Abstract
Retinitis Pigmentosa (RP) is an incurable retinal disease that leads to blindness. One puzzling aspect concerns the progression of the disease. While most mutations that cause RP are in rod photoreceptor (PR) -specific genes, cone PRs die as well. To understand the mechanism of non-autonomous cone death, four mouse models harboring mutations in rod-specific genes were analyzed. Changes in the insulin/mTOR pathway that coincided with the activation of autophagy during the period of cone death were found. We thus either increased or decreased the insulin level and measured the survival of cones in one of the models. Mice treated systemically with insulin had prolonged cone survival, while depletion of endogenous insulin had the opposite effect. These data suggest that the non-autonomous cone death in RP could, at least in part, be due to the starvation of cones.
Introduction
Retinitis Pigmentosa is a family of inherited retinal degenerations (RD) that is currently untreatable and frequently leads to blindness. Affecting roughly 1 in 3,000 individuals, it is the most prevalent form of RD caused by a single disease allele (RetNet, http://www.sph.uth.tmc.edu/Retnet/). The phenotype is characterized by an initial loss of night vision due to the malfunction and death of rod PRs, followed by a progressive loss of cones1. Since cones are responsible for color and high acuity vision, it is their loss that leads to a reduction in the quality of life. In many cases, the disease-causing allele is expressed exclusively in rods; nonetheless, cones die too. Indeed, to date there is no known form of RD in humans or mice where rods die, and cones survive. In contrast, mutations in cone-specific genes result only in cone death. Several theories have been proposed to explain this finding. For example, cone death could be due to the release of a toxin produced by dying rods, or the loss of a trophic factor that is produced by healthy rods2–7. Alternatively, cone death could be caused by microglia mobilized initially during rod death8 or by oxidative stress9,10. Oxidative stress also might directly harm cones. There is a constant flow of oxygen through the retinal pigmented epithelium (RPE) to PRs, and the loss of rods, which are 95% of the PRs in human and mouse, may result in an overload of oxygen to the remaining cones11. Evidence for all of these mechanisms exist in mouse, yet none are able to fully explain why cones may survive for many years in the absence of rods in humans. Nonetheless, rodents are a very good model for this type of RD. While the rodent retina lacks a macula, which is the cone-rich and rod-free area present in humans, the macula plays no part in the initial phase of the disease. In humans, RP starts outside of the macula, where the distribution of rods and cones is similar in humans and mice.
To determine the common underlying mechanism for cone death in RP, we compared 4 mouse models harboring mutations in rod-specific genes (PDE-β−/−12, PDE-γ-KO13, Rho-KO14 and P23H15, see Material & Methods). Affymetrix arrays were used to identify common changes in gene expression that accompany cone death. Changes in a significant number of genes involved in cellular metabolism coincided with the onset of cone death. These changes were suggestive of cones suffering from a shortage of nutrients. Cones were then shown to exhibit signs of autophagy, a cellular self-digestion process, which is consistent with prolonged starvation. We also found that several aspects of the insulin/mTOR pathway, a key pathway that regulates cellular metabolism, were affected during the period of cone degeneration. As a result of this finding, we either increased or decreased the insulin level and measured the survival of cones in one of the models. Mice treated systemically with insulin had prolonged cone survival, while depletion of endogenous insulin had the opposite effect. Therefore, cone starvation is a likely contributor to the slow demise of cones in humans with RP. Treatments aimed at improving nutrition of cones are thus a plausible therapeutic avenue.
Results
Rod and cone death kinetics
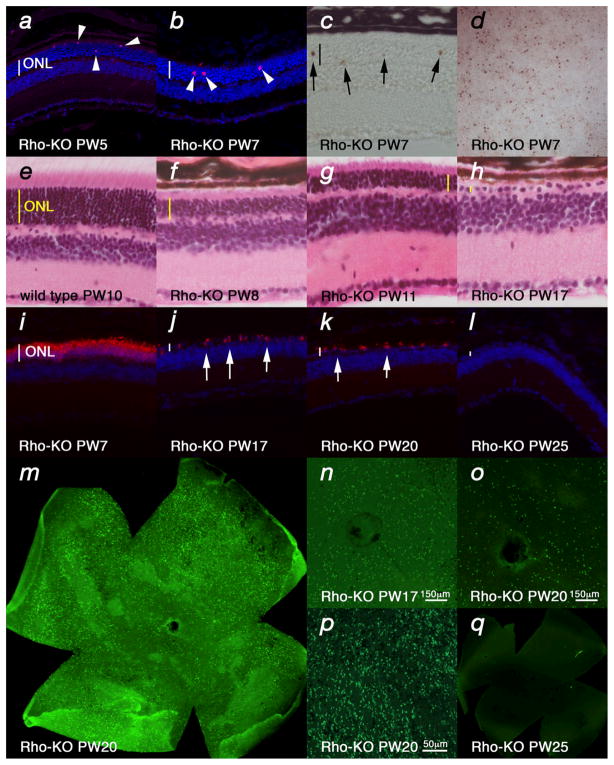
To establish a framework for comparing gene expression in 4 different models of RP, the equivalent stages of disease pathology were established through examination of the kinetics of rod (Fig. 1) (see also Supplementary Fig. 1 & 2 online) and cone (Fig. 2) (see also Supplementary Fig. 3 online) death. Rod death kinetics were established by determining the onset, progression and end phase of rod death (Fig. 1). The time from the onset of rod death to the time when the outer nuclear layer (ONL) was reduced to 1 row of cells will be referred to as the major rod death phase. The time thereafter until rod death was complete will be referred to as the end phase of rod death. To determine the beginning of the major phase of rod death, cleavage of the nuclear envelope protein LaminA (Fig. 1a), and of the apoptotic protease Caspase3 (Fig. 1b), as well as TUNEL (Fig. 1c, d) were used. The continuation of the major rod death phase was monitored by these assays, as well as inspection of histological sections (Fig. 1e–h), as rods account for more than 95% of all PRs. Once the ONL reached one row of cells, the major phase of rod death was over. The end phase of rod death was determined using rod-specific markers to perform either in situ hybridization (see Supplementary Fig. 1 online) or immunohistochemistry (Fig. 1i–l) on retinal sections. However, unless every section of a single retina is collected it is difficult to determine if any rods remain. Thus, retinal flat mounts also were used to allow a comprehensive analysis of the end phase of rod death (Fig. 1m–q). Interestingly, while in the two PDE mutants and in the Rho-KO mutant the end phase of rod death was clearly defined, in the P23H mutant, rods died so slowly that even 50 weeks (latest time point analyzed) after the end of the major phase of rod death, some rods were still present (see Supplementary Fig. 2 online).
