Abstract
Histone deacetylase 4 (HDAC4) shuttles between the nucleus and cytoplasm, and serves as a nuclear corepressor that regulates bone and muscle development. We report that HDAC4 regulates the survival of retinal neurons in the mouse in normal and pathological conditions. Reduction in HDAC4 expression during normal retinal development led to apoptosis of rod photoreceptors and bipolar (BP) interneurons, whereas overexpression reduced naturally occurring cell death of the BP cells. HDAC4 overexpression in a murine model of retinal degeneration prolonged photoreceptor survival. The survival effect was due to the activity of HDAC4 in the cytoplasm, and relied at least in part upon the activity of hypoxia inducible factor 1α (HIF1α). These data provide evidence that HDAC4 plays an important role in promoting the survival of retinal neurons.
Results
Blindness often results from the death of rod and cone photoreceptor cells, which respond directly to light and thus mediate the first step in vision. HDACs regulate the structure of chromatin through deacetylation of histones, though they also have other targets (1). HDAC4 is expressed in the nervous system and is predominantly cytoplasmic in neurons (2). The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of the transcription factor MEF2, which forms a complex with HDAC4, suggesting a role of HDAC4 in supporting neuronal survival (3). HDAC4 shuttles between the nucleus and cytoplasm (4, 5). In muscle and bone, it interacts with MEF2 to inhibit differentiation as a transcription repressor in the nucleus (6–9).
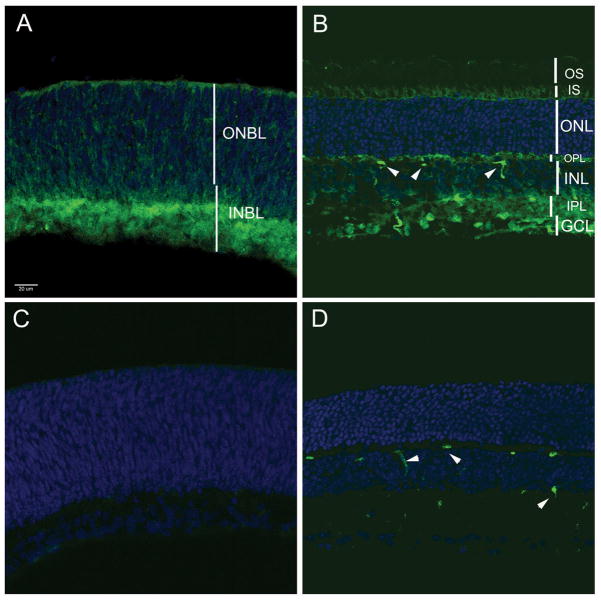
We examined HDAC4 expression by immunohistochemistry, in the postnatal day 1 (P1) murine retina (Fig. 1A), where it was seen in the inner neuroblastic layer (INBL). It was mostly excluded from the nuclei of developing amacrine cells (AC) and ganglion cells in the INBL (Fig. S1). It also was seen in the cytoplasm of cells in the outer neuroblastic layer (ONBL), which contains mitotic progenitor cells and newborn photoreceptor cells. HDAC4 immunoreactivity was lower in the mature retina (Fig. 1B), in the inner segment (IS) of photoreceptor cells, but not in the outer segments (OS), or in cell bodies located in the outer nuclear layer (ONL). In the inner nuclear layer (INL), faint immunoreactivity was detected in the cytoplasm of BP cells and AC. Nuclear staining was seen in the ganglion cell layer (GCL), and in the two plexiform layers where processes are located. HDAC4 immunoreactivity was abolished by preincubation of the antibody with the cognate blocking peptide (Fig. 1, C and D).
Fig. 1.
(A, B) HDAC4 immunohistochemistry on murine retinal sections at P1 and P21, respectively. (C, D) Preincubation of the anti-HDAC4 antibody with blocking peptide led to lack of staining at P1 and P21. Arrowheads (B, D), blood vessels labeled by the mouse antibody.
