Abstract
New primer pairs of genomic DNA microsatellite markers were tested to assess the genetic diversity of eleven date palm genotypes. The results indicated that out of thirty, only seven primers (23.3%) failed to amplify the expected PCR fragments, while thirteen primers (43.3%) amplified monomorphic banding patterns and the remaining ten primers (33.4%) generated polymorphic banding patterns. A total of 77 alleles have been observed with a mean of 7.7 alleles per locus. The average of gene diversity was 0.80 ranging from 0.6 (in marker DP168) to 0.9 (in two markers DP157 and DP175). These new co-dominant markers will be a starting point for researchers making use of the markers for genetic mapping and diversity analysis of date palm.
Keywords: Phoenix dactylifera, Microsatellite marker, Simple sequence repeats
Introduction
Date palm (Phoenix dactylifera L.) belonging to the Arecaceae family, originated in Mesopotamia and thousands of cultivars have been reported (Hanachi et al. 1998). Date palms is a diploid (2n = 2x = 36), and the predicted genome size is estimated to be approximately between 550 and 650 Mbp long (Malek 2010). Date palms have always been clonally propagated to ensure the identity and uniformity of the cultivars and the morphological markers of many traits are often unreliable or extremely difficult to be estimated correctly, especially because of the influence of environmental factors (Elhoumaizi et al. 2002). Therefore, marker technology for DNA fingerprinting has become increasingly important in recent years to discriminate among closely related cultivars. Several marker systems have been used to study the genetic diversity of date palm. In brief, randomly amplified polymorphic DNA (RAPD) fingerprints have been used to identify date palm accessions in Algeria (Benkhalifa 1999), in Morocco (Sedra et al. 1998), in Tunisia (Trifi et al. 2000), in Saudi Arabia (Al-Khalifah and Askari 2003), and in Egypt (Soliman et al. 2003; Adawy et al. 2006). Amplified fragment length polymorphic (AFLP) markers have been applied to study the genetic diversity of date palm cultivars in Egypt and California (Cao and Chao 2002; El-Assar et al. 2005; Adawy et al. 2006).
Microsatellite or simple sequence repeat (SSR) marker has been used in plant diversity analysis; the popularity of these markers is due to their ease of amplification by polymerase chain reaction (PCR), their co-dominant nature and their typically high levels of allelic diversity at different loci. There are numerous reports suggesting the usefulness of microsatellite markers for measuring the genetic variability in a wider taxonomic range (Ryberg et al. 2002; Li et al. 2007; Kawka et al. 2007; Chan et al. 2008; Banhos et al. 2008).
Microsatellite markers have been developed and used to investigate genetic diversity in Phoenix dactylifera (Billotte et al. 2004). They used (GA)n microsatellite-enriched library to develop 16 microsatellite markers. More recently further 17 microsatellite loci were developed by constructing two microsatellite-enriched libraries of date palm using (GA)n and (GT)n repeats (Akkak et al. 2009). These microsatellite markers have been used to assess the genetic diversity and relationships of date palm varieties in Tunisia (Zehdi et al. 2004), in Sudan (Elshibli and Korpelainen, 2007), in Oman (Al-Ruqaishi et al. 2008), and in Qatar (Ahmed and Al-Qaradawi 2009). However, still 33 microsatellite markers are not enough to cover the whole genome and give a comprehensive assessment of biodiversity in date palm. This creates an urgent need to develop more of microsatellite markers in this genome. The objective of this research is to use recently developed new microsatellite markers in date palm genome to assess the genetic diversity in date palm.
Materials and methods
DNA extraction
Eleven cultivars were chosen randomly from different locations in Qatar namely Khalas, Shaishi, Barhi, Hilali, Sukari, Khnaizi, Shahil, Khasab, Rzaiz and Lulu as females and one male Phahel.
The frozen young leaf tissues of date palm collected from each of the cultivars were first cleaned carefully with distilled water to remove the waxy layer and then 1 g of leaf sample was cut into small pieces and grinded into fine powder using liquid nitrogen. DNeasy Plant Maxi kit protocol (QIAGEN) was used to extract DNA by following the manual instructions of the kit (DNeasy Plant Handbook). The obtained DNA were quantified and qualified using Nanodrop Spectrophotometer. For further estimation of the DNA quantity 2 μl was loaded on 0.85% agarose gel at 100 V for 30 min. The gels were stained in ethidium bromide and visualized under UV light.