Figure 1.
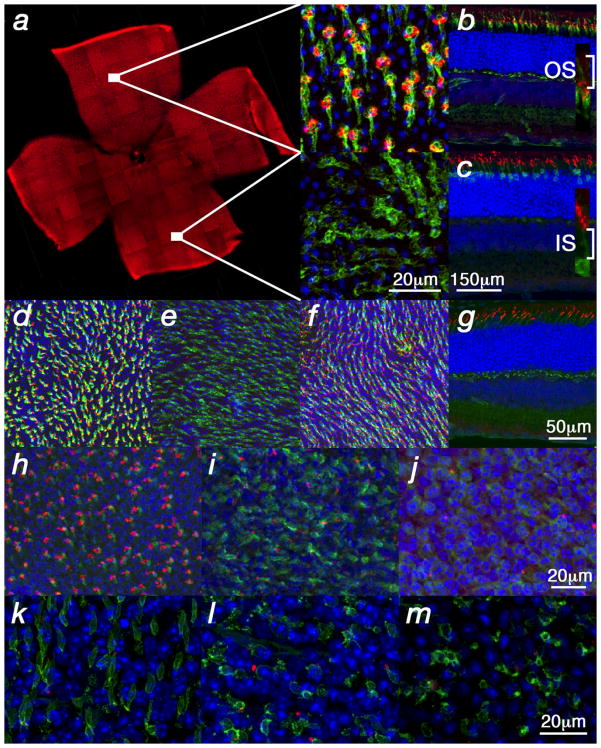
Rod death kinetics in the Rho-KO mutant. (a–d) Onset of rod death seen by cleaved nuclear envelope protein LaminA (a), Cleaved Caspase3 (b) (arrowheads: magenta and red signal respectively) as well as TUNEL (c, d) (arrows: brown signal) (blue in a, b shows nuclear DAPI staining). (d) Shows a retinal flat mount with view onto the photoreceptor layer. (e–h) Progression of rod death determined by the reduction of the ONL as seen by HE staining. (i–q) End phase of rod death assessed by section analysis (i–l) or by retinal flat mounts (m–q). In the Rho-KO the onset of rod death is around PW5 (a) and progresses up to PW25 (l). By PW17 the ONL is reduced to one row of cells (h, j) and in the following 8 weeks the remaining rods die (j–q) as seen by immunofluorescence with an antibody directed against guanine nucleotide protein alpha transducin (Gnat1) on sections of progressively older animals (j–l). (m–q) Retinal flat mounts showing rods visualized by immunofluorescence with an antibody directed against Gnat1. (m) Shows entire retina while (n, o) show higher magnification around the optic nerve head and (p) shows peripheral region. (q) Shows no signal at PW25 where on sections rods were also not detected (l). Age (in postnatal weeks (PW)) is indicated in the panels. Vertical bar in (ac, e–l) indicates thickness of the ONL.
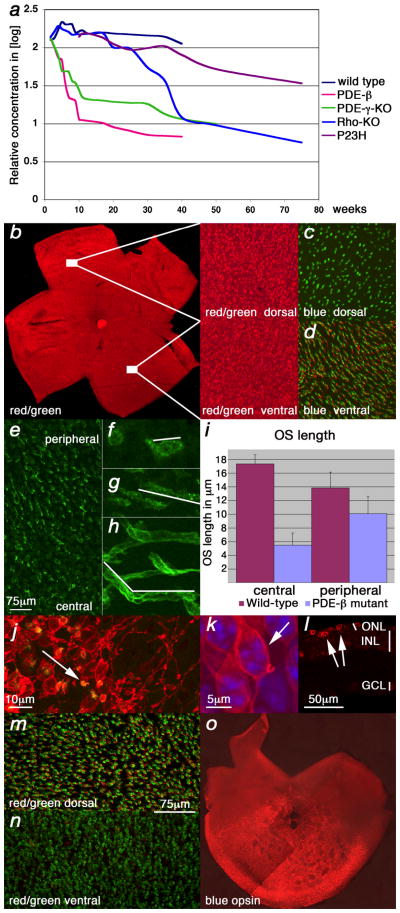
Figure 2.
Cone death kinetics. (a) qRT-PCR analysis for Opn1sw during cone degeneration. Changes are in indicated as the logarithm of the relative concentration over time on the Y-axis while X-axis indicates postnatal weeks. (b–h, j, k, m–o) Show retinal flat mounts. (k) Shows a retinal section. Green signal shows PNA expression, red signal shows red/green opsin expression (b, j–n) or blue opsin expression (c, d, o). (b–d) Wild type retina at P35. Red/green opsin (b) and PNA (c, d) expression were detected dorsal and ventral while blue opsin (c, d) was detected only ventrally. (e–g, j–o) Analysis in the PDE-β mutant. (e–g) Central to peripheral gradient of PNA and shortening of cone outer segments (OS). At P20, prior to the major cone death phase, there were fewer elongated OS in the center (e) as compared to the periphery. (f) High magnification of a central or peripheral (g) OS from (e). (h) Wild type OS (white line in f–h marks the OS). (i) Quantification of OS length in central and peripheral regions. The data represents an average of 15 measurements on 3 different retinae of 3 week old mice. With the shortening of OSs during degeneration, red/green opsin was localized throughout the membrane of the cell body and PNA, which detects an extracellular protein(s), was reduced to a small dot attached to the residual OS (j) (arrow: yellow shows red/green and PNA overlap). (k) High magnification of a cone showing red/green localization at the membrane of the main cell body (arrow). (l) Cross section showing red/green in cell body (arrows; j–l P70). Red/ green opsin was detected mainly dorsal (l) during degeneration while PNA (m, n) or blue opsin (o) were not altered (m, n: P21, same scale bar; o: P49).