To investigate the function of HDAC4 in vivo, we transfected a plasmid with an shRNA directed to HDAC4 (Fig. S2), or a control shRNA against GAPDH, into the postnatal day zero (P0) mouse retina by in vivo electroporation (10). To label transfected cells, a plasmid pCAG-GFP, encoding green fluorescent protein (GFP) driven by a broadly active promoter, CAG, was also electroporated. Retinas electroporated with the GAPDH shRNA showed that the majority of transfected cells were rod photoreceptors, with a minority being BP cells, AC, and Müller glial cells, located in the INL((10) and Fig. 2A). When HDAC4 was targeted, the number of cells expressing GFP declined rapidly (Fig. S3, H) with very few seen at P6 (Fig. 2B). Thus HDAC4 appeared to be required for survival. The RNAi phenotype observed was not due to an off-target effect as transfection of a pCAG vector expressing an allele of HDAC4 that is resistant to HDAC4 shRNA rescued the cells (Fig. 2C). A terminal dUTP nick-end labeling (TUNEL) assay showed that cells receiving HDAC4 shRNA died from apoptosis (Fig. S3, A–C), as did an assay for caspase activation in P5 retinal sections (Fig. S3, D–G).
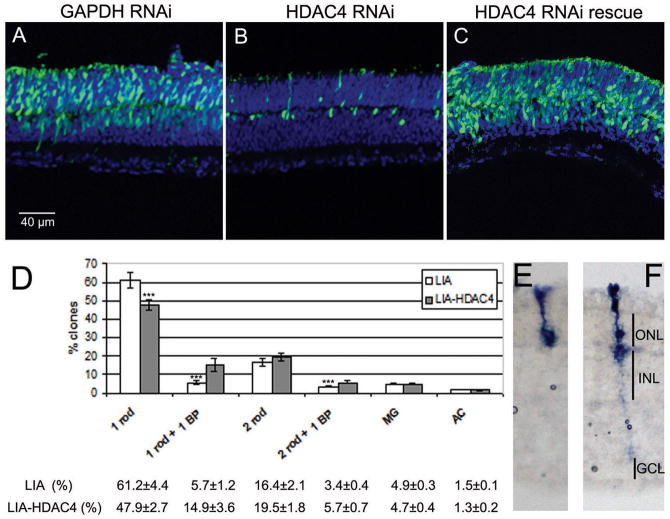
Fig. 2.
HDAC4 is required for retinal cell survival and rescues BP cells from naturally occurring cell death. RNAi vectors targeting GAPDH or HDAC4 were electroporated into the wild type mouse retina at P0, with assay at P6 (A–C). (A) Cryosections from retinas with GAPDH RNAi, (B) HDAC4 RNAi, (C) HDAC4 shRNA and an RNAi resistant HDAC4-expressing construct. (D) Clonal analysis at P21 using replication-incompetent retrovirus LIA, or LIA-HDAC4, injected at P0. LIA-HDAC4 clones exhibited an increase in the percentage of BP cell-containing clones, and a reduction in the percentage of 1 rod clones. 800–1000 clones were analyzed on each of 3 retinas infected by LIA or LIA-HDAC4. *** p<0.05 by Student’s t-Test. (E) Retinal section showing a typical 1 rod clone that comprises the majority of the clone types. (F) Retinal section showing a typical 1 rod plus 1 BP clone that was the most increased clone type labeled by LIA-HDAC4.
We overexpressed HDAC4 in the P0 mouse retinas. Electroporated retinas were collected at P14 when retinal neurogenesis is complete (11). HDAC4 overexpression led to more GFP-expressing BP cells in the upper INL than those observed in pCAG-GFP electroporation (Fig. S4). During retinal development, BP cells are overproduced, and many are eliminated by P14 (12). The higher number of BP cells after HDAC4 electroporation suggested that HDAC4 might protect BP cells from naturally occurring cell death. Previous lineage analyses showed that a P0 retinal progenitor can give rise to a rod photoreceptor and a BP cell, as well as many clones with only a single rod (13). Because many BP cells die naturally, it is likely that some rod-only clones result from loss of a BP cell from clones that initially have both a rod and a BP cell. We therefore did clonal analysis with a replication-incompetent retrovirus, LIA, expressing human placental alkaline phosphatase to mark infected cells, and HDAC4, or without HDAC4 (Fig. 2D–F). There was a statistically significant increase in the number of clones containing 1 rod and 1 BP cell or 2 rods and 1 BP following LIA-HDAC4 infection. The total BP cell-containing clones increased from 9.1% for LIA infection to 20.6% for LIA-HDAC4 infection. These results are consistent with increased survival of BP cells, and are complementary to the findings from the HDAC4 RNAi treatment.