Primer design and PCR amplification
The sequences of 30 new primer pairs of genomic DNA of date palm were obtained from the International Center for Agricultural Research in the Dry Areas (ICARDA) recently published by Hamwieh et al. (2010). These primers were made in (IDT) Integrated DNA Technologies, Inc and derived from an assembly draft of the date palm genome generated by whole genome shotgun next generation DNA sequencing issued by researchers in the Weill Cornell Medical College in Qatar (WCMC-Q).The length of the primers, sequence information, repeat motifs, allele size and melting temperature (Tm) are shown in (Table 1). Polymerase chain reaction was performed in a total reaction mixture of 25 μl containing 2 μl (20–30 ng) of total genomic DNA, 12.5 μl of AmpliTaq Gold® 360 Mastermix (Applied Biosystems), 1 μl (5 pmol/μl) of primers each and 8.5 μl of nuclease free water. Amplification was carried out in a Veriti 96 Well Fast Thermal cycler (Applied Biosystems) under the following conditions: initial denaturation 95 °C for 10 min, 35 cycles (denaturation 95 °C for 30 s, annealing temperature depending on primer for 30 s, extension 72 °C for 1 min), final extension 72 °C for 7 min.
Table 1.
Forward and reverse primer sequences, repeat motifs and expected sizes of microsatellite loci and its status of amplification of SSR loci of date palm
| Primer name | Primer’s sequence | Tm (°) | Expected size | Motif repeat | Status of amplification | |
|---|---|---|---|---|---|---|
| DP150 | F | CTGCGCCAATCTAAACCATT | 52.6 | 177 | (GAA)9 | − |
| R | GCAAATTGCAACAAATCCTTG | |||||
| DP151 | F | TTGCTGGTTGAAATGGTGTT | 53.3 | 168–186 | (AC)37 | ++ |
| R | GCAACAGATGCTCTTGCTCA | |||||
| DP152 | F | ACGAGTTTTTGGGAGAGCAA | 53.6 | 224 | (TAT)8 | − |
| R | GCAAGTTGCCAACATTCTTGT | |||||
| DP153 | F | TCATCACAGGCAATGGCTAA | 51.6 | 204 | (TCA)9 | + |
| R | GCAGATGGCCATTGAACC | |||||
| DP154 | F | ACACACACACACCGCGAAT | 53.1 | 249 | (AC)19 | + |
| R | GCATGTGAGGCGCATATCTA | |||||
| DP155 | F | CCCCCTCTCTCTCTCTCTCTC | 55.8 | 200 | (TC)51 | − |
| R | GCCAAGAGGATTGGAGATTG | |||||
| DP156 | F | TGTGTGTGTGTGTGTGTGTGA | 54.6 | 221 | (GA)17 | + |
| R | GCCATTGTTTGTGTGGACTG | |||||
| DP157 | F | TGGACAATGACACCCCTTTT | 54.6 | 180–244 | (TC)19 | ++ |
| R | GCCCACACAACAACCTCTCT | |||||
| DP158 | F | TCATTGGCTAATCCACACACA | 54.3 | 204 | (GA)29 | − |
| R | GCCTTGTGGTCATGAGAGGT | |||||
| DP159 | F | AGCTCCAATTTGCTGCAGAG | 54.3 | 156–172 | (TC)27 | ++ |
| R | GCTGACCTGGAGTCCAAAAC | |||||
| DP160 | F | AAGAGCGACAATCATGACCA | 57.7 | 108–136 | (GAAA)5 | ++ |
| R | GGAAATTGAAGGGCATCTTG | |||||
| DP161 | F | TGGTTGCTGCTTATCTGCTG | 54.9 | 211 | (CT)13 | + |
| R | GGAGGGAACCGAGAGAGAGA | |||||
| DP162 | F | TGGACTTCAAGAAGTGCGAAT | 57.3 | 183 | (TACA)9 | − |
| R | GGCAGTCACATTTTGCTTCA | |||||
| DP163 | F | GTGCGTGTGTGTGTGTGTGT | 53.4 | 215 | (GA)19 | + |
| R | GGCTGTTTGGGTTCGTACTG | |||||
| DP164 | F | GGACCAAGAACCGACAGTTG | 57.1 | 200 | (ATAG)6 | + |
| R | GGGAAGGTGAGGTGGTATGA | |||||
| DP165 | F | AAGCATCCTATGGCTTTGACA | 54.8 | 222 | (AATA)5 | + |
| R | GGGCTGTATGTGATGCATTG | |||||
| DP166 | F | CAATTTCTTCTCGCCTGGAG | 54.5 | 210 | (GAAA)5 | + |
| R | GGGGTTTCTTTTCCTTCTGC | |||||
| DP167 | F | ACATCCAATGGCATCCAAAT | 57.6 | 243 | (GAAA)6 | + |
| R | GGGTTTCCAGGTTTTCTTCTC | |||||
| DP168 | F | GCAGCAAAGCCCTTAGGC | 54.3 | 163–175 | (CAT)8 | ++ |
| R | GGTGTTATGTGCAGCCAATG | |||||
| DP169 | F | GCATGGACTTAATGCTGGGTA | 57.1 | 129–223 | (AAT)12 | ++ |
| R | GGTTTTCCTGCCAACAACAT | |||||
| DP170 | F | TCTTTGGGCTTACGACAACC | 55.9 | 195–227 | (AGGG)5 | ++ |
| R | GTATGGCCCAAGATGCAGAT | |||||
| DP171 | F | GTGGGAGTAGCGAGGTATGG | 56 | 197–218 | (TTC)10 | ++ |
| R | GTCCGGCACTTTAGGAAGTT | |||||
| DP172 | F | GGTGTTTGGGCCTATTTCCT | 54.2 | 199–235 | (AGG)11 | ++ |
| R | GTCCTCCTCCTCCTCTGTCC | |||||
| DP173 | F | CCACATGCAATTTCCAAAAG | 51.9 | 198 | (TC)27 | + |
| R | GTGCGTATCGGGAGAGAGAG | |||||
| DP174 | F | CTCTGTCGTACGGAGGAAGG | 55.6 | 187 | (CGTG)5 | − |
| R | GTGGCACTATCACGCTCTCA | |||||
| DP175 | F | ACACACACACACACACACACC | 57.6 | 196–274 | (CA)19 | ++ |
| R | GTGGCTTCTTTTTGGCTGTC | |||||
| DP176 | F | GCCATTAACGAAATGGCTTG | 54.5 | 198 | (CAA)9 | + |
| R | GTTTGCACATAGCGCTTCAA | |||||
| DP177 | F | TTCCTTGGGCTCACTTCAAC | 54 | 216 | (AGGC)6 | + |
| R | TAACATGCCAGCAAAGGTGA | |||||
| DP178 | F | AGTTTGTCAGGCCATTTGGT | 58.7 | 186 | (TC)19 | + |
| R | TACATGTGCGTATCGGGAGA | |||||
| DP179 | F | GGTTAGCCATCCAAAAGTGG | 56.2 | 183 | (ATTT)5 | − |
| R | TATGTAGCCTCCACCGCATC |
−, Non-amplification; +, amplification of monomorphic band; ++, amplification of polymorphic band of desired PCR product with Qatari date palm
In order to achieve better resolution, electrophoresis was performed using the Spreadex® EL600 Mini gel (Elchrom Scientific). These gels are manufactured from a novel monomer and have 3× higher resolving power than any other synthetic gels with working separation range 40–600 bp and optimal separation range 150–350 bp. The amplified DNA fragments 2 μl were diluted with 3 μl distilled water and 2 μl of loading dye making a total volume of 7 μl were loaded on to the gel. Electrophoresis was carried out at 150 V for 150 min in 1× TAE buffer (30 mM). The gel was stained with ethidium bromide (EtBr) solution (1 mg/ml) for 30 min at 350 rpm and destained with double distilled water for 45 min at 400 rpm. The DNA banding patterns were visualized on a UV transilluminator and documented using Gel Documentation System (Alpha Innotech).
Data analysis. Microsatellite bands were precisely measured by gel documentation system AlphImager EC by Alpha View software V.3.0.0.0 and scored for each genotype. Each polymorphic DNA band at particular position on the gel was treated as a separate character and scored as allele size. Data were then computed with the PowerMarker software V3.0 (Liu and Muse 2005) to detect the percentage of heterozygosity and the phylogenetic relationship among the genotypes on the basis of the allele’s size. The phylogenetic diagram was drawn by TreeView V.1.6.6 (Page 1996).
Results and discussion
Thirty new primer pairs of genomic DNA microsatellite markers of date palm were tested to assess the genetic diversity of eleven date palm genotypes. The results indicate that ten of the thirty markers (33.4%) generate polymorphic banding patterns at expected size (Fig. 1). These primers are: DP151, DP157, DP159, DP160, DP168, DP169, DP170, DP171, DP172 and DP175 (Table 1). These new co-dominant markers will be a starting point for researchers making use of the markers for genetic mapping and diversity analysis of date palm. However, these results are relatively lower when compared to Akkak et al. (2009), who selected seventeen markers (41%) as polymorphic, after screening of forty-one simple sequence repeats. The results also show that thirteen markers (43.3%) amplified monomorphic banding patterns (Fig. 1), while the remaining seven primers (23.3%) failed to amplify the expected PCR fragments.
Fig. 1.
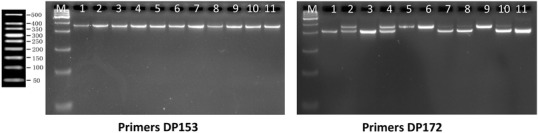
Markers amplified monomorphic and polymorphic banding patterns at expected size: M is 50 bp Genplot marker, date palm sample Cultivar No. 1 Khakas, 2 Shaishi, 3 Barhi, 4 Hilali, 5 Sukari, 6 Khanaizi, 7 Shahil, 8 Khasab, 9 Razaiz, 10 Lulu and 11 Male
The microsatellites examined were highly polymorphic possessing a great number of alleles, a total of 77 alleles with a mean of 7.7 alleles per locus were scored, however, the number of alleles varied between 4 in primer DP168 and 12 in primers DP157 and DP175 (Table 2). The number of alleles per locus detected in this study were higher than those scored by Ahmed and Al-Qaradawi (2009) who marked 40 different alleles with a mean of 4 alleles per locus by examining 15 Qatari date palm cultivars.
Table 2.
Genetic biodiversity information of Qatari date palm using 10 SSR markers
| Marker | Major allele frequency | Genotype no. | Allele no. | Gene diversity | Heterozygosity | PIC |
|---|---|---|---|---|---|---|
| DP151 | 0.36 | 7.00 | 7.00 | 0.79 | 0.00 | 0.77 |
| DP157 | 0.18 | 9.00 | 12.00 | 0.90 | 0.45 | 0.89 |
| DP159 | 0.36 | 7.00 | 6.00 | 0.78 | 0.27 | 0.75 |
| DP160 | 0.27 | 9.00 | 8.00 | 0.84 | 0.45 | 0.82 |
| DP168 | 0.59 | 4.00 | 4.00 | 0.60 | 0.36 | 0.55 |
| DP169 | 0.31 | 8.00 | 8.00 | 0.80 | 0.36 | 0.77 |
| DP170 | 0.27 | 8.00 | 7.00 | 0.80 | 0.91 | 0.79 |
| DP171 | 0.36 | 8.00 | 7.00 | 0.79 | 0.45 | 0.76 |
| DP172 | 0.36 | 8.00 | 6.00 | 0.77 | 0.18 | 0.74 |
| DP175 | 0.13 | 10.00 | 12.00 | 0.90 | 0.73 | 0.89 |
| Mean | 0.32 | 7.80 | 7.70 | 0.80 | 0.42 | 0.77 |
| SD | 0.12 | 1.62 | 2.50 | 0.10 | 0.26 | 0.09 |
| CV | 0.38 | 0.21 | 0.33 | 0.11 | 0.62 | 0.12 |
The numbers of allele per locus detected in this study were almost equivalent to those graded by Zehdi et al. (2004) who recognized 7.14 alleles per locus when examining 46 Tunisian date palm accessions using 14 microsatellite loci.
Elshibli and Korpelainen (2007) identified 21.4 alleles per locus, which were more than the numbers of alleles per locus detected in this study; it may be due to using more number of microsatellite loci (16) in addition to using different genotype area 68 Sudan and Morocco date palm accessions.
The ten primers used in this study successfully produced clear amplified SSR band sizes ranging from 108 bp (marker DP160) to 274 bp (marker DP175), similar to Ahmed and Al-Qaradawi (2009) results which ranged from 100–300 bp.
The sequences flanking microsatellite sites are generally conserved within species and also often in closely related species (Gupta and Varshney 2000). Interestingly, the ten SSR markers tested in this study formed mean of 7.8 genotype numbers (Table 2), however, the highest were 10 different genotypes scored in marker DP175 (206/262 bp, 198/198 bp, 236/274 bp, 196/196 bp, 200/258 bp, 198/234 bp, 226/226 bp, 196/236 bp, 226/250 bp and 234/270 bp) represented in Barhi, Hilali, Khalas, Khasab, Khnaizi, Lulu, Male, Razaiz, Shahil, and Shaishi, respectively while cultivar Sukari share the same genotype with cultivar Khnaizi.
On the other hand, only 4 different genotypes was renowned in marker DP186 (166/175 bp with Barhi, Hilali and Shaishi, 172/172 with Khalas, 163/163 with Khasab, Khnaizi, Lulu, Male, Shahil and Sukari, 163/172 bp with Razaiz cultivar.
The mean of gene diversity was 0.80 (Table 2) ranging from 0.60 (for locus DP168) to high diversity 0.90 (for two loci DP157 and DP175). This high level of gene diversity is similar to 0.83 reported in date palms germplasm from Sudan (Elshibli and Korpelainen 2007). This high level of diversity is expected because of the unique mechanism responsible for generating SSR allelic diversity by replication slippage. Replication slippage is thought to occur more frequently than single nucleotide mutations and insertion/deletion events, which generated the polymorphisms detected by RAPD analysis (Powell et al. 1996).
The heterozygosity was 0.42%, much lower than 0.84 and 0.82 in Sudan and Morocco date palm cultivars, respectively (Elshibli and Korpelainen 2007). The average major allele frequency was 0.32 ranging from 0.13 in marker DP175 to 0.59 in marker DP168. Similarly, the polymorphism information content PIC value which is commonly used in genetics as a measure of polymorphism for a marker locus used in linkage analysis, ranged between 0.55 in DP168 to 0.89 in DP157 and DP175, with an average of 0.77 per marker. Allelic variation might be correlated with the number of repeats within a particular microsatellite locus. A positive relationship was found between the number of repeats and the PIC of earlier reports in tomato (Smulders et al. 1997; Areshchenkova and Ganal 1999; He et al. 2003, Pritesh et al. 2010). In this study (Table 2), DP168 with the lower PIC (0.55) has eight repeats compared to DP157, which has PIC (0.89) with nineteen repeats, which is in agreement with the finding of Pritesh et al. (2010). Marker with lower PIC (0.08) has three repeats compared to marker which has PIC (0.40) with seven repeats. Moreover, similar to the report of He et al. (2003) no relationship was found between PIC and the number of nucleotides per repeat. However, there are reports that the polymorphism level in trinucleotide repeats is lower than that in di-nucleotide repeats for rice (Blair et al. 1999) and ryegrass (Jones et al. 2001).
A dissimilarity matrix between Qatari date palm cultivars showed an average dissimilarity coefficient ranging from 0.00 to 1.00. The cultivars studied here were highly divergent at the DNA level. The lowest dissimilarity coefficient value was observed between Khnaizi and Sukari cultivars (0.25) which seem to be the nearest two varieties and can be closely regrouped (Fig. 2). Ahmed and Al-Qaradawi (2009) found that the similarity coefficient value of Qatari date palm ranged from 0.00 to 0.75 while Zehdi et al. (2004) found that the similarity coefficient value of Tunisian date palm ranged from 0.3008 to 0.7885. The next closet cultivars with 0.43 dissimilarity coefficient value were obtained between Khasab and Shahil. Dissimilarity coefficient value of 1.00 was obtained between Barhi and each of Khasab, Khnaizi, Shahil and Sukarii cultivars, indicating that how far is the relationship between Barhi cultivar and those cultivars (Fig. 2). Molecular markers can provide an effective tool for efficient selection of desired agronomic traits because they are based on the plant genotypes and thus, are independent of environmental variation. Several molecular markers are being currently employed, of which simple sequence repeats (SSRs) or microsatellites are the most widely used types. SSRs are not only very common but also hypervariable for number of repetitive DNA motifs in the genomes of eukaryotes (Hamada et al. 1984; Edwards et al. 1991; Vosman and Arens 1997; Rallo et al. 2000; van der Schoot et al. 2000). All the other cultivars displayed different levels of dissimilarity but still were grouped with each others (Fig. 2). It is suggested that the variation or polymorphism of SSRs are a result of polymerase slippage during DNA replication or unequal crossing over (Levinson and Gutman 1987).
Fig. 2.
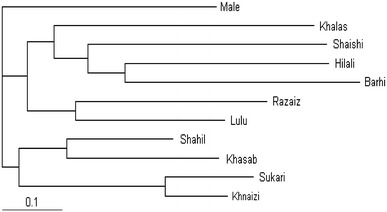
Phylogram Tree of Qatari date palm based on Distance of Nei 1983 by NJ method
The phylogenetic diagram (Phylogram) illustrates the divergence between the studied Qatari date palm cultivars (Fig. 2) and suggests their tree branching. Male (Phahil) cultivar was clustered separately from the rest of other cultivars. Mainly the phylogenetic tree showed two major clusters, the first included four cultivars (Shahil, Khasab, Sukari and Khnaiz) and the second cluster contained six cultivars (Khalas, Shaishi, Hilali, Barhi, Razaiz and Lulu); however, Lulu and Razaiz were grouped in a separate sub cluster (Fig. 2).
Genetic diversity is desirable for long-term crop improvement and reduction of vulnerability in plants to important crop diseases. Measurements of genetic diversity can be used in breeding programs to increase the genetic variation in base populations by crossing cultivars with a high level of genetic distance as well as for the introgression of exotic germplasm. Molecular genetic diversity estimates are extremely useful for intellectual property protection, particularly in the determination of essential derivation. The genetic diversity estimates based on molecular marker data may be compared to a minimum genetic distance which indicates that two cultivars are not essentially derived (Lefebvre et al. 2001).
References
- Adawy SS, Hussein EHA, Ismail SEME, El-Itriby HA (2006) Genomic diversity in date palm (Phoenix dactylifera L.) as revealed by AFLPs in comparison to RAPDs and ISSRs. In: Abstracts of 3rd international date palm conference, Abu Dhabi, United Arab Emirates, 19–21 February
- Ahmed TA, Al-Qaradawi A. Molecular phylogeny of Qatari date palm genotypes using simple sequence repeats markers. Biotechnology. 2009;8:126–131. doi: 10.3923/biotech.2009.126.131. [DOI] [Google Scholar]
- Akkak A, Scariot V, Torello Marinoni D, Boccacci P, Beltramo C, Botta R. Development and evaluation of microsatellite markers in Phoenix dactylifera L. and their transferability to other Phoenix species. Biologia Plantarum. 2009;53:164–166. doi: 10.1007/s10535-009-0026-y. [DOI] [Google Scholar]
- Al-Khalifah NS, Askari E. Molecular phylogeny of date palm (Phoenix dactylifera L.) cultivars from Saudi Arabia by DNA fingerprinting. Theor Appl Genet. 2003;107:1266–1270. doi: 10.1007/s00122-003-1369-y. [DOI] [PubMed] [Google Scholar]
- Al-Ruqaishi IA, Davey M, Alderson P, Mayes S. Genetic relationships and genotype tracing in date palm (Phoenix dactylifera L.) in Oman based on microsatellite markers. Plant Genet Resour. 2008;61:70–72. [Google Scholar]
- Areshchenkova T, Ganal MW. Long tomato microsatellites are predominantly associated with centromeric regions. Genome. 1999;42:536–544. doi: 10.1139/g98-155. [DOI] [PubMed] [Google Scholar]
- Banhos A, Hrbeck T, Gravena W, Sanaiotti T (2008) Genomic resources for the conservation and management of the harpy eagle (Harpia harpyja, Falconiformes, Accipitridae). Genet Mol Biol 31(1):146–154
- Benkhalifa A (1999) Gestion de la diversité génétique du palmier dattier en Algérie. Paper presented at the workshop constitution et ÿharacteriza d’équipes de ÿharacter scientifique dans les ÿharact de foresterie et des arbres fruitiers, Marrakech, Morocco, 13–15 April
- Billotte N, Marseilla N, Brottier P, Noyer JL, Jacquemoud-Collet JP, Moreau C, Couvreur T, Chevallier MH, Pintaud JC, Risterucci AM. Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): characterization, utility across the genus Phoenix and in other palm genera. Mol Ecol Notes. 2004;4:256–258. doi: 10.1111/j.1471-8286.2004.00634.x. [DOI] [Google Scholar]
- Blair MW, Panaud O, McCouch SR. Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.) Theor Appl Genet. 1999;98:780–792. doi: 10.1007/s001220051135. [DOI] [Google Scholar]
- Cao BR, Chao CT. Identification of date cultivars in California using AFLP markers. Hortic Sci. 2002;37:966–968. [Google Scholar]
- Chan CH, Zhao Y, Cheung MY, Chambers GK. Isolation and characterization of microsatellites in the kakerori (Pomarea ÿharacter) using feathers as source of DNA. Conserv Genet. 2008;9:1067–1070. doi: 10.1007/s10592-007-9456-2. [DOI] [Google Scholar]
- Edwards A, Civitello A, Hammond HA, Caskey CT. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 1991;49:746–756. [PMC free article] [PubMed] [Google Scholar]
- El-Assar AM, Krueger RR, Devanad PS, Chao CT. Genetic analysis of Egyptian date (Phoenix dactylifera L.) accessions using AFLP markers. Genet Resour Crop Evol. 2005;52:601–607. doi: 10.1007/s10722-004-0583-z. [DOI] [Google Scholar]
- Elhoumaizi MA, Saaidi M, Oihabi A, Cilas C. Phenotypic diversity of date-palm cultivars (Phoenix dactylifera L.) from Morocco. Genet Resour Crop Evol. 2002;49:483–490. doi: 10.1023/A:1020968513494. [DOI] [Google Scholar]
- Elshibli S, Korpelainen H. Microsallite markers reveal high genetic diversity in date palm (Phoenix dactylifera L.) germplasm from Sudan. Genetica. 2007;134:251–260. doi: 10.1007/s10709-007-9232-8. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Hamada H, Petrino MG, Kakunaga T, Seidman M, Stollar BD. Characterization of genomic poly(dT-dG) poly(dC-dA) sequences: structure, organization and conformation. Mol Cell Biol. 1984;4:2610–2621. doi: 10.1128/MCB.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamwieh A, Farah J, Moussally S, Al-Shamaa K, Elmeer K, Khierallah H, Udupa S, Lababidi S, Malek JA, Aaouine M, Baum M. Development of 1000 microsatellite markers across the date palm (Phoenix dactylifera L.) genome. Acta Hortic. 2010;882:269–277. doi: 10.17660/ActaHortic.2010.882.29. [DOI] [Google Scholar]
- Hanachi S, Benkhalifa A, Khirtri D, Brac de la Perrière RA (1998) Inventaire Varietal de la Palmeraie Algerienne. [Varietal inventory of the Algerian palms.] CDARS, Anep Rouiba. (In French)
- He C, Poysa V, Yu K. Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theor Appl Genet. 2003;106:363–373. doi: 10.1007/s00122-002-1076-0. [DOI] [PubMed] [Google Scholar]
- Jones ES, Dupal MP, Kolliker R, Drayton MC, Forster JW. Development and characterisation of simple sequence repeat (SSR) markers for perennial ryegrass (Lolium perenne L.) Theor Appl Genet. 2001;102:405–415. doi: 10.1007/s001220051661. [DOI] [Google Scholar]
- Kawka M, Horbanczuk JO, Sacharczuk M, Zieba G. Genetic characteristics of the Ostrich population using molecular methods. Poult Sci. 2007;86:277–281. doi: 10.1093/ps/86.2.277. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Goffinet B, Chauvet JC, Caromel B, Signoret P, Brand R, Palloix A. Evaluation of genetic distances between pepper inbred lines for cultivar protection purposes: comparison of AFLP, RAPD and phenotypic data. Theor Appl Genet. 2001;102:741–750. doi: 10.1007/s001220051705. [DOI] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li Y, Wongprasert K, Shekhar M, Ryan J. Development of two microsatellite multiplex systems for black tiger shrimp Penaeus monodon and its application in genetic diversity study for two populations. Aquaculture. 2007;266:279–288. doi: 10.1016/j.aquaculture.2007.01.038. [DOI] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Malek JA. Next generation DNA sequencing applied to the Date palm tree (Phoenix dactylifera) Acta Hortic. 2010;882:249–252. doi: 10.17660/ActaHortic.2010.882.27. [DOI] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breeding. 1996;2:225–238. doi: 10.1007/BF00564200. [DOI] [Google Scholar]
- Pritesh P, Vishal PO, Vaishnavi C, Patel AD, Kathiria KB, Subramanian RB. Genetic diversity and DNA fingerprint study of tomato discerned by SSR markers. Int J Biotechnol Biochem. 2010;6(5):657–666. [Google Scholar]
- Rallo P, Dorado G, Martin A. Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.) Theor Appl Genet. 2000;101:984–989. doi: 10.1007/s001220051571. [DOI] [Google Scholar]
- Ryberg WA, Fitzgerald LA, Honeycutt RL, Cathey JC. Genetic relationships of American alligator populations distributed across different ecological and geographic scales. J Exp Zool. 2002;294:325–333. doi: 10.1002/jez.10207. [DOI] [PubMed] [Google Scholar]
- Sedra MH, Lashermes P, Trouslot P, Combes M, Hamon S. Identification and genetic diversity analysis of date palm (Phoenix dactylifera L.) varieties from Morocco using RAPD markers. Euphytica. 1998;103:75–82. doi: 10.1023/A:1018377827903. [DOI] [Google Scholar]
- Smulders MJM, Bredemeijer G, Rus-Kortekaas W, Arens P, Vosman B. Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicon esculentum cultivars and accessions of other Lycopersicon species. Theor Appl Genet. 1997;97:264–272. doi: 10.1007/s001220050409. [DOI] [Google Scholar]
- Soliman SS, Ali BA, Ahmed MMM. Genetic comparisons of Egyptian date palm cultivars (Phoenix dactylifera L.) by RAPD-PCR. Afr J Biotechnol. 2003;2:86–87. [Google Scholar]
- Trifi M, Rhouma A, Marrakchi M. Phylogenetic relationships in Tunisian date-palm (Phoenix dactylifera L.) germplasm collection using DNA amplification fingerprinting. Agronomie. 2000;20:665–671. doi: 10.1051/agro:2000158. [DOI] [Google Scholar]
- van der Schoot J, Pospiskova M, Vosman B. Development and characterization of microsatellite markers in black poplar (Populus nigra L.) Theor Appl Genet. 2000;101:317–322. doi: 10.1007/s001220051485. [DOI] [Google Scholar]
- Vosman B, Arens P. Molecular characterization of GATA/GACA microsatellite repeats in tomato. Genome. 1997;40:25–33. doi: 10.1139/g97-004. [DOI] [PubMed] [Google Scholar]
- Zehdi S, Sakka H, Rhouma A, Salem AOM, Marrakchi M, Trifi M. Analysis of Tunisian date palm germplasm using simple sequence repeat primers. Afr J Biotechnol. 2004;3:215–219. doi: 10.5897/AJB2004.000-2040. [DOI] [Google Scholar]