Two methods were used to determine the onset and progression of cone death. First, the overall time frame of cone demise was determined by quantitative real-time polymerase chain reaction (qRT-PCR) (Fig. 2a) for the ventral16 cone specific transcript Opn1sw (opsin1 short-wave-sensitive: blue cone opsin). This allowed for an initial quantitative comparison among different strains, but was not adequate to determine the number of cones as transcript levels could vary prior to cell death. Next, whole mount immunohistochemistry for red/green opsin (Opn1mw: opsin1 medium-wave-sensitive) and peanut agglutinin lectin (PNA) were used (Fig. 2b–n). Both markers are expressed throughout the murine retina allowing for the visualization of cones (Fig. 2b–d). Interestingly, the onset of cone death always occurred at the equivalent stage of rod death, namely after the major rod death phase, when the thickness of the ONL was reduced to only a single row of cells. Cone death was found to proceed from the center to the periphery in all 4 models, as seen by staining with PNA (Fig. 2e). It was preceded by a gradual reduction of the outer segment (OS) length (Fig. 2f–i) and by opsin localization from the OS to the entire cell membrane (Fig. 2j–l). In addition, red/green opsin (Opn1mw) protein, which is normally detected throughout the mouse retina (Fig. 2b), was detected mainly dorsally during cone degeneration (Fig. 2m, n). However, PNA staining showed no appreciable difference across the dorsal/ventral axis (Fig. 2m, n). Similarly, blue opsin expression, which is normally detected only ventrally16 (Fig. 2c, d), was not affected during degeneration (Fig. 2o). Shortening of cone OSs and loss of cone-specific markers has also been described in human cases of RP17.
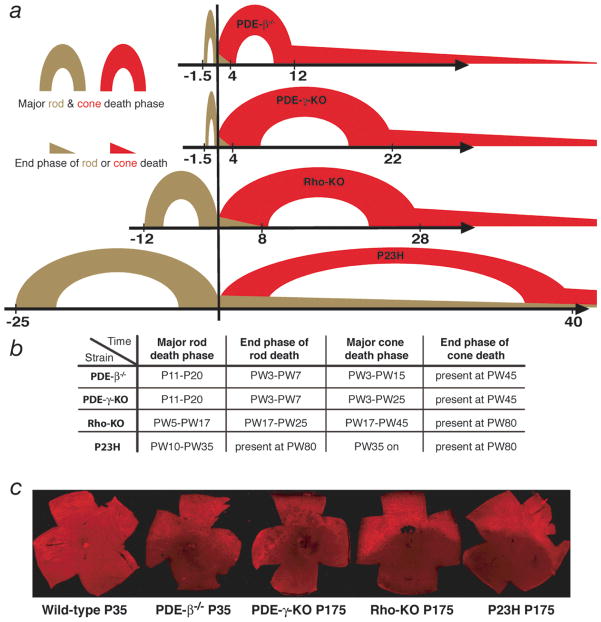
In summary, the kinetics and histological changes that accompanied rod and cone death shared several features across the 4 models. First, cone degeneration always started after the major rod death phase (Fig. 3a, b). This point was reached at very different ages in three of the 4 mutants, as the overall kinetics of rod death were quite different. Second, cone death was always central to peripheral and was preceded by a reduction in OS length. Third, in all 4 mutants, red/green opsin protein levels were detectable mainly dorsally during cone degeneration (Fig. 3c). These common features suggested that there might be a common mechanism(s) of cone death, and that clues about this mechanism(s) might be suggested by gene expression changes that were common across the 4 models at the onset of cone death.
Figure 3.
(a) Schematic representation of the rod and cone death kinetics found in the 4 mouse models of RP. The onset of cone death is set as time zero. The corresponding time windows on the x-axis are given in weeks. For the major rod death phase see text. The major cone death phase was the time until roughly 85% of cones had died. The end phase of cone death was the time thereafter. (b) Summary of rod and cone death kinetics. Time is indicated in postnatal days (P) or postnatal weeks (PW). (c) Immunofluorescence on retinal flat mounts showing the ventral reduction of red/green opsin expression found in the 4 mutants during cone degeneration. Strain and time, in postnatal days, are indicated below each image. (For higher magnification of wild type see figure 2b).
Microarray analysis
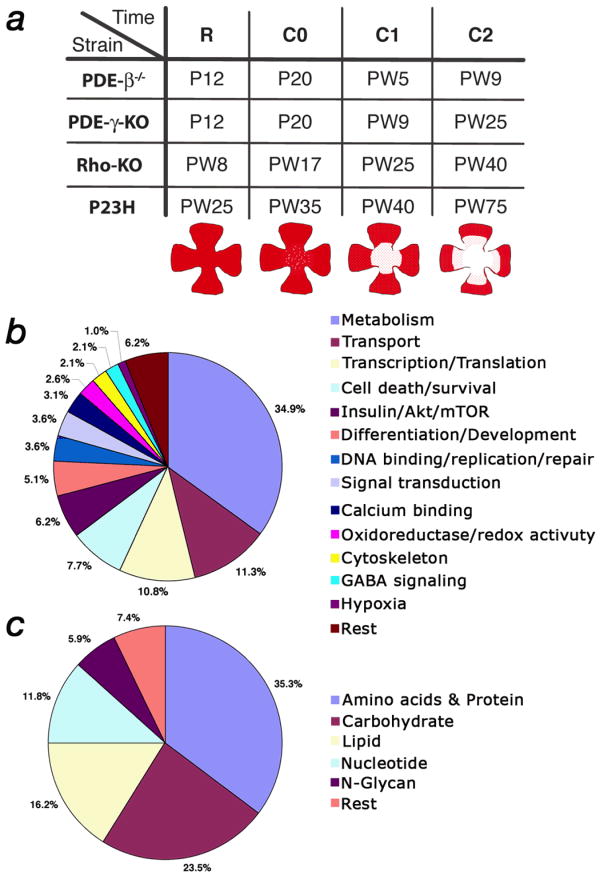
To determine common gene expression changes, RNA samples from all 4 models were collected halfway through the major phase of rod death, at the onset of cone death, and from two time points during the cone death phase (Fig. 4a). The RNA was then hybridized to an Affymetrix 430 2.0 mouse array. Gene expression changes were compared within the same strain across the 4 time points. Two criteria had to be fulfilled to select a gene for cross comparison among the 4 strains. First, the change over time had to be statistically significant (see Material & Methods). Second, a gene had to be upregulated at least 2 fold at the onset of cone death compared to the other three time points. This second criterion removed rod-specific changes that were still occurring at the onset of cone death while at the same time enriched for changes at the onset of cone death. A total of 240 Affymetrix IDs were found that satisfied both criteria within each of the 4 strains. The 240 IDs matched to 230 genes (see Supplementary Table 1 online). Of the 195 genes that could be annotated, 34.9% (68 genes) were genes involved in cellular metabolism (Fig. 4b, c). The signaling pathway with the highest number of hits (12 genes) was the insulin/mTOR (mammalian target of rapamycin) signaling pathway (Fig. 4b), a key pathway in regulating many aspects of cellular metabolism. Thus, the data suggested that events at the onset of cone death coincided with changes in cellular metabolism likely to be regulated by the insulin/mTOR pathway.