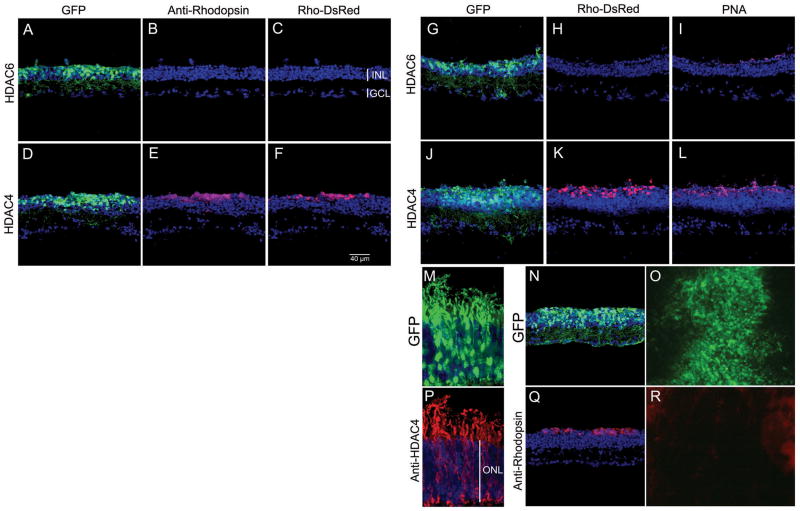
There is little naturally occurring rod death in normal development (14). Therefore, we assayed rod survival following introduction of HDAC4 in a mouse model of retinal degeneration. The rd1 mutation is in the rod-specific gene, PDE6β (phosphodiesterase 6β subunit), which encodes a key enzyme in the phototransduction cascade. This mutation causes photoreceptor degeneration (15, 16). In rd1 mice, nearly all the rod photoreceptors die by P21, and cone death follows over the next few months (17). By P35, ~ 20% of cone photoreceptors remain (18). Electroporation at P0 results in transfection of many rods, but few cones (10). We electroporated pCAG-HDAC4 or pCAG-HDAC6 into P0 rd1 mice. HDAC6 was included for a test of specificity because it rescues retinal degeneration in Drosophila (19). Retinas were analyzed at P50 with an antibody to Rhodopsin, a rod-specific marker. In the transfected areas labeled by co-electroporated pCAG-GFP, many rods were present in retinas overexpressing HDAC4, but not in retinas overexpressing HDAC6 (Fig. S5), or HDAC5 (Fig. S6). We also assayed the activity of the Rhodopsin promoter with a reporter plasmid for rhodopsin, Rho-DsRed (10), in which a 2.2 kb Rhodopsin promoter drives the expression of DsRed, a red fluorescent protein. Rho-DsRed was electroporated with pCAG-HDAC4 into the P0 rd1 retina. Tissues harvested at P70 showed that rods, marked by anti-Rhodopsin immunohistochemistry (Fig. 3E), expressed DsRed (Fig. 3F) in the GFP-labeled electroporated area (Fig. 3D). The HDAC6-electroporated retina, did not exhibit rhodopsin immunoreactive cells or Rhodopsin promoter activity (Fig. 3, A–C). We scored 24.3±7.2 Rhodopsin+ cells per 100 μm section for HDAC4 treated retinas and 0 rhodopsin+ cells in the HDAC6 electroporated retinas (n=3). However, HDAC6 seemed to be as effective as HDAC4 in protecting BP cells from naturally occurring cell death in the rd1 retina (Fig. 3, A and G), as well as in the wild type retina (Fig. S7).
Fig. 3.
