRiC subunits, 1 and 7, that are contacted by the isolated peptide containing the Box1 motif. We conclude that the specificity of Box1 for these TRiC subunits is autonomous to the sequence of the motif and is not modified by the flanking sequences in the intact protein.
The Chaperonin Apical Domains Contain the Substrate Binding and Specificity Determinants
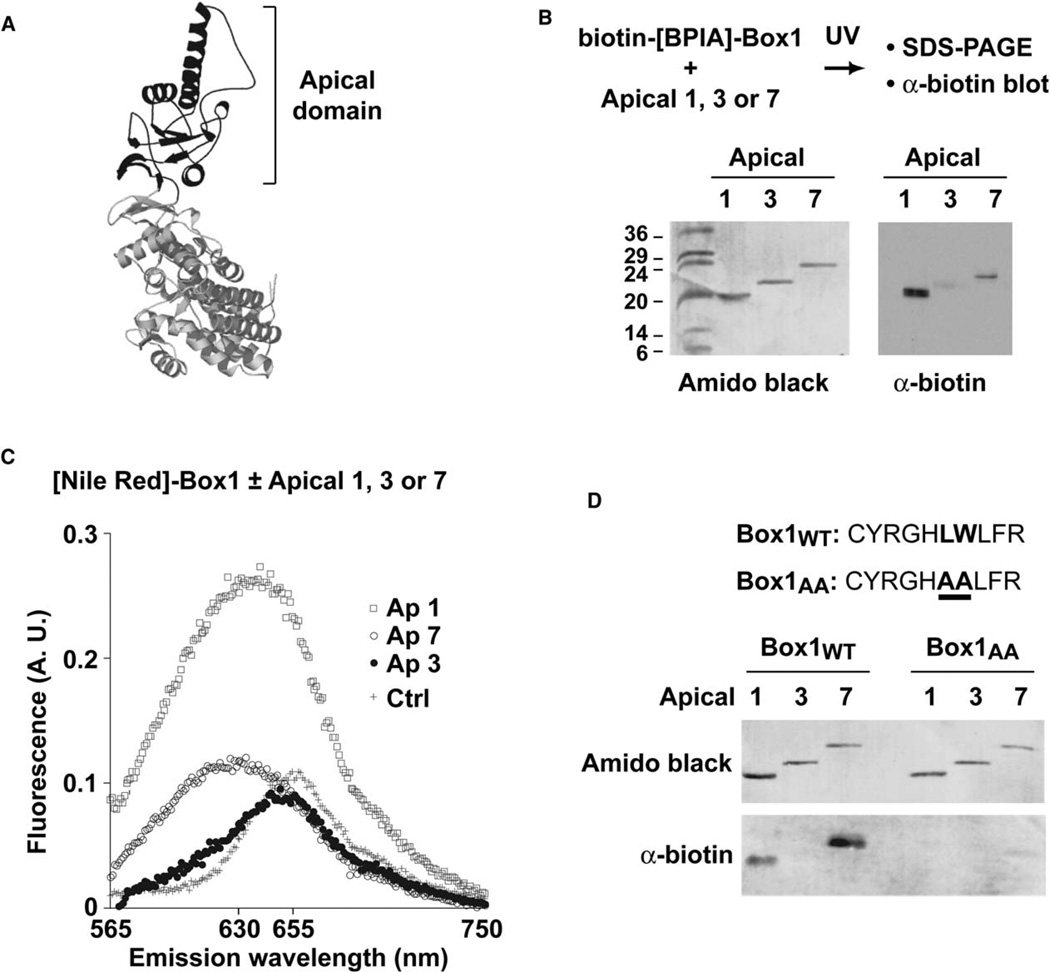
The crosslinks between Box1 and subunits 1 and 7 could result from direct binding of this motif to each of these subunits. Alternatively, only one of these subunits may contain the binding site, whereas the other would merely be located in close proximity to it. These scenarios could be distinguished by directly testing the interaction of Box1 with each subunit. Although the location of the substrate binding sites in TRiC has not yet been defined, we hypothesized by analogy to GroEL that substrate recognition occurs within the apical domain (Figure 4A). We expressed and purified the isolated apical domains of subunits 1 and 7 (herein Ap1 and Ap7, respectively). As a control, we also expressed and purified the apical domain of subunit 3 (herein Ap3), which does not crosslink to Box1 within the intact TRiC complex (Figure 3). Importantly, the isolated apical domains were correctly folded, soluble, and monomeric, as determined by circular dichroism spectroscopy and gel filtration chromatography (Figure S6; data not shown).
Figure 4. The TRiC/CCT Apical Domains Contain the Substrate Binding and Specificity Determinants.
(A) Ribbon diagram of a model of an individual TRiC/CCT subunit. The apical domain, which contains the built-in lid and the putative substrate binding sites, is highlighted in black.
(B) Box1 crosslinks to purified apical domains of TRiC subunits 1 and 7, but not to apical domain 3. The biotin tag on crosslinked adducts was detected as in Figure 2. Total protein on the membrane was visualized by staining with Amido black.
(C) Box1 binds to apical domains 1 and 7 in a hydrophobic environment similar to that of intact TRiC. The emission spectra of NR-Box1 incubated with apical domains 1,7, and 3 was recorded as described for Figure 1E. No spectral change is observed upon incubation with apical domain 3.
(D) Mutations in the Box1 motif that abolish VHL-TRiC binding in vivo impair interaction with the isolated apical domains 1 and 7. Crosslinking was carried out as in (B) but comparing Box1 with Box1AA, a variant containing alanine replacements at positions corresponding to Leu116 and Trp117 in full-length VHL.
Initially, we used the crosslinking assay to assess binding of Box1 to the isolated apical domains. Box1 crosslinked to the apical domains of subunit 1 and subunit 7, indicating that indeed both subunits interact directly with this motif. The apparent molecular weight of the adducts corresponds to a single peptide mass addition, indicating that Box1 binds at a single site within these apical domains, which presumably corresponds to the substrate binding site of these TRiC subunits. In contrast, Box1 did not crosslink to the apical domain of subunit 3 (Figure 4B). Thus, the apical domains recapitulate binding to Box1 with the subunit specificity observed for the intact TRiC complex.
We next explored the binding environment of Box1 in the isolated apical domains by using the Nile Red-labeled peptide (Figure 4C). Addition of Ap3 did not produce a significant change in the fluorescent signal compared to free NR-Box1 in solution, consistent with the fact that Box1 does not bind to this apical domain. On the other hand, as observed with intact TRiC complex, binding of Box1 peptide to Ap1 and Ap7 caused the fluorescent shift characteristic of binding in a hydrophobic environment. These findings further indicate that the interaction of the peptide with the isolated apical domains recapitulates the binding observed in the intact TRiC complex.
In full-length VHL, a single tryptophan to alanine substitution at position 117 within the Box1 binding motif prevents association of VHL with TRiC (Feldman et al., 2003). We thus tested whether the isolated apical domains interact with peptide Box1AA, which contains two alanine substitutions at positions corresponding to W117 and L116 in intact VHL. As expected, if the interaction of Box1 with Ap1 and Ap7 resembles the interaction of full-length VHL with TRiC, the Box1AA peptide does not crosslink to either apical domain (Figure 4D).
We conclude that the apical domains contain the substrate binding domains of TRiC. Furthermore, the isolated apical domains maintain the specificity observed in the intact chaperonin and recognize the same structural elements that determine VHL association with TRiC in vivo, supporting the physiological relevance of our ensuing analyses.
Helix 11 in the Apical Domains Imparts Substrate Binding and Specificity
Identification of the substrate binding sites in TRiC has proven difficult so far because no experimental system has yet provided either a genetic or biochemical solution to approach the problem. Having established that isolated apical domains bind substrate-derived motifs with specificity mirroring that of intact TRiC, we next used a mutagenesis approach to identify their binding and specificity determinants. Two distinct surfaces within the apical domain have been considered as putative substrate binding sites (Spiess et al., 2004). Based on cryo-EM and evolutionary analyses, it was proposed that the substrate binding sites reside in the region lining the inner face of the cavity in the closed, ATP-induced, state (herein PS site, for Polar Strands) (Gomez-Puertas et al., 2004; Pappenberger et al., 2002). Because this region consists of several strands containing charged and polar amino acids, this view implies that TRiC/CCT recognizes surface-exposed charged and polar regions in substrate proteins (Pappenberger et al., 2002). Alternatively, substrate binding could be mediated by structural elements homologous to those in GroEL, namely helices H and I, which in group II chaperonins, correspond to helices 10 and 11 in the apical domain (herein HL site, for HeLical) (Ditzel et al., 1998).
We directly addressed the contribution of either site to substrate binding by introducing alanine replacements in either region of Ap1 and evaluating their effect on the interaction with VHL-Box1 (Figures 5A and 5B). For the PS site, we followed the analysis of Pappenberger et al. (2002) and replaced the central amino acids in the polar strands, R319, R320, K321, and K322 with four alanines (Figure 5B). The mutations introduced in the HL site were based on studies of GroEL (Chen and Sigler, 1999; Kobayashi et al., 1999). To guide our mutagenesis, we created homology models for the TRiC apical domains by using the thermosome crystal as a template and performed pair-wise alignments between the HL sites in apical domains Ap1 and Ap7, which bind to Box1, and that in Ap3, which does not. This analysis identified a hydrophobic patch flanked by a negatively charged region shared by Ap1 and Ap7, but not Ap3 (Figure 7A and data not shown). We thus generated two sets of alanine-substituted mutants in the HL site of apical domain Ap1 (Figure 5A): one targeting the proximal part of both helices (Ap1H10/11) and one targeting the distal region of helix 11 (Ap1H11).
Figure 5. Helix 11 in the Apical Domains Contains Substrate Binding and Specificity Determinants.
(A and B) The relative contribution to substrate binding of (A) Helices 10/11 (HL site) and (B) the Polar Strands site (PS site) was evaluated by introducing alanine replacements at the positions highlighted in red on the ribbon diagrams of apical domain 1. Binding of Box1 to wild-type Ap1 and the indicated variants was assessed as in Figure 4. Mutation of the distal part of helix 11 (Ap1H11) impairs crosslinking to Box1. Of note, the corresponding amino acids in GroEL are also involved in substrate binding.