Figure 4.
Affymetrix microarray analysis. (a) Equivalent time points in the 4 different mutants at which the microarray analysis was performed (R: approximately halfway through the major phase of rod death; C0: onset of cone death; C1 & C2 first and second time point during cone death respectively). Time is indicated in postnatal days (P) or postnatal weeks (PW). Cartoons depicting the progression of cone death are shown below the corresponding time points. (b) Distribution in percentage of the 195 genes that were annotated. (c) Distribution in percentage of the 68 genes (34.9%) that are part of metabolism in (b).
mTOR in wild type and degenerating retinae
Based on the findings of the microarray analysis, the insulin/mTOR signaling pathway was examined during the period of cone death. The kinase, mTOR, is a key regulator of protein synthesis and ribosome biogenesis18. When cellular energy levels are high, mTOR allows energy consuming processes, such as translation, and prevents autophagy, while nutrient poor conditions have the reverse effect. Therefore, glucose, which increases cellular ATP levels, and amino acid availability, especially that of leucine, positively affect mTOR activity. To understand if cellular energy levels or amino acid availability might be compromised in cones during degeneration, levels of phosphorylated mTOR (p*-mTOR) were examined by immunofluorescence. Phosphorylation of mTOR increases kinase activity, and therefore levels of p*-mTOR can serve as an indicator of its activity level. Since every eukaryotic cell expresses mTOR, a certain level of p*-mTOR is likely to be found in every cell. Surprisingly, high levels of p*-mTOR were detected only in dorsal cones of wild type retinae (Fig. 5a–c). This phosphorylation pattern was reminiscent of the red/green opsin pattern seen during cone degeneration (Fig. 3c). Since mTOR is a key regulator of translation, we investigated whether the ventral red/green opsin downregulation that occurred during cone degeneration could be mimicked by a reduction in mTOR activity. To this end, wild type mice were treated with rapamycin, an mTOR inhibitor18. This treatment resulted in ventral downregulation of red/green opsin, without affecting blue opsin or PNA staining or the dorsal phosphorylation of mTOR itself (Fig. 5d–g). Thus, inhibition of mTOR in wild type recapitulated the expression of red/green opsin and blue opsin, as well as the pattern of PNA staining, in the mutants during degeneration, suggesting that the ventral downregulation of red/green opsin seen during degeneration might be due to reduced mTOR activity. As expected for mTOR function, the downregulation of red/green opsin did not occur at the RNA level, but at the protein level, in untreated mutant mice, as well as in wild type mice treated with rapamycin (see Supplementary Fig. 4 online). Finally, analysis of mutant retinae showed a decrease of p*-mTOR levels in dorsal cones during cone degeneration (Fig. 5h–m). To test whether the high level of p*-mTOR found in dorsal wild type cones was glucose-dependent, retinal explants of wild type mice were cultured in media for 4 hours in the presence or absence of glucose. Dorsal p*-mTOR was abolished in the absence of glucose even when leucine concentrations were increased in the medium (see Supplementary Fig. 5 online). Thus, the data on mTOR establish a link between mTOR activity, the expression changes of red/green opsin seen during degeneration, and the microarray data, which suggested metabolic changes at the onset of cone death. Those changes may be caused by compromised glucose uptake in cones.
Figure 5.
p*-mTOR in wild type and degenerating retinae. All panels show immunofluorescence on retinal flat mounts (photoreceptor side up) with the exception of (b, c, g) which show retinal sections. Blue shows the nuclear DAPI stain. (a–c) p*-mTOR levels in wild type retinae. (a) Dorsal (up) enrichment of p*-mTOR. Higher magnification of dorsal and ventral region is shown to the right showing p*-mTOR in red and cone segments in green as detected by PNA. (b, c) Dorsal retinal sections stained for p*-mTOR (red signal) and PNA (b) (green signal) or α–β-galactosidase (c) (green signal). The β-galactosidase is under the control of the human red/green opsin promoter and is expressed in all cones48 (see Material & Methods). The insets in (b, c) show higher magnification of the cone segments suggesting that the p*-mTOR signal is located in the lower part of the outer segment (OS; IS: inner segment). (d–g) Rapamycin treatment of wild type mice leads to downregulation of red/green opsin ventrally (e) but not dorsally (d) (red signal). Ventral blue opsin (f) (red signal) remains unaffected, as does PNA (d–g) (green signal). Rapamycin treatment does also not affect mTOR phosphorylation in wild type (g) (red signal). (h–m) Reduced levels of dorsal p*-mTOR during photoreceptor degeneration (red signal). (h) Wild type control. (i, j) PDE-β mutant. The reduction starts during rod death at P15 (i) as the OSs (green signal: PNA) start to detach from the retinal pigmented epithelium. (i) By P30 only few cones (green signal: α-β-galactosidase) show high levels of p*-mTOR (red signal). (k–l) A similar reduction is seen in dorsal cones of the other three mutants (cones marked in green by PNA). (k) PDE-γ-KO P35. (l) Rho-KO PW20. (m) P23H PW70.