Rescue of retinal degeneration by HDAC4. (A–F) Co-electroporation of pCAG-GFP and pCAG-HDAC6 (A–C) or pCAG-HDAC4 (D–F) and a reporter construct for the rhodopsin promoter, Rho-DsRed, at P0 with assay at P70 for anti-Rhodopsin (B, E) or reporter activity (C, F). (G–L) HDAC4 electroporation increased cone photoreceptor density. (J) In the HDAC4 electroporated area, PNA-labeled cone photoreceptors (L) were more abundant in the rods-saved area (K), compared to the HDAC6 electroporated area (G–I). (M–R) Analysis of the activity of cytoplasmic and nuclear forms of HDAC4 on retinal degeneration. (M, P) Retinas were co-electroporated at P0 with pCAG-GFP (M) and pCAG-HDAC4 and examined at P14 by anti-HDAC4 immunohistochemistry (P). (N, Q) Cytoplasmic HDAC4-L175A was co-electroporated with pCAG-GFP into rd1 at P0 and cryosections were assayed at P70 with anti-Rhodopsin (Q), with GFP signal shown in (N). Co-electroporation of nuclear HDAC4-3SA and pCAG GFP (O) failed to rescue rd1 rods at P50, assayed by anti-Rhodopsin immunohistochemistry on the flat-mount retina (R).
Because the survival of cone photoreceptors depends upon the survival of rod photoreceptors, HDAC4 treated retinas were collected at P70 and stained with peanut agglutinin (PNA), which labels cones. HDAC4 overexpression (Fig. 3J) led to increased cone density (Fig. 3L) in the area with rescued rods (Fig. 3K), compared to retinas overexpressing HDAC6 (Fig. 3, G–I).
HDAC4 shuttles between the nucleus and cytoplasm. Retention of HDAC4 in the nucleus requires its interaction with MEF2 transcription factors (4). Phosphorylated HDAC4 moves into the cytoplasm, terminating its nuclear function (5). In wild type photoreceptors, endogenous (Fig. 1B) or overexpressed HDAC4 (Fig. S8) was predominantly located in the cytoplasm, including the photoreceptor IS and OS, as well as axonal endings (Fig. 3, M and P). Thus HDAC4-mediated photoreceptor survival in rd1 mice might result from its cytoplasmic function. To investigate whether cytoplasmic or nuclear HDAC4 mediated the photoreceptor survival effect, HDAC alleles modified to be targeted primarily to the cytoplasm or nucleus were tested. The cytoplasmic HDAC4-L175A mutant doesn’t interact with MEF2 transcription factors and thus stays primarily in the cytoplasm ((4) and Fig. S9). The HDAC4-3SA mutant, which has its 3 serine phosphorylation sites mutated to alanine, stays primarily in the nucleus ((5, 7) and Fig. S9). The transfected cytoplasmic HDAC4-L175A mutant preserved rd1 rods out to at least P70 (Fig. 3, N and Q), but the nuclear HDAC4-3SA mutant failed to rescue even until P50 (Fig. 3, O and R).
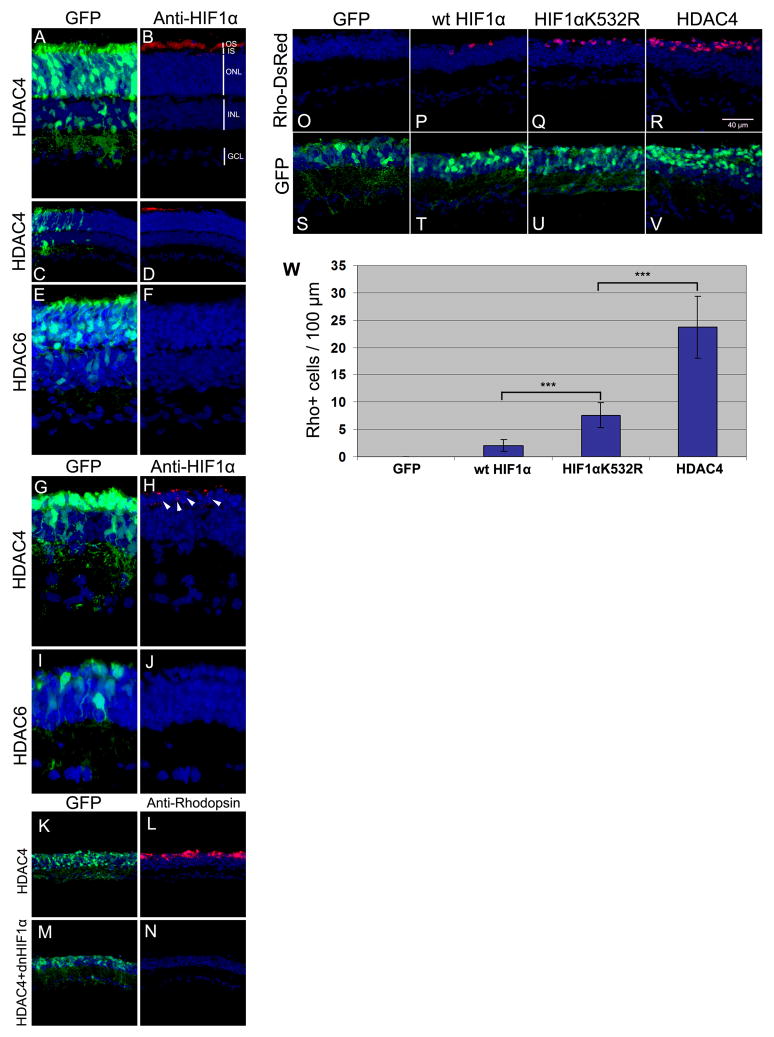
Hypoxia inducible factor 1α (HIF1α) plays a central role in the regulation of oxygen homeostasis (20). HIF1α protein is not detectable in the mature mouse retina (21). Exposure of retinas to hypoxia causes stabilization of HIF1α and protects photoreceptors from light-induced retinal degeneration (21). HIF1α protein stability is decreased by lysine acetylation. Acetylation of HIF1α by the acetyl-transferase, ARD1, enhances its degradation (22). HIF1α stabilization thus might provide a mechanism for HDAC4-induced photoreceptor protection in rd1 mice. HIF1α protein was detected by immunohistochemistry in the OS of wild type photoreceptors after HDAC4 electroporation (Fig. 4, A–D). No HIF1α immunoreactivity was detected after overexpression of HDAC6 (Fig. 4, E and F). Likewise, expression of HDAC4, but not that of HDAC6 (Fig. 4, I and J), led to the detection of HIF1α that appeared nuclear or perinuclear in the photoreceptors of the rd1 retina (Fig. 4, G and H). Expression of a dominant negative HIF1α (dnHIF1α) construct (23) with pCAG-HDAC4 in the rd1 retina negated the photoreceptor survival effect of HDAC4 (Fig. 4, K-N). A plasmid with an shRNA directed to HIF1α also blocked HDAC4-mediated photoreceptor survival (Fig. S10). Thus HIF1α appears to be required for the HDAC4 survival effect.
Fig. 4.
Role of HIF1α in HDAC4 survival activity. (A–F) Wild type retinas electroporated at P0 with pCAG-HDAC4 and pCAG-GFP, or pCAG-HDAC6 and pCAG-GFP, and assayed at P14 by anti-HIF1α immunohistochemistry on retinal sections. (A) HIF1α protein in the photoreceptor OS by anti-HIF1α in HDAC4 electroporated retina (B), only in the HDAC4 electroporated area (C, D). (E) In the HDAC6 electroporated area, no HIF1α immunoreactivity (F) was detected. (G–J) HIF1α in the rd1 retina. rd1 retinas were co-electroporated at P0 with pCAG-HDAC4 and pCAG-GFP (G, H), or pCAG-HDAC6 and pCAG-GFP (I, J), and assayed at P22 by anti-HIF1α immunohistochemistry. Arrow heads (H), HIF1α nuclear immunoreactivity. (K–N) HIF1α protein is required for rescue of photoreceptors by HDAC4. rd1 retinas were co-electroporated at P0 with pCAG-HDAC4 (K, L) or pCAG-HDAC4 plus pCAG-dnHIF1α (M, N), and pCAG-GFP, and assayed at P70 for rod photoreceptor survival by Rhodopsin immunohistochemistry. (O–V) HIF1α acetylation mutant rescues retinal degeneration. rd1 retinas were electroporated at P0 with pCAG-GFP, Rho-DsRed, and pCAG-wt HIF1α (P, T), or pCAG-HIF1αK532R (Q, U), or pCAG-HDAC4 (R, V), and assayed at P70 for rod photoreceptor survival. (W) Quantification of photoreceptor survival in rd1 mice at P70. The number of Rho-DsRed+ cells was counted per 100 μm section in transfected areas. Three retinas were scored for each treatment. *** p<0.05 by Student’s t-Test.
To test whether acetylation of HIF1α might influence rod death in the rd1 retina, we expressed HIF1αK532R, an acetylation mutant of HIF1α, that is more stable than its wild type form (22), and the wild type HIF1α in the rd1 retina. Wild type HIF1α preserved a few rods (Fig. 4, P and T) and HIF1αK532R preserved more (Fig. 4, Q and U). HDAC4 was the most effective in saving rod photoreceptors (Fig. 4, R, V, and W). No additive effects were seen when HDAC4 was co-expressed with HIF1αK532R (Fig. S11). The greater efficacy of HDAC4 relative to that of HIF1αK532R might result from HDAC4 having target(s) in addition to HIF1α, or HIF1αK532R could be less effective than deacetylated wild type HIF1α.
In the mouse retina HDAC4 has an essential role in neuronal survival. More importantly from a therapeutic perspective, HDAC4 prolonged photoreceptor survival in mice undergoing retinal degeneration. HDAC6 did not lead to increased abundance of HIF1α protein or promote rod survival in mice, although it rescued degeneration in Drosophila (19). Therefore, more than one pathway for neuronal survival may be regulated by HDACs.
Supplementary Material
Acknowledgments
We thank JR Lu for providing HDAC4, HDAC5, and HDAC6 expression constructs, TP Yao for providing HDAC4-3SA mutant, XJ Yang for providing HDAC4-L175A mutant, GL Semenza for providing dnHIF1α constructs, and KW Kim for providing HIF1α and HIF1αK532R constructs, and T. Matsuda for assistance with the various techniques. Supported by NIH EYO 9676 and the Howard Hughes Medical Institute. CLC is an Investigator of HHMI and BC is a postdoctoral fellow of HHMI. U.S. Pending patent application: Prevention of neurodegeneration by HDACs or stabilized HIF1alpha.
Contributor Information
Bo Chen, Email: bochen@genetics.med.harvard.edu.
Constance L. Cepko, Email: cepko@genetics.med.harvard.edu.
References
- 1.Yang XJ, Seto E. Oncogene. 2007;26:5310–8. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 2.Bolger TA, Yao TP. J Neurosci. 2005;25:9544–53. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolger TA, Zhao X, Cohen TJ, Tsai CC, Yao TP. J Biol Chem. 2007;282:29186–92. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 4.Wang AH, Yang XJ. Mol Cell Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, et al. J Biol Chem. 2001;276:35042–8. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 6.Miska EA, et al. Embo J. 1999;18:5099–107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vega RB, et al. Cell. 2004;119:555–66. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, McKinsey TA, Zhang CL, Olson EN. Mol Cell. 2000;6:233–44. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 9.Arnold MA, et al. Dev Cell. 2007;12:377–89. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda T, Cepko CL. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young RW. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 12.Voyvodic JT, Burne JF, Raff MC. Eur J Neurosci. 1995;7:2469–78. doi: 10.1111/j.1460-9568.1995.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 13.Turner DL, Cepko CL. Nature. 1987;328:131–6. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 14.Young RW. J Comp Neurol. 1984;229:362–73. doi: 10.1002/cne.902290307. [DOI] [PubMed] [Google Scholar]
- 15.Bowes C, et al. Nature. 1990;347:677–80. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 16.McLaughlin ME, Sandberg MA, Berson EL, Dryja TP. Nat Genet. 1993;4:130–4. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 17.Carter-Dawson LD, LaVail MM, Sidman RL. Invest Ophthalmol Vis Sci. 1978;17:489–98. [PubMed] [Google Scholar]
- 18.Komeima K, Rogers BS, Lu L, Campochiaro PA. Proc Natl Acad Sci U S A. 2006;103:11300–5. doi: 10.1073/pnas.0604056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey UB, et al. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 20.Semenza GL. J Appl Physiol. 2000;88:1474–80. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 21.Grimm C, et al. Nat Med. 2002;8:718–24. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 22.Jeong JW, et al. Cell. 2002;111:709–20. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 23.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. J Biol Chem. 1996;271:17771–8. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.