Abstract
When not transporting cargo, kinesin-1 is autoinhibited by binding of a tail region to the motor domains, but the mechanism of inhibition is unclear. We report the crystal structure of a motor domain dimer in complex with its tail domain at 2.2 Å and compare it with a structure of the motor domain alone at 2.7 Å. These structures indicate that neither an induced conformational change nor steric blocking is the cause of inhibition. Instead, the tail cross-links the motor domains at a second position, in addition to the coiled-coil. This ‘double lockdown’, by cross-linking at two positions, prevents the movement of the motor domains that is needed to undock the neck linker and release ADP. This autoinhibition mechanism could extend to some other kinesins.
Kinesin-1 (previously named conventional kinesin and hereafter called kinesin) is a molecular motor that uses energy from ATP hydrolysis to move cargos processively towards the plus end of microtubules (MTs)(1–3). When not transporting cargo, kinesin is autoinhibited to prevent squandering of ATP. Although it is widely accepted that the tail domain binds to the motor domain to keep it in a folded autoinhibited state(4–6), the molecular mechanism remains unclear and several autoinhibitory mechanisms have been proposed, including steric and allosteric inhibition(5, 7). An unexpected recent discovery revealed that only one of the two identical tail domains in a heavy chain dimer binds to the motor domain dimer to inhibit ADP release from both motor domains(8).
We report here the crystal structure of a kinesin motor domain dimer in complex with one of its tail domains at 2.2 Å resolution (Fig. 1a) and compare it with the structure of a dimer alone at 2.7 Å resolution (Fig. S1). Data collection and refinement statistics are shown in Table S1. The conformations of the catalytic core of the motor domains in the free motor domain dimer and in the dimer-tail complex are similar, only differing in the angle of attachment of the coiled-coil (Fig. S2, mean r.m.s.d. = 0.765 Å). The major difference between the free dimer and the dimer-tail complex is the rotation of the two motor domains towards each other to be bridged and cross-linked by a single tail peptide.
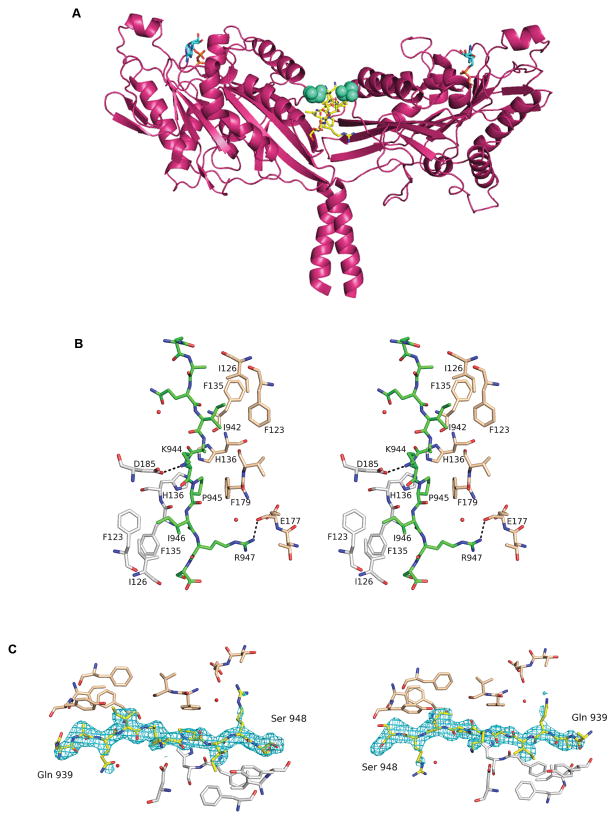
Figure 1. Structure of motor-tail complex.
(A) D. melanogaster kinesin motor domain dimer complexed with tail domain (yellow). (Cyan sticks: Mg2+ADP, green spheres: Ser 181). (B) Stereoplot showing C—H-π and hydrogen bond interactions (dotted lines) between the tail (green) and motor domains (chain A: grey, B: beige). (C) Dimer-tail interface showing residues of motor domain (chain A: grey, B: beige), tail domain (yellow) and omit map (Fo-Fc) (contoured at σ=3.00) for tail binding in both directions.
The tail peptide used here (residues 937–952) contains the core IAK region that is both necessary and sufficient for inhibition(8), although addition of the neighboring positively charged auxiliary binding site (ABS) region increases affinity(4). The final model of the tail domain comprises residues Gln 939 to Ser 948, with the rest of the tail domain being disordered and not modelled. The stoichiometry is one tail peptide per motor domain dimer in agreement with biochemical measurements(8). The asymmetric unit comprises four molecules of the kinesin motor domain, with two molecules (chains C & D) forming a dimer via interactions of the coiled-coil domains and the other two molecules (chains A & B) forming dimers with their symmetry-related molecules (A’ and B’ respectively). Each monomer has one molecule of Mg2+ADP bound to it.
The tail peptide observed in the structure has an extended and linear conformation, forming an expansion of the eight-stranded ß-sheet of the monomeric motor domain (Fig. S3). We also observed a hydrogen bond interaction between the side chains of the tail domain residue Lys 944 (NZ) and the motor domain residue Asp 185 (OD2), and hydrophobic interactions between Ile 942, Ile 946 of the tail domain and Phe 123, Ile 126, and Phe 135 of each motor domain. Finally, there is also a C—H-π-p interaction of the pyrrolidine ring of Pro 945 with Phe 179 (Fig. 1b).
Owing to its amino acid sequence, the tail peptide is almost symmetrical, in terms of side chain properties, about the Lys 944 residue. This allows it to bind in two directions between the motor domain dimer, which also exhibits a two-fold symmetry (Fig. 1c). As such, we observe pseudosymmetry in the crystal structure and the tail domains binding to the AA’ and BB’ dimers lie on the two-fold symmetry axis. The tail peptide binds simultaneously to β4 of both motor domains in the dimer and the total buried surface area is approximately 1000 Å2. This interaction surface is supported by the high evolutionary conservation, in both metazoan and fungal kinesins, of the IAK region and of the motor domain residues that interact in the complex (Fig. S4).
Additional support for the structure is provided by mutagenesis studies. Asp 185 of the motor domain directly interacts with Lys 944 of the IAK region and the His 136 side chains of each motor domain are in close proximity in the interface (~2.9 Å) (Fig. 1b). These positions are far removed from the nucleotide binding pocket and the MT binding surface and the D185R and H136E substitutions do not inhibit the rate of MT-stimulated ADP release (Table S2). However, both the D185R and H136E substitutions yield motor domain dimers that are not significantly inhibited by the tail domain (Fig. S5).
Several possible models for the mechanism of inhibition by tails are eliminated by the structure of the dimer-tail complex. The binding site for the tail domain is on the opposite side of the motor domain from the nucleotide binding site, ruling out models in which the tail sterically blocks the release of ADP or interacts with switch I or switch II(7) that play a critical role in coupling conformational changes to nucleotide binding(9). The tail also does not interact with the neck linker or coiled-coil domain. Steric interference with MT binding is also ruled out because the MT binding sites on the motor domains of the complex are fully exposed. The structural similarity between the free motor domain dimer and the dimer-tail complex (Fig. S2) argues against an allosteric model, in which tail binding induces a conformation change in the motor domain that propagates to the nucleotide binding site and switch I and II to reduce the rate of ADP release.
One significant difference between the free dimer and the dimer-tail complex is that the relative movement of the two motor domains is highly restricted in the latter because the motor domains are cross-linked at two positions: coiled-coil and tail interface. In the absence of tail binding, the motor domains likely have considerable freedom of movement, as evident from the different orientation of the published dimer structure(10) as compared to our dimer structure. In the dimer-tail complex, however, the ‘double lockdown’ at both the neck coil and the tail interface freezes out major relative movements of the motor domains. This could prevent conformational changes such as undocking of the neck linker and overlying N-terminal strand(11, 12).
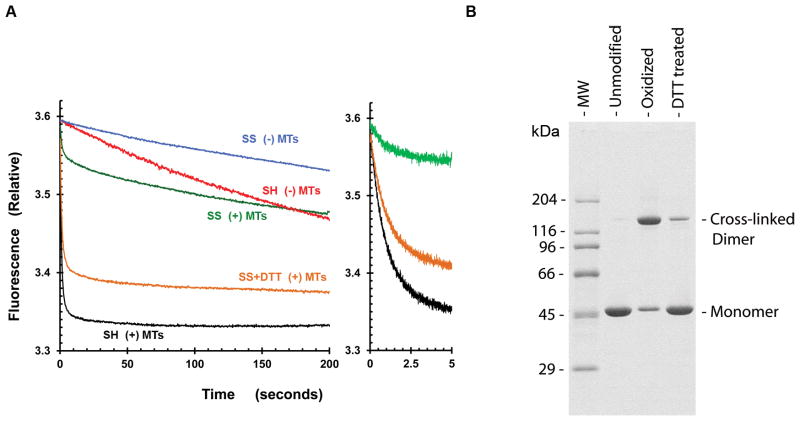
The ‘double lockdown’ mechanism predicts that introduction of a covalent cross-link between motor domains at the tail interface should mimic inhibition in the absence of tails. Ser 181 residues in the two motor domains (Fig. 1a, green) were substituted with Cys (in a background with the other reactive cyteines removed). A covalent cross-link between them could be readily formed by oxidation to a disulfide and reversed by reduction with DTT (Fig. S6). MT-stimulated ATPase activity was lost and regained in parallel with the extent of cross-linking (Fig. S6), indicating that cross-linking strongly inhibited hydrolysis and introduced redox control of activity. In this preparation, about 10 % of the motor domain monomers were refractory to cross-linking.
Single turnover experiments (Fig. 2a) allow direct evaluation of the high extent of inhibition of the cross-linked fraction. In this experiment, a dimer with S181C substitutions was equilibrated with the fluorescent ADP analogue mant-ADP before oxidation. Mant-ADP is released from kinesin with kinetics that are similar to those of unmodified ADP and gives a fluorescence decrease(13). In the absence of MTs, ADP release from the uncross-linked dimer is slow (red trace) and further decreased by cross-linking (blue trace). MTs greatly accelerate the rate of mant-ADP release from uncross-linked dimers (black trace), but produces a biphasic transient with cross-linked dimers (green trace). The relative amplitude of the fast phase is small and similar to the active fraction that is refractory to cross-linking in this preparation (Fig. 2b). However, the majority of the cross-linked preparation exhibits no stimulation of mant-ADP release by MTs (the slow phase of the green trace is approximately parallel to the blue trace). Partial reversal of cross-linking by DTT (orange trace) restores the amplitude of the fast phase in parallel to regain of uncross-linked protein. Thus, the covalent cross-link, which mimics the double lockdown of the dimer, inhibits ADP release from the motor domains.
Figure 2. Disulfide mimic in mant-ADP release experiment.
Cys-lite motor domain dimer with a S181C substitution was oxidized to the covalent disulfide dimer and then reduced back to the sulfhydryl form by addition of dithiothreitol (DTT). (A) Time course for release of mant-ADP from unmodified (SH), oxidized (SS), and DTT treated (SS + DTT) species after mixing with a chase of excess unlabeled ATP. Concentrations after mixing were 0.15 3M kinesin motor domain (0.075 3M dimer) with or without 0.3 3M tubulin (dimer) as indicated. (B) Formation of the covalently linked disulphide dimer was analyzed by denaturing SDS-PAGE in the absence of reducing agents.
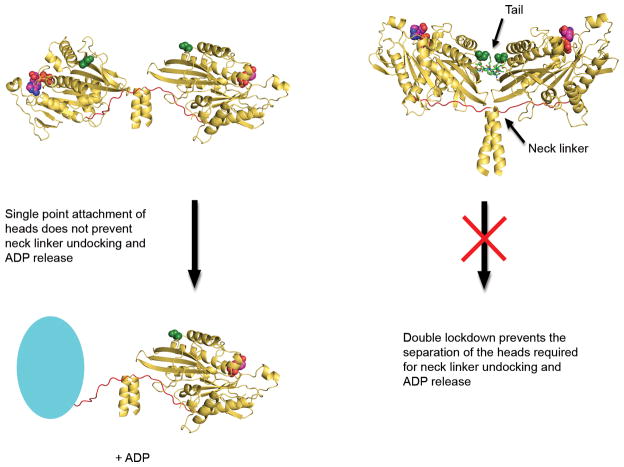
Release of ADP has been hypothesized to be coupled to neck linker undocking(11, 12, 14). Our crystal structure and covalent mimic provide support for this hypothesis and suggest that the inhibition of ADP release by tails could be due to blocking of neck linker undocking through a ‘double lockdown’ mechanism (Fig. 3). Coupling of neck linker undocking and ADP release is also supported by the inhibition of ADP release produced by introducing a cross-link between the neck linker and core motor domain(15). In short, we propose a ‘double lockdown’ autoinhibition mechanism, whereby cross-linking at the coiled-coil and tail interface prevents the movement of the motor domains that is needed to undock the neck linker and release ADP. This opens up the possibility that other kinesins may be regulated by a common autoinhibitory mechanism.
Figure 3. Proposed model for ‘double lockdown’ mechanism of autoinhibition.
Based on the crystal structures, the motor domains (yellow, cartoon) have considerable freedom of movement for neck linker (red) undocking and ADP (red, spheres) release in the absence of tail (green, sticks) binding. Cyan oval represents nucleotide-free motor domain with neck-linker undocked. However, when the tail binds, the motor domains are also cross-linked at the tail interface. In light of the cross-linking experiment results and based on the neck linker undocking hypothesis, we propose a ‘double lockdown’ mechanism, which prevents the separation of the motor domains that is required for neck linker undocking and ADP release. Ser 181 residues (green, spheres) are far apart in the free dimer but close together in the dimer-tail complex, and are on opposite sides of the tail interface.
Supplementary Material
Acknowledgments
We thank David Flot of ESRF and EMBL-Grenoble, and Marcus Muller of SLS for assistance and support in using beamlines ID23-2 and PXI respectively. We also thank Eleanor Dodson for valuable crystallography advice, Oliver Rath for data collection and Venkat Ulaganathan for useful discussion. We also thank Young Yeo and Margret Kim for assistance in cloning and preparation of mutant kinesin motor domains and Avin Snyder for assistance in preparation of the tail fusion peptide. Kristal Kaan holds a National Science Scholarship, financed by A*STAR (Singapore), and this publication contains part of her doctoral thesis. We thank CR-UK, the National Institutes of Health (NS058848) and the National Science Foundation (MCB-0615549) for financial support. Coordinates and structure factor files for the dimer and dimer-tail complex have been deposited in the Protein Data Bank under accession numbers 2Y5W and 2Y65 respectively. The authors declare no competing financial interests.
Footnotes
Materials and Methods
SOM Text Figs. S1 to S6
References and Notes
- 1.Vale RD, Reese TS, Sheetz MP. Cell. 1985 Aug;42:39. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirokawa N. Science. 1998 Jan 23;279:519. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 3.Endow SA, Kull FJ, Liu H. J Cell Sci. 2010 Oct 15;123:3420. doi: 10.1242/jcs.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackney DD, Stock MF. Nat Cell Biol. 2000 May;2:257. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 5.Stock MF, et al. J Biol Chem. 1999 May 21;274:14617. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 6.Verhey KJ, Hammond JW. Nature Reviews Molecular Cell Biology. 2009 Nov;10:765. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich KA, et al. Proc Natl Acad Sci U S A. 2008 Jul 1;105:8938. doi: 10.1073/pnas.0803575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackney DD, Baek N, Snyder AC. Biochemistry. 2009 Apr 21;48:3448. doi: 10.1021/bi8022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kull FJ, Endow SA. J Cell Sci. 2002 Jan 1;115:15. doi: 10.1242/jcs.115.1.15. [DOI] [PubMed] [Google Scholar]
- 10.Kozielski F, et al. Cell. 1997 Dec 26;91:985. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 11.Hwang W, Lang MJ, Karplus M. Structure. 2008 Jan;16:62. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Rice S, et al. Nature. 1999 Dec 16;402:778. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 13.Cheng JQ, Jiang W, Hackney DD. Biochemistry. 1998 Apr 14;37:5288. doi: 10.1021/bi972742j. [DOI] [PubMed] [Google Scholar]
- 14.Sindelar CV, Downing KH. Proc Natl Acad Sci U S A. 2010 Mar 2;107:4111. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahlen K, et al. J Biol Chem. 2006 Jul 7;281:18868. doi: 10.1074/jbc.M508019200. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, Stock MF, Li X, Hackney DD. J Biol Chem. 1997 Mar 21;272:7626. doi: 10.1074/jbc.272.12.7626. [DOI] [PubMed] [Google Scholar]
- 17.Kaan HY, Ulaganathan V, Hackney DD, Kozielski F. Biochem J. 2010 Jan 1;425:55. doi: 10.1042/BJ20091207. [DOI] [PubMed] [Google Scholar]
- 18.Huang TG, Suhan J, Hackney DD. J Biol Chem. 1994 Dec 23;269:32708. [PubMed] [Google Scholar]
- 19.Leslie AGW. 1992;26 [Google Scholar]
- 20.Evans PR. Acta Cryst. 2005;D62:72. [Google Scholar]
- 21.N. Collaborative Computational Project. Acta Cryst. 1994:760. [Google Scholar]
- 22.McCoy AJ, et al. J Appl Crystallogr. 2007 Aug 1;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Nature. 1996 Apr 11;380:550. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997 May 1;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Acta Crystallographica Section D-Biological Crystallography. 2004 Dec;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Chen VB, et al. Acta Crystallographica Section D-Biological Crystallography. 2010 Jan;66:12. [Google Scholar]
- 27.Krissinel E, Henrick K. Journal of Molecular Biology. 2007 Sep 21;372:774. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Schrodinger, LLC. 2010 [Google Scholar]
- 29.Kobashi K. Biochim Biophys Acta. 1968 May;158:239. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- 30.Hackney DD, Stock MF. Biochemistry. 2008 Jul 22;47:7770. doi: 10.1021/bi8006687. [DOI] [PubMed] [Google Scholar]
- 31.Khalil AS, et al. Proc Natl Acad Sci U S A. 2008 Dec 9;105:19247. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sablin EP, et al. Nature. 1998 Oct 22;395:813. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- 33.Kozielski F, De Bonis S, Burmeister WP, Cohen-Addad C, Wade RH. Structure. 1999 Nov 15;7:1407. doi: 10.1016/s0969-2126(00)80030-1. [DOI] [PubMed] [Google Scholar]
- 34.Yun M, et al. EMBO J. 2003 Oct 15;22:5382. doi: 10.1093/emboj/cdg531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yonekura H, et al. Biochem Biophys Res Commun. 2006 May 5;343:420. doi: 10.1016/j.bbrc.2006.02.169. [DOI] [PubMed] [Google Scholar]
- 36.Seeger MA, Rice SE. J Biol Chem. 2010 Mar 12;285:8155. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seiler S, et al. Nat Cell Biol. 2000 Jun;2:333. doi: 10.1038/35014022. [DOI] [PubMed] [Google Scholar]
- 38.Wong YL, Rice SE. Proc Natl Acad Sci U S A. 2010 Jun 29;107:11781. doi: 10.1073/pnas.1005854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.