Responses of cones to nutritional imbalance
The data on mTOR suggested a nutritional imbalance in cones during cone degeneration, possibly caused by reduced glucose levels in cones. To test this idea, the level of the heterodimeric transcription factor, Hypoxia inducible factor 1 (HIF-1α/β), which improves glycolysis under stress conditions such as low oxygen, was examined. HIF-1 and mTOR are tightly linked as low oxygen results in low energy due to reduced oxidative phosphorylation, and therefore in reduced mTOR activity18–23. An upregulation of the regulated subunit HIF-1α would likely reflect low glucose levels in cones, and not hypoxic conditions, as oxygen levels are increased due to the loss of rods11. Immunofluorescence analysis of HIF-1α during cone degeneration revealed an upregulation of the protein in cones in all 4 mouse models (Fig. 6a–f; & Supplementary Fig. 6a–d online). Consistent with the upregulation of HIF-1α, glucose transporter 1 (GLUT1), a HIF-1α target gene24,25 also was found to be upregulated in cones, again in all 4 mouse models (Fig. 6g–j; & Supplementary Fig. 6e–h online). Thus HIF-1α and GLUT1 upregulation are consistent with a response in cones to overcome a shortage of glucose. It also provides a link to the decreased p*-mTOR levels found during degeneration as well as the sensitivity of p*-mTOR to glucose.
Figure 6.
Upregulation of Hif-1α and GLUT1 in cones. All panels show immunofluorescent staining. Left column (a, d, g, h,) shows retinal flat mounts and right column (b, c, e, f, i, j) retinal sections. Blue shows nuclear DAPI staining and green shows cones marked with PNA. (a–f) Staining for HIF-1α (red signal). (a) Wild type (PW10) (inset) showing higher magnification. (b, c) Cross sections in wild type (PW10). (c) DAPI overlap of (b). (d–f) During cone degeneration in PDE-β−/− (PW10) increased levels of HIF-1α are found in cones (d, inset). (e, f) Cross sections show that the increase of Hif-1α occurs mainly in cones (arrows point to cones that at this stage are located within the top layer of the inner nuclear layer). (f) DAPI overlap of (e). (g) GLUT1 expression in wild type (PW10) (red signal). Most of the signal in between the cones reflects expression in rods. (h–j) Increased expression of GLUT1 in cones during degeneration seen in flat mounts (h) and sections (i–j). (i) Overlap of (j) with PNA.
To ascertain if cones are nutritionally deprived, autophagy within cones was assessed. Two types of autophagy are inducible by various degrees of nutrient deprivation: macroautophagy and chaperone mediated autophagy (CMA)26–29. Macroautophagy is non-selective, targets proteins or entire organelles, and is marked by de novo formation of membranes that form intermediate vesicles (autophagosomes) that fuse with the lysosomes. The machinery required for macroautophagy has been shown to be present in PRs30. In contrast, CMA is selective and targets individual proteins for transport to the lysosomes. The presence of macroautophagy was assessed by infection with a viral vector encoding a fusion protein of green fluorescent protein (GFP) and light chain 3 (LC3), an autophagosomal membrane marker 31–33. No difference was observed in GFP distribution in cones of wild type and mutant mice, suggesting that formation of autophagosomes was absent during cone death (see Supplementary Fig. 7a–f online). Additionally, high levels of phosphorylated ribosomal protein S6 were found in all, or most, cones (see Supplementary Fig. 7g–h online) reflecting an increased activity of ribosomal S6 kinase 1 (S6K1), an inhibitor of macroautophagy28. Consistent with these findings is the fact that macroautophagy reflects an acute short-term response to nutrient deprivation or cellular stress conditions26,27. Prolonged non-selective degradation of newly synthesized proteins to overcome the stress condition would not be favorable to cells and would likely result in the relatively rapid death of most cones, rather than the slow death seen in RP.
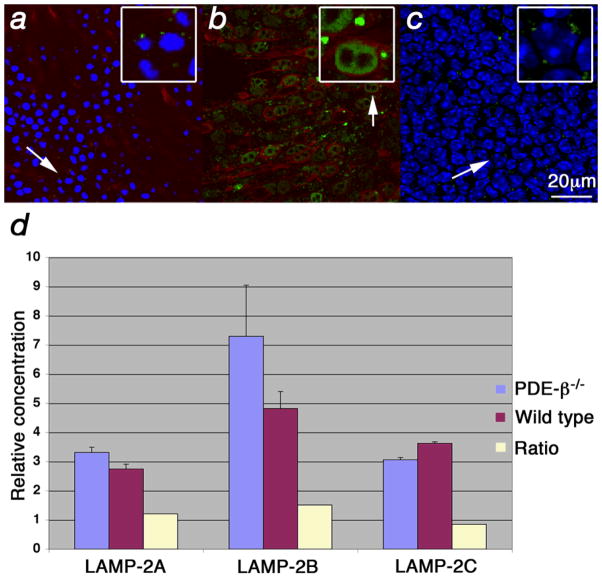
CMA is normally activated over extended periods of starvation and results in increased levels of lysosomal-associated membrane protein (LAMP) type 2A at the lysosomal membrane26,27,34. Both starvation and oxidative stress can induce CMA26. Starvation increases LAMP-2A by preventing its degradation while oxidative stress results in de novo synthesis of LAMP-2A35. A LAMP-2 antibody that recognizes the proteins resulting from all 3 splice isoforms36 (A, B, C) showed high levels of LAMP-2 at the lysosomal membrane in all 4 mutants during cone degeneration (Fig. 7a–c; data only shown for PDE-β−/−). The high levels were specific to cones and were not seen in cells of the inner nuclear layer (Fig. 7b, c), which might reflect the possibility that cones are the only starving cells in the RP retina. qRT-PCR for the three splice isoforms showed only a minor increase in mRNA levels of LAMP-2A (1.2x) and a decrease in LAMP-2C (Fig. 7d) suggesting that the increase seen in protein at the membrane is mainly due to nutritional deprivation and only to a lesser extent to oxidative stress9,10,35. Taken together, the data suggest that nutritional imbalance in cones leads to the activation of CMA, a process that is consistent with prolonged starvation.