(C) Helix 11 controls binding specificity for Box1. The surface-exposed residues of helix 11, required for the Ap1-Box1 interaction, were engineered onto the backbone of the apical domain of subunit 3, which does not bind to Box1 (Ap3HX1). Conversely, the same region of helix 11 in Ap3 was grafted into Ap1 (Ap1HX3). Exchanging this region of helix 11 switched the specificity of these apical domains for Box1.
Molecular structures in (A)–(C) are models of the apical domain of TRiC subunit 1 based on PDB ID 1E0R.
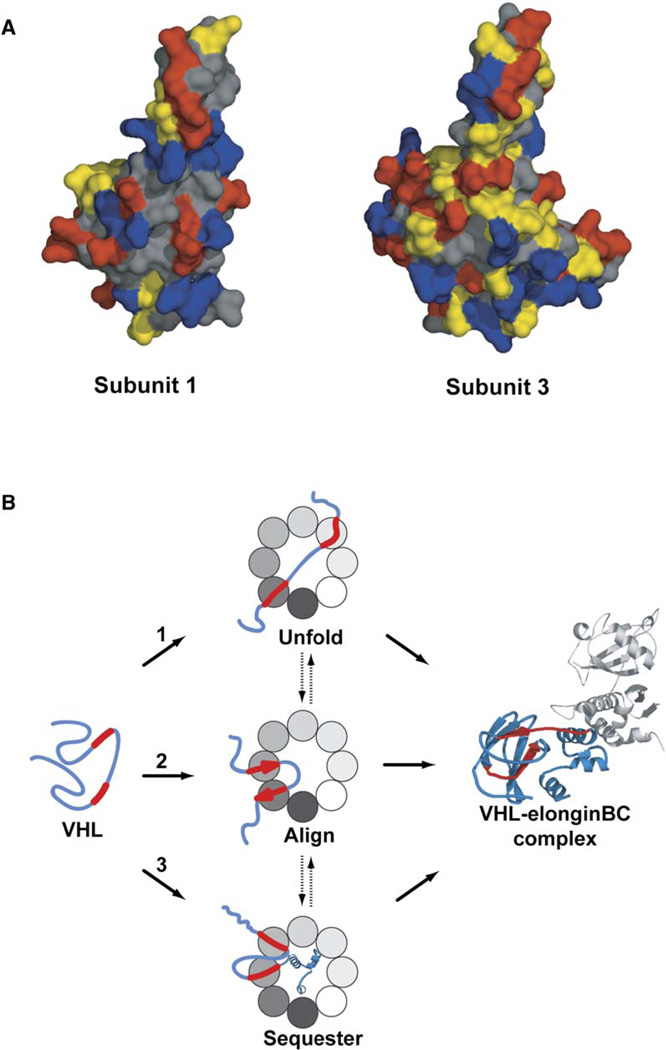
Figure 7. Structural and Mechanistic Implications of the Hetero-Oligomeric Nature of TRiC/CCT.
(A) Surface properties of the helical region in apical domains of TRiC subunits 1 and 3. Residues are colored according to their side-chain character (negatively charged in red, positively charged in blue, uncharged polar in yellow, and uncharged nonpolar in gray). The groove delimited by helix 10 and helix 11 (H10 and H11, respectively) in TRiC subunit 1 has hydrophobic character, whereas in subunit 3, the hydrophobic elements are paired with polar ones (see Figure S8 for further details).
(B) Possible scenarios for how the heterooligomeric nature of TRiC may influence the folding landscape of VHL. Folded VHL contains a β-barrel domain and an α-helical domain that contacts the elongin BC complex (PDB ID 1VCB). Binding of the Box1 and Box2 VHL motifs to a subset of TRiC subunits can (1) unfold or destabilize kinetically trapped intermediated, (2) align the strands corresponding to Box1 and Box2 to favor a topology that primes formation of the β-sheet domain in VHL, or (3) sequester sequences containing the Box1 and Box2 binding sites to facilitate folding of the C-terminal VHL domain, thus preventing intradomain aggregation. Because the binding determinants in VHL are recognized by multiple TRiC subunits, these possible scenarios may occur sequentially during the folding reaction.
All isolated alanine variants of Ap1 were analyzed for their effect on the interaction with Box1. Although we observed a significant reduction in Box1 crosslinks to the apical domain variant Ap1H11, carrying distal Ala replacements in helix 11, neither mutation of the PS site nor the other mutation in the HL site, Ap1H10/11, substantially decreased Box1 interaction (Figures 5A and 5B). These results suggest that the PS region in the TRiC/CCT apical domains does not contribute to VHL-Box1 binding, whereas the HL site, and in particular helix 11, contains important substrate binding determinants.
We hypothesized that if helix 11 contacts polypeptides directly, it may control the substrate specificity of the TRiC/CCT apical domains. Accordingly, we swapped the surface-exposed residues of helix 11 in Ap1 with those of Ap3, which does not bind Box1 (Figure 5C). Strikingly, introduction of helix 11 residues from Ap3 into Ap1 reduced the substrate interaction. Conversely, engineering helix 11 from Ap1 onto Ap3 conferred binding of Ap3 to Box1. We conclude that helix 11 contains a major specificity determinant for polypeptide binding in the apical domains of TRiC. Together, the above results suggest that the polypeptide substrate interaction sites in the group II chaperonin TRiC are structurally homologous to those of group I chaperonins such as GroEL.
Helix 11 of TRiC Subunit 1 Is Important for VHL Folding In Vivo
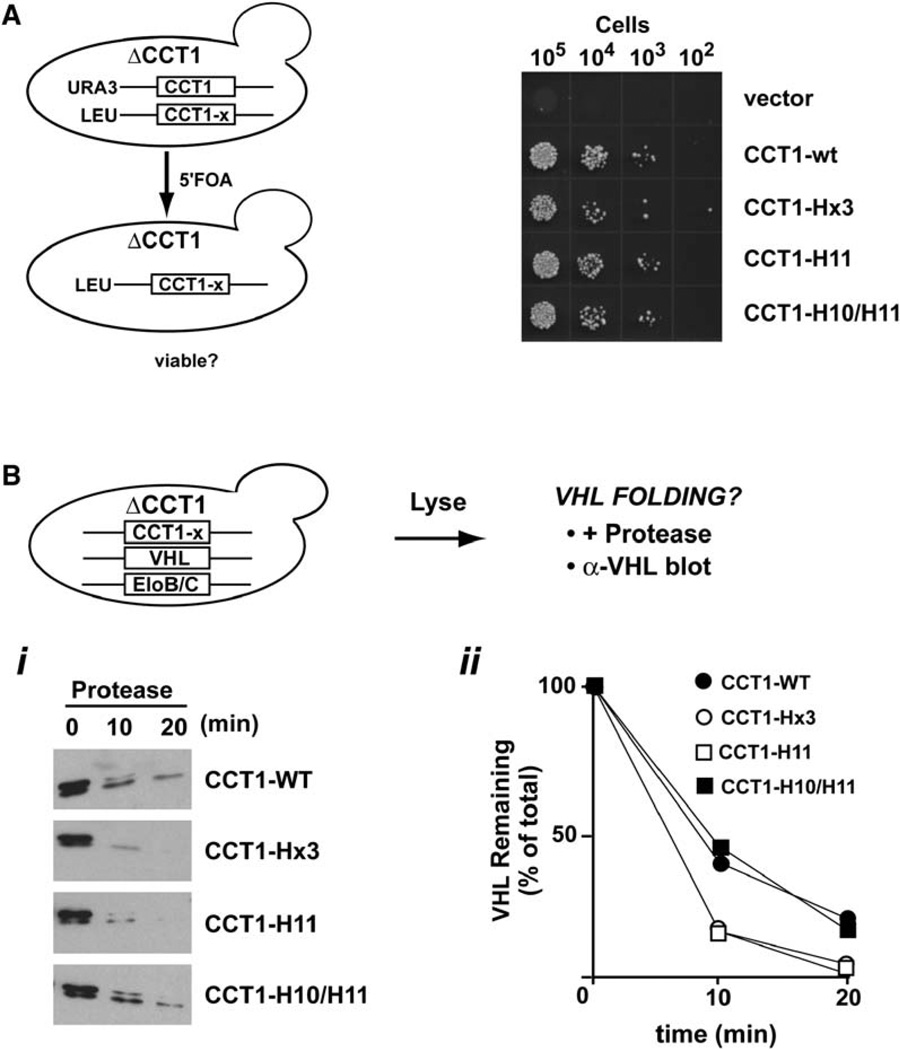
We next examined whether the residues of helix 11 that contribute to the interaction between VHL Box1 and the apical domain of subunit 1 are also important for VHL folding in vivo. To this end, we used a previously described assay for VHL folding in the yeast S. cerevisiae (Melville et al., 2003) and introduced the HL mutations tested above into the CCT1 gene coding for subunit 1 of yeast TRiC/CCT. These CCT1 variants, herein termed CCT1-H10/H11, CCT1-H11, and CCT1-Hx3, as well as control plasmids expressing the wild-type gene, herein CCT1-WT, were introduced as the single source of Cct1p protein by plasmid shuffling in a haploid yeast strain carrying a chromosomal deletion of the CCT1 gene (Figure 6A). Because all TRiC/CCT subunits are essential, we initially tested whether the CCT1 variants could support cell viability by using a serial dilutions assay (Figure 6A). As expected, the empty vector could not support growth; however, all CCT1 variants tested complemented the Δcct1 null to a level comparable to CCT1-WT. This result is consistent with the idea that there is substantial redundancy between the different subunits in the chaperonin complex, whereby the plasticity of TRiC/CCT may provide folding substrates with possible alternative sites to bind to the chaperonin.
Figure 6. Helix 11 in the Apical Domain of Subunit 1 Is Important for VHL Folding In Vivo.
(A) CCT1 mutations in helices H10 and H11 support cell viability. The ability of the yeast subunit 1 gene CCT1 carrying the indicated mutations in the HL region to support cell viability was tested in a haploid strain with a chromosomal CCT1 deletion by plasmid shuffling on 5′FOA plates.
(B) Helix 11 mutations that affect Box1-Ap1 binding in vitro also impair VHL folding in vivo. VHL and elongin BC were expressed in cells containing either CCT1-WT or the indicated CCT1 mutants as their sole source of CCT1. After lysis, VHL folding was assessed by using a previously established protease sensitivity assay, followed by immunoblot analysis of VHL protein. Expression in cells carrying CCT1-H11 and CCT1-Hx3, with mutations in helix 11 that affect Box1 binding in vitro, impaired VHL folding in vivo (open symbols). In contrast, VHL folded to wild-type levels in cells expressing CCT1-H10/11 (closed symbols), with a mutation that does not affect Box1-Ap1 binding in vitro.
We next investigated whether mutations in the CCT1 HL region affect VHL folding (Figure 6B). Previous work established a yeast-based VHL folding assay that exploits the enhanced protease resistance of folded VHL, which is coupled to its assembly into a complex with elongin BC (herein VBC) (Feldman et al., 1999; Melville et al., 2003). Accordingly, VHL and elongin BC were expressed in cells carrying the indicated CCT1 variants; VHL protease sensitivity was then tested by thermolysin treatment, as described (Melville et al., 2003) (Figure 6B). In striking agreement with our in vitro analysis, we observed that mutations that abolished the interaction with Box1 peptide in vitro also reduced VHL folding in vivo (Figure 6B, compare with Figures 5A and 5C). Thus, the protease sensitivity of VHL was enhanced upon expression in cells carrying CCT1 mutation H11 or Hx3 (Figure 6B). In contrast, VHL expressed in cells carrying CCT1-H10/11, a mutation that does not affect Box1 binding to Ap1 in vitro, exhibited WT levels of protease resistance (Figure 6B). These experiments indicate that the helix 11 binding determinants identified in vitro are also important for VHL folding in vivo.
Discussion
Identification of the Substrate Binding Sites in the Chaperonin TRiC/CCT
We defined here basic principles of substrate recognition by the eukaryotic chaperonin, taking advantage of the well-characterized TRiC recognition sequences in the VHL tumor suppressor. Individual VHL motifs interacted with a subset of TRiC/CCT subunits. The TRiC substrate binding determinants were mapped to a helical region within the apical domains that is analogous to the substrate binding determinant of bacterial chaperonins (Figure S8). Thus, the structural architecture of the substrate binding site appears to be similar in group I and group II chaperonins, as expected from their evolutionary conservation of structure and function.
Our finding that, upon binding to TRiC, the VHL binding determinants are proximal to a hydrophobic binding environment is inconsistent with the proposal that substrate binding to TRiC/CCT is generally mediated by polar and charged interactions with the PS site (Pappenberger et al., 2002). However, it is consistent with both mutagenesis analysis of the VHL TRiC binding determinants (Feldman et al., 2003) as well as with the presence of a putative hydrophobic patch in the HL site of subunits CCT1 and CCT7, implicated by our results in substrate binding (Figure 7A, Figure S8, and data not shown).
Our conclusions, based on the analysis of VHL-TRiC interactions, help explain previous observations for other substrates. For instance, the growing number of proteins identified as cellular TRiC/CCT substrates lack any shared consensus motifs with each other or with actin or tubulin, as would be predicted if substrate binding is mediated by highly specific polar interactions (Spiess et al., 2004). Furthermore, analysis of a few of these substrates identified TRiC/CCT binding motifs in buried regions with hydrophobic character, similar to our findings with VHL (Kubota et al., 2006; Camasses et al., 2003; Dobrzynski et al., 2000; Rommelaere et al., 1999).
Subunit Diversity Provides Plasticity and Specificity in Substrate Binding
Most protein-protein interactions have evolved sequence-specific interaction surfaces. In contrast, chaperones such as GroEL or Hsp70 must recognize the folding intermediates of a broad array of substrates. To achieve this, their substrate binding sites rely on the nonspecific recognition of hydrophobic regions that are most likely surface exposed in nonnative proteins (Bukau and Horwich, 1998; Hartl and Hayer-Hartl, 2002). We propose that subunit heterogeneity in TRiC creates a unique mode of substrate selection that combines specificity and plasticity in substrate binding. Homology modeling of the TRiC/CCT apical domains indicates that the helical region in different subunits contains different combinations of hydrophobic and polar residues (e.g., compare CCT1 with CCT3, Figure 7A). The specific configuration of polar and hydrophobic residues in the helical region of each subunit may underlie its individual binding specificity. As a result, the nature of the substrate-TRiC interactions may vary between TRiC subunits, thus expanding the range of possible motifs recognized by the complex. Notably, we could transfer specificity from one apical domain to another by reengineering a few residues within the helical binding site. This opens the way for the design of new chaperonin variants with novel substrate specificities.
Future studies should define the precise interactions that govern TRiC-substrate specificity. Based on homology models, CCT1 (Figure 7A and Figure S8) and CCT7 (data not shown) contain a central hydrophobic patch in the HL site flanked by a negatively charged amino acid in the distal region of helix 11. A binding surface with these chemical properties would be well suited to interact with the Box1 motif of VHL, which consists of hydrophobic residues flanked by a positively charged arginine. In principle, a charged interaction could impart specificity and directional binding of VHL Box1 to promote a desired substrate topology. Of note, CCT3, which does not bind to VHL Box1, contains a positively charged amino acid at the same position (Figure 7A and Figure S8). The idea that TRiC-substrate recognition combines polar and hydrophobic interactions resonates with previous observation that both hydrophobic and electrostatic interactions contribute to VHL binding to TRiC (Feldman et al., 2003).
A possible evolutionary illustration of how changes in the HL region may generate a blend of specificity and plasticity may be found in the sequence changes distinguishing CCT6-1 from its highly homologous testis-specific isoform CCT6-2, which is presumably optimized for folding of testis proteins (Kubota et al., 1997) (Figure S7). Most surface-accessible amino acid replacements in the apical domain are located in the distal part of helices 10 and 11. Based on our results, it is tempting to speculate that these residues change the binding specificity and thus the functionality of the testis isoform (Figure S7).
A New Model for TRiC/CCT-Mediated Folding
An intriguing aspect of TRiC/CCT-mediated folding is its unique ability to fold several eukaryotic proteins (Spiess et al., 2004). TRiC/CCT uses ATP to promote closure of a built-in lid, thereby confining the substrate in the central cavity (Meyer et al., 2003). It is unclear how lid closure affects the substrate environment and leads to folding. An important implication of identifying the helical region in the apical domains as the TRiC/CCT substrate binding site is that, based on the crystal structure of a related group II chaperonin, this region is no longer exposed to the central cavity in the closed conformation (Ditzel et al., 1998). Accordingly, lid closure should produce a change in the chemical properties of the chamber, a prediction borne out by our fluorescence studies (C.S., unpublished data). These findings do not support a model that postulates that the chaperonin binds to the substrate through the PS sites, which would be exposed in both the open and closed TRiC/CCT conformations (Llorca et al., 2000; Pappenberger et al., 2002). In this model, folding results from a mechanical push of substrate domains toward the center of the cavity (Gomez-Puertas et al., 2004). Instead, our results with VHL support an alternative and more general model for chaperonin function, whereby lid closure changes substantially the substrate environment in the cavity. Our results imply that TRiC/CCT-mediated folding does not occur by mechanical pushing but by exploiting the distinct physicochemical properties of both the open and closed states to modulate the folding landscape of substrates. Significantly, the presence of different binding surfaces in different subunits may allow TRiC to modify the landscape in a far more complex manner than can be accomplished by homo-oligomeric chaperonins such as GroEL (Tang et al., 2006). Importantly, the model we propose here provides a compelling basis for how the chaperonin distinguishes between native and nonnative states and releases the substrate upon folding.
Mechanistic Advantages of Folding in a Hetero-Oligomeric Chaperonin
The combination of multiple substrate binding sites distinguishes TRiC/CCT-substrate binding from other chaperones with more generic binding specificities. It is tempting to speculate that this unique feature helped solve specific problems posed by folding in the eukaryotic cytosol. For instance, eukaryotic proteins tend to have more complex topologies than prokaryotic proteins (Koonin et al., 2002). Distinct specificities among chaperonin subunits may allow for a more complex ability to modulate folding intermediates. This mechanistic gain might explain why TRiC/CCT can fold certain proteins, such as actin, that GroEL can bind, but not fold. From an evolutionary point of view, the ability of TRiC to diversify its substrate recognition domains may be related to its reliance on a built-in lid to close the folding chamber. In group I chaperonins, the binding site for GroES overlaps with the substrate binding site, which may impose a strong selective pressure against changes in the substrate binding site (Kawe and Pluckthun, 2006). In contrast, the built-in lid of TRiC may allow for a greater flexibility in their substrate binding sites, enabling the appearance of paralog subunits with diverging binding specificities.
What could be the advantages of folding in a hetero-oligomeric chaperonin? We envision several possibilities for how TRiC/CCT may influence the folding landscape of VHL and maybe other substrates (Figure 7B). For instance, TRiC could keep the polypeptide chain unfolded by binding to specific kinetically trapped intermediates (Figure 7B1). Also, binding the substrate in a defined orientation could allow the chaperonin to favor a topology that facilitates folding (Figure 7B2). Additionally, because many substrates of TRiC are multidomain proteins, the chaperonin could prevent inappropriate interactions between the domains by sequestering a portion of the chain to allow a domain to fold (Figure 7B3). Because TRiC/CCT interacts with many of its substrates cotranslationally, the availability of subunits with overlapping binding specificities could impart directionality to the chaperonin-mediated folding pathway. For instance, substrates could bind unfolded upon translation, fold partially, and rebind to a different set of subunits, as suggested by the analysis of the interaction of actin with TRiC during cotranslational folding (Etchells et al., 2005).
In conclusion, although substrate binding by group I and group II chaperonins is more similar than previously thought, these distinct chaperonin classes exhibit significant differences in terms of the mechanism of lid closure as well as in the principles of substrate recognition. Future studies should clarify how these mechanistic differences, together with the expansion to paralogous subunits, bestowed TRiC/CCT with its unique ability to fold eukaryotic proteins.
Experimental Procedures
Plasmids and Reagents
All plasmids were constructed by PCR amplification of the respective DNA. Detailed information about the oligonucleotides used for cloning can be obtained from the authors. pCS71 is a pET28a (Novagen) derivative coding for amino acids L218–K379 of CCT1 of S. cerevisisae, pCO40 is a pPROEX HTa (Amersham) derivative coding for amino acids G202–G376 of TCPH of H. sapiens, and pJMS1 is a pET28a derivative coding for amino acids E210–S380 of TCPG of H. sapiens. Amino acid substitutions were generated by site-directed mutagenesis of pCS71 and pJMS1 by the Quik-Change kit (Stratagene). The following mutations were made on pCS71: Ap1H11 (K308A, E309A, and E312A), Ap1H10/11 (V281A, L282A, D304A, and L305A), and Ap1Hx3 (K308H, E309Y, and E312R); pJMS1: Ap3Hx1 (H301K, Y302E, and R305E). The same mutations were introduced by site-directed mutagenesis into pESC-LEU-CCT, which carries CCT1 of S. cerevisiae under the copper promoter. pVHL-121C is a pET28a derivative coding for the VHL gene. By site-site directed mutagenesis, the following mutations were introduced: C77A, C162A, and D121C.
Peptides
Peptides Box1 and C-Box2 were synthesized by ResGen (Huntsville, AL), and Box2-C and Box1AA were synthesized by Genemed Synthesis Inc. (South San Francisco, CA). Peptides have either an N-terminal biotin or a free amino terminus, and an amidated C terminus. N-terminal labeling with fluorescein was carried out with Fluorescein-5-EX succinimidyl ester (Molecular Probes) followed by quenching with Tris/HCl (pH 8.0).
Protein Purification
TRiC/CCT from bovine testis and recombinant VHL-121C was purified as described (Feldman et al., 2003). Apical domains corresponding to subunits 1, 3, and 7 were respectively expressed from plasmids pCS71, pJMS1, and pCO40 in BL21STAR cells (Invitrogen). The proteins were purified with Co-TALON affinity resin (BD Biosciences) according to the manufacturer’s recommendations and dialyzed versus buffer A (25 mM HEPES/KOH [pH 7.5], 300 mM NaCl, 10% glycerol, and 1 mM DTT). All apical domains were correctly folded as judged by their Circular Dichroism (CD) spectra. Purity of all proteins was determined by SDS-PAGE and silver staining to be >95%.
Fluorescence Binding Studies
All peptides and VHL-121C were labeled at the unique cysteine with the maleimide derivative of the respective dye (Molecular Probes). After reaction was complete, free dye was quenched with β-mercaptoethanol.
Fluorescence binding studies were carried out in buffer X (25 mM HEPES/KOH [pH 7.5], 100 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 1 mM DTT). Spectra were recorded on a SPEX Fluorolog, scanning from 540 to 635 nm after excitation at 530 nm for Alexa Fluor 546 and from 560 to 750 nm after excitation at 550 nm for Nile Red.
Crosslinking of PBox1/Box2 to TRiC and Apical Domains of Subunits 1, 3, and 7
Box1 or Box2 peptides were reacted with benzophenone-4-iodacetamide (Molecular Probes) for 90 min at room temperature; unreacted BPIA was quenched by addition of DTT for another 30 min. BPIA peptides were incubated with TRiC in buffer X or apical domains in buffer A for 30 min at room temperature. Symmetrically closed TRiC complexes were generated as described (Meyer et al., 2003). Photolysis was carried out for 10 min under filtered UV light (lamp: UVGL-25, UVP).
TRiC subunits were separated by RP-HPLC (column: 214TP54, Vydac; TRiC subunits elute at 50%–60% acetonitrile/0.1% TFA). Individual column fractions were analyzed on 12%SDS-PAGE, transferred to an Immobilon-P membrane (Millipore), and probed with Streptavidin-HRP conjugate (Zymed). Crosslinks to apical domains were detected as above after 15% SDS-PAGE analysis.
Crosslinking of VHL-121C to TRiC
BPIA-labeled VHL-C121 was crosslinked to TRiC as described for Box1 peptide.
For immunoblot analysis of the crosslinks, samples were resolved by 12% SDS-PAGE, transferred to nitrocellulose membranes, and probed with either α-VHL (Melville et al., 2003) or α-TCP-1 α antibody (Stressgen, CTA-191). For mass-spectrometry identification of VHL-crosslinked TRiC subunits, VHL adducts were isolated by immunoprecipitation as described (Melville et al., 2003), followed by 12% SDS-PAGE and silver staining. Unique bands were excised from the gel and analyzed by MALDI-MS.
In Vivo Analysis of CCT1 Mutants
Complementation assays to test viability of various CCT1-mutations and VHL folding assays for VHL expressed in yeast cells were carried out essentially as described (Melville et al., 2003).
Molecular Modeling
Homology models of TRiC subunits were obtained by using the SWISS-Model software (http://swissmodel.expasy.org). TRiC apical domains were modeled by using the β-apical domain of the thermosome (1E0R) as a homology template and full-length TRiC subunits using 1A6E, the closed crystal structure of the thermosome. Models were visualized and figures prepared with MacPyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Dr. W.E. Moerner and Soyeon Kim for discussion of fluorescence experiments; Dr. Robert Twieg and Alexander Semyonov for providing the Nile Red-maleimide; Jill Lin and Dr. A.L Burlingame from the UCSF Mass Spectrometry Facility for mass-spectrometry analysis of TRiC subunits after HPLC; Dick Winant from the Stanford PAN facility for analysis of TRiC-VHL crosslinks; and Stephen Tam for providing the strain used in the in vivo experiments. We also thank Raul Andino and members of the Frydman lab for useful discussions and comments on the manuscript. This work was supported by National Institutes of Health grant GM74074. C.S. was partially supported by grants R21-GM75166 and the Nanomedicine Roadmap Initiative.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, and eight figures and can be found with this article online at http://www.molecule.org/cgi/content/full/24/1/bxs/DC1/.
References
- Archibald JM, Blouin C, Doolittle WF. Gene duplication and the evolution of group II chaperonins: implications for structure and function. J. Struct. Biol. 2001;135:157–169. doi: 10.1006/jsbi.2001.4353. [DOI] [PubMed] [Google Scholar]
- Ashcroft AE, Brinker A, Coyle JE, Weber F, Kaiser M, Moroder L, Parsons MR, Jager J, Hartl UF, Hayer-Hartl M, Radford SE. Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies. J. Biol. Chem. 2002;277:33115–33126. doi: 10.1074/jbc.M203398200. [DOI] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Buskiewicz I, Deuerling E, Gu SQ, Jockel J, Rodnina MV, Bukau B, Wintermeyer W. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl. Acad. Sci. USA. 2004;101:7902–7906. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Chen L, Sigler PB. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- Dobrzynski JK, Sternlicht ML, Peng I, Farr GW, Sternlicht H. Evidence that beta-tubulin induces a conformation change in the cytosolic chaperonin which stabilizes binding: implications for the mechanism of action. Biochemistry. 2000;39:3988–4003. doi: 10.1021/bi992110s. [DOI] [PubMed] [Google Scholar]
- Etchells SA, Meyer AS, Yam AY, Roobol A, Miao Y, Shao Y, Carden MJ, Skach WR, Frydman J, Johnson AE. The cotranslational contacts between ribosome-bound nascent polypeptides and the subunits of the hetero-oligomeric chaperonin TRiC probed by photocross-linking. J. Biol. Chem. 2005;280:28118–28126. doi: 10.1074/jbc.M504110200. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol. Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Puertas P, Martin-Benito J, Carrascosa JL, Willison KR, Valpuesta JM. The substrate recognition mechanisms in chaperonins. J. Mol. Recognit. 2004;17:85–94. doi: 10.1002/jmr.654. [DOI] [PubMed] [Google Scholar]
- Gutsche I, Essen LO, Baumeister W. Group II chaperonins: new TRiC(k)s and turns of a protein folding machine. J. Mol. Biol. 1999;293:295–312. doi: 10.1006/jmbi.1999.3008. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hynes GM, Willison KR. Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of beta-actin. J. Biol. Chem. 2000;275:18985–18994. doi: 10.1074/jbc.M910297199. [DOI] [PubMed] [Google Scholar]
- Kawe M, Pluckthun A. GroEL walks the fine line: the subtle balance of substrate and co-chaperonin binding by GroEL. A combinatorial investigation by design, selection and screening. J. Mol. Biol. 2006;357:411–426. doi: 10.1016/j.jmb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Kim S, Willison KR, Horwich AL. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Kim SY, Semyonov AN, Twieg RJ, Horwich AL, Frydman J, Moerner WE. Probing the sequence of conformationally induced polarity changes in the molecular chaperonin GroEL with fluorescence spectroscopy. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2005;109:24517–24525. doi: 10.1021/jp0534232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Freund SM, Chatellier J, Zahn R, Fersht AR. NMR analysis of the binding of a rhodanese peptide to a minichaperone in solution. J. Mol. Biol. 1999;292:181–190. doi: 10.1006/jmbi.1999.3042. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes GM, Kerr SM, Willison KR. Tissue-specific subunit of the mouse cytosolic chaperonin-containing TCP-1. FEBS Lett. 1997;402:53–56. doi: 10.1016/s0014-5793(96)01501-3. [DOI] [PubMed] [Google Scholar]
- Kubota S, Kubota H, Nagata K. Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc. Natl. Acad. Sci. USA. 2006;103:8360–8365. doi: 10.1073/pnas.0600195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack EA, Rohman MJ, Willison KR. Mutational screen identifies critical amino acid residues of beta-actin mediating interaction between its folding intermediates and eukaryotic cytosolic chaperonin CCT. J. Struct. Biol. 2001;135:185–197. doi: 10.1006/jsbi.2001.4389. [DOI] [PubMed] [Google Scholar]
- Melville MW, McClellan AJ, Meyer AS, Darveau A, Frydman J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 2003;23:3141–3151. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- Pappenberger G, Wilsher JA, Roe SM, Counsell DJ, Willison KR, Pearl LH. Crystal structure of the CCTgamma apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J. Mol. Biol. 2002;318:1367–1379. doi: 10.1016/s0022-2836(02)00190-0. [DOI] [PubMed] [Google Scholar]
- Rommelaere H, De Neve M, Melki R, Vandekerckhove J, Ampe C. The cytosolic class II chaperonin CCT recognizes delineated hydrophobic sequences in its target proteins. Biochemistry. 1999;38:3246–3257. doi: 10.1021/bi9815905. [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Gierasch LM. First glimpses of a chaperonin-bound folding intermediate. Proc. Natl. Acad. Sci. USA. 2005;102:13715–13716. doi: 10.1073/pnas.0506510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Tian G, Vainberg IE, Tap WD, Lewis SA, Cowan NJ. Specificity in chaperonin-mediated protein folding. Nature. 1995;375:250–253. doi: 10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.