Figure 7.
Increased levels of LAMP-2 at the lysosomal membrane. (a–c) Immunofluorescence on retinal flat mounts where LAMP-2 is shown in green, red/green opsin in red and blue signal shows nuclear DAPI stain. Insets in upper right corner (with box) show enlarged cells (arrow). (a) Wild type retinae at PW5 showing lysosome (small green dots) with normal LAMP-2 distribution. Weak red/green opsin signal is detected at the level of the PR nuclei since in wild type it is mainly found in the OSs. (b, c) PDE-β mutant at PW5. (b) Enlarged lysosomes (dots) due to accumulation of LAMP-2 at the lysosomal membrane are seen specifically in cones. (c) Confocal section of same field as in (b) taken at the level of the inner nuclear layer showing levels of LAMP-2 similar to those in wild type (a). (d) qRT-PCR for the 3 different LAMP-2 splice forms showing the relative concentration and the ratios between the PDE-β mutant and wild type.
Stimulation of the insulin receptor pathway prolongs cone survival
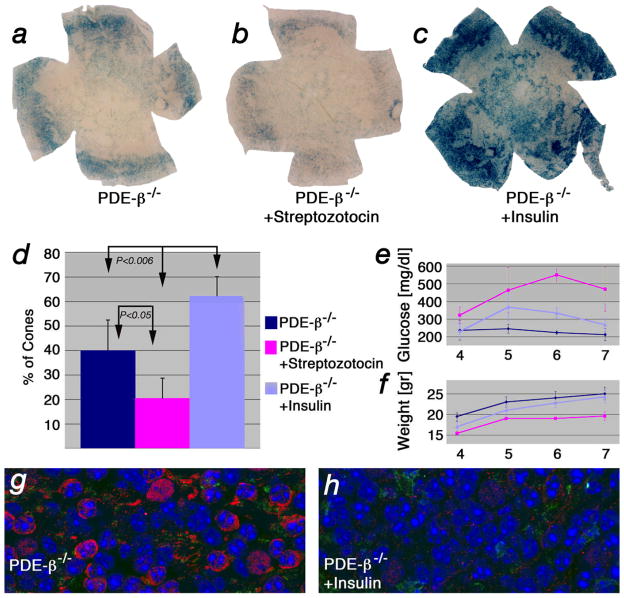
The data on mTOR, HIF-1α, GLUT1 and the induction of CMA suggested a shortage of glucose in cones resulting in starvation and suggested that the insulin/mTOR pathway plays an important role during cone death. To determine if the insulin/mTOR pathway can influence cone survival, we stimulated the pathway by systemic treatment of PDE-β−/− mice with insulin. The PDE-β mutant was chosen over the other three mutants due to its faster cone death kinetics, allowing for a better read-out of cone survival. Mice were treated with daily intraperitoneal injections of insulin over a 4 week period, starting at the onset of cone death. To reduce insulin, a single injection of streptozotocin, a drug that kills the insulin-producing beta cells of the pancreas, also was examined. Systemic administration of insulin results in a desensitized insulin receptor due to a feedback loop in the pathway, which causes an increase in blood glucose levels. Injection of streptozotocin, which also results in increased blood glucose levels, served as a control for the effect of elevated blood glucose, and also provided animals with reduced levels of insulin. PDE-β−/− mice injected with insulin showed improved cone survival compared to uninjected control mice. PDE-β−/− mice injected with Streptozotocin showed a decrease in cone survival (Fig. 8a–d). Improved cone survival was therefore due to insulin and not to the increased blood glucose levels (Fig. 8e). Additionally, cones in mutant mice treated with insulin did not show the upregulation of HIF-1α seen normally in cones during degeneration, consistent with the notion that cones were responding to insulin directly (Fig 8g, h).
Figure 8.
Insulin levels affect cone survival. (a–c) Retinal flat mounts of PDE-β mutants at PW7 stained for lacZ49,48 to detect cones (see Material & Methods and Supplementary Fig. 8 online). (a) Example of untreated control. (b) Example of mouse injected with streptozotocin. (c) Example of mouse injected daily with insulin. (d) Quantification of cone survival after 4 weeks of treatment. Data represents an average of at least 8 retinae and indicates on the y-axis percentage of cone surface area versus surface area of entire retina (see Supplementary Fig. 9 & 10 online). (e) Measurements of blood glucose levels and body weight (f) performed weekly over the time span of the experiment. (g, h) Immunofluorescent staining on retinal flat mounts for HIF-1α (red signal) and PNA (green signal) in untreated control PDE-β−/− (g) and PDE-β−/− mice treated for 4 weeks with insulin (h). Blue shows nuclear DAPI.
Discussion
Here we report that cones show signs of nutritional imbalance during the period of cone degeneration in RP mice. The microarray analysis suggested that there are changes in cellular metabolism involving the insulin/mTOR pathway at the onset of cone death. We showed that inhibition of mTOR in wild type mice resulted in the same pattern of loss of red/green opsin as seen during degeneration. In accord with changes in p*-mTOR, and its sensitivity to glucose, we saw an upregulation of HIF-1α and GLUT1, suggesting that glucose uptake, and/or the intracellular levels of glucose, may be compromised in cones of RP mice. Additionally, systemic administration of insulin prolonged cone survival, whereas depletion of endogenous insulin had 7the reverse effect. The systemic treatment with insulin prevented the upregulation of HIF-1α in cones seen normally during cone degeneration, suggesting that insulin was directly acting on cones. Interestingly, a prolonged treatment of insulin during a time span of 7 weeks instead of 4 weeks did not show any significant improvement of cone survival (see Supplementary Fig. 10 online). This may reflect the feedback loop of the pathway in which S6K1 acts directly onto the insulin-receptor substrate (IRS). While the cross talk in cones between the insulin receptor, mTOR, HIF-1α, S6K1 and IRS remains to be investigated, the results strengthen the notion that nutrient availability in cones may be altered during the period of cone degeneration and that the insulin/mTOR pathway plays a crucial role. A recent report showed that constitutive expression of proinsulin in the rd10 mouse model of RP delays photoreceptor death, both of rods and cones37. However, proinsulin seems not to act through the insulin receptor as mice treated with proinsulin did not develop hyperglycemia. Proinsulin blocks developmental cell death and thus may interfere with the apoptotic pathway in the postnatal retina. Although downstream effectors, such as PI3K/Akt, are regulated signaling by insulin and proinsulin, the relationship between the results of the studies reported here on insulin and the proinsulin effects remains to be determined.
Macroautophagy, which is controlled by mTOR through its downstream target S6K1, was not detected during cone degeneration, while CMA appeared to be activated. Increased LAMP-2A levels at the lysosomal membrane are suggestive of an activation of CMA. In addition, the observations concerning mTOR, HIF-1α, and GLUT1 are consistent with starvation and CMA. However, additional experiments are needed to prove that starvation and CMA are indeed occurring, and further, to prove that they are an important contributor to cone death. The lack of detectable macroautophagy does not rule out the possibility that macroautophagy might occur for a short period of time (e.g. 24 hours) prior to the activation of CMA. The data only show that macroautophagy is not the main form of autophagy over an extended period of time, which is consistent with the notion that macroautophagy is a short-term response. The prolonged inhibition of macroautophagy is likely due to increased S6K1 activity as seen by increased p*-S6 levels. S6K1 is positively regulated by mTOR and AMP-activated protein kinase (AMPK)28, which reads out cellular ATP levels. Therefore, while mTOR may report metabolic problems with respect to glucose uptake, and reduce energy consuming processes and improve glycolysis through HIF-1α, AMPK may report normal cellular ATP levels and inhibit macroautophagy. This may represent a specific response to the energy requirements of cones. Most of the glucose taken up by PRs never enters the Krebs cycle38. Thus the shortage of glucose may not cause a shortage of ATP. Lactate, provided by Muller glia, can generate ATP via the Krebs cycle39. However, glucose is needed to generate NADPH in the pentose phosphate cycle, and NADPH is required for synthesis of phospholipids, the building blocks of cell membranes. PRs constantly shed their membranes at the tip of the OSs. Since reduced levels of glucose would result in reduction of membrane synthesis, the rate of OS phagocytosis by the RPE may be higher than the rate of membrane synthesis by cones. Consistent with this, OS shortening preceded cell death in these 4 models, as is also observed in human cases of RP17. Additionally, changes that affect lipid metabolism were also seen by the microarray analysis.
Why does the loss of rods result in cone death in RP? The previous hypotheses mentioned in the Introduction have in common that they can’t fully explain the pathology found in humans. The rod and cone death kinetics shown here clearly argue against a toxin produced by dying rods as a cause for cone death since the onset of cone death always occurred after the major rod death period. If a rod toxin caused cone death, then the onset of cone death should have either coincided with the onset of rod death or should have started shortly thereafter, since this would be the period of peak toxin production. Interestingly, the lack of a trophic factor produced by healthy rods and required for cone survival would agree with the onset of cone death seen in all four models as one would expect the onset of cone death during the end stages of rod death. However, the progression of cone death and the end phase of rod death make this unlikely hypothesis as the sole reason for cone death. In the two PDE mutants and in the Rho-KO mutant, cones were dying for many weeks after the end phase of rod death, suggesting that they could survive quite awhile in the absence of rods. In addition, in the P23H model, rods died so slowly during the end phase of rod death, that during the entire period of cone death, rods were still present. The hypothesis that a lack of a rod trophic factor being the main cause for cone death seems unlikely given these discrepancies. However, the present investigation cannot rule out the lack of rod trophic factor given that the 4 mouse models used are in 4 different genetic backgrounds. Background differences could result in differences, such as different levels of such a factor, which might account for the observed differences in the progression of cone death.
How could our observations of nutritionally deprived cones explain the dependence of cones on rods? The OS-RPE interactions are vital since the RPE shuttles nutrition and oxygen from the choroidal vascaulture to PRs. Roughly 95% of all PRs in mouse and human are rods and approximately 20–30 OSs contact one RPE cell40,41. Thus, only 1–2 of those RPE-OS contacts are via cones. During the collapse of the ONL, the remaining cone:RPE interactions are likely perturbed. If these interactions drop below a threshold required for the proper flow of nutrients, the loss of rods might result in a reduced flow of nutrients to cones. In all 4 mouse models, the onset of cone death occurred when the ONL reached one row of cells. This cell density could therefore represent the critical threshold. Then, while the remaining rods die due to a mutation in a rod-specific gene, cone death may begin due to nutrient deprivation. In accord with this notion, cone death progressed more slowly when the remaining rods died slowly. This mechanism would also explain why the loss of cones does not lead to rod death42,43. Since in humans and mouse, cones are less than 5% of all PRs, the critical threshold that perturbs OS-RPE interactions would not be reached. Further support for this idea is provided by studies in zebrafish where the overall ratio of rods to cones is reversed (1:8). Additionally, the distribution of rods and cones in zebrafish is uneven such that certain regions are cone-rich whereas other regions are rod-rich. A recently isolated mutation in a cone-specific gene resulted in rod death, but only in regions of high cone density44, leading Stearns and co-workers to conclude that cell density is the crucial determinant. We propose that once a critical threshold of cell density is breached, improper OS-RPE interactions result in reduced flow of nutrients (e.g. glucose). This results in reduced OS membrane synthesis, which in turn further contributes to a reduced uptake of nutrients from the RPE. Ultimately, prolonged starvation, as suggested by the activation of CMA, leads to cell death. Since starvation can occur slowly over extended periods of time, and because the rate may fluctuate due to fluctuations in nutrient uptake, the slow and irregular demise of cones observed in humans may be expected. Therefore, this study not only provides a new mechanism of cone death in RP that should direct future therapeutic approaches, but also consolidates the data from the literature with respect to the death kinetics of rods and cones seen in mice and patients with different RP mutations.
Material and Methods
Animals
Wild type (wt) mice (C57Bl/6N) and PDE-β−/− mice (normally referred as rd1 or FVB/N) were purchased from Taconic Farms. The PDE-β−/− mice have a mutation in the β-subunit of cGMP phosphodiesterase12 (PDE). The PDE-γ knock-out (PDE-γ-KO) lacks the γ-subunit of PDE and was provided by Steve Tsang13 (UCLA). The rhodopsin knock-out (Rho-KO) lacks the rod-specific opsin gene and was provided by Janis Lem13,14 (Tufts Medical School). The P23H mouse has a proline-23 to histidine mutation in the rhodopsin gene and was provided by Muna Naash15 (University of Oklahoma). As this mouse carries a transgene the strain was always crossed back to C57Bl/6N to ensure that none of the progeny would carry two alleles of the transgene. The transgene is specifically expressed in rods45–47 and carries 3 mutations in the rhodopsin gene (Val-20 to Gly, Pro-23 to His, Pro-27 to Leu). In this study it is referred as the P23H mutant. The cone-lacZ strain was provided by Jeremy Nathans48 (Johns Hopkins School of Medicine). All procedures involving animals were in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Affymetrix array analysis
RNA was extracted as described previously49. Three to 4 retinae were used per extraction. A minimum of two arrays were analyzed per time point. The statistical significance of each gene expression profile was determined by a Jonckheere-Terpstra test of the hypothesized cone-death patterned alternative, using exact p-values calculated by the Harding algorithm50.
qRT-PCR was performed as described previously with the same primers and conditions for Opn1sw and gapdh49. The following primers and conditions were used for the three LAMP-2 splice forms: LAMP-2 forw. ctgaaggaagtgaatgtctacatg; LAMP-2A rev. gctcatatccagtatgatggc; LAMP-2B rev. cagagtctgatatccagcatag; LAMP-2C rev. gacagactgataaccagtacg. Conditions for all three PCRs: 95° for 3 sec, 52° for 15 sec, 72° for 25 sec. The data in figure 2a and 7d represent an average of 3 measurements corrected for gapdh.
Retinal explant cultures
The retina was dissected free from other ocular tissues in DMEM, and then incubated in conditions according to the chart in Supplementary Fig. 5a online. Regular DMEM was at 4.5g/L glucose, low glucose was at 1g/L, leucine was added at 200μM and FCS at 10%. Incubation was performed for 4h and the retinae were fixed and processed for antibody staining as described below.
TUNEL, X-gal histochemistry and In Situ Hybridizations were performed as described previously49. For the double labeling of cones (see Supplementary Fig. 8 online), retinae were first fixed in 2% PFA for 15min. then processed for the X-GAL reaction and then post fixed in 4% PFA for 15min. A biotin-PNA was used in an antibody staining procedure (see below) and detected with Streptavidin-POD (1:500, Roche) by a DAB stain (Sigma) according to the manufacture’s instructions. The following ESTs were used for the red/green opsin and blue opsin probes respectively: red/green opsin (BE950633); blue opsin (BI202577). Probe for rhodopsin was generated by sub-cloning the coding sequence of the gene into pGEM-T Easy (Promega). The following primers were used for amplification of the coding sequence: forw. agccatgaacggcacagaggg; rev. cttaggctggagccacctggct. The antisense RNA was generated with T7 RNA polymerase.
Viral injections were performed as described previously33. Mice were injected at embryonic day 10 and harvested at postnatal week 10. The fusion protein was generated with a NotI site at the 5′ end followed by GFP, then LC3, and then an XhoI site at the 3′ end and cloned into pQCXIX (Clonetech: cat.# 631515). The following primers were used for the fusion protein: 5′NotI-GFP atgcgggccgccaccatggtgagcaagggcgaggagc, 3′GFP-LC3 aggtcttctcggacggcatcttgtacagctcgtccatgc-cgag, 5′LC3 atgccgtccgagaagaccttcaagc, 3′LC3-XhoI atctcgagttacacagccattgctgtcccgaatg.
Rapamycin, Streptozotocin and Insulin treatments were performed as follows. Rapamycin was diluted to 10mg/ml in ethanol. The stock was diluted to 0.015mg/ml in drinking water over a period of 2 weeks. A single intraperitoneal injection of 150μl (12mg/ml in 0.1M citric acid, ph4.5) of Streptozotocin was injected at postnatal day (P) 21. Insulin was injected intraperitoneally daily starting at P21. The concentration was increased weekly such that the first week, 10U/kg body weight, the second 15U/kg, the third 20U/kg and fourth 30U/kg body weight, were injected. In the treatment that lasted 7 weeks 30U/kg body weight were injected for the remaining 3 weeks. Blood glucose levels were measured by collecting a drop of blood from the tail directly onto a test strip from TrueTrack smart system (CVS pharmacy). Eye bleeds were avoided due to the fact that cell survival in the retina was being assayed.
Quantification of cone survival was performed as follows. The colors of the bright light image were inverted and processed with Imaris software (Bitplane Inc) to calculate the percentage of blue surface area versus the total retinal surface area (see also Supplementary Fig. 9 online). A minimum of 8 retinae per treatment, and for the control, were analyzed. P-values were calculated by the student’s t-test. The cone lacZ transgene was chosen over PNA as a cone marker since the transgene labels cones more persistently, since, due to the shortening of the cone OS, PNA was found to stain less reliably than lacZ (see Supplementary Fig. 8 online).
Whole mount and section antibody staining were performed as previously33 described with the following modifications. Antibody staining for LAMP-2: Triton was replaced with 0.01% Saponin. Antibody staining for p*-mTOR and p*-S6: PBS was replaced by TBS in every step of the procedure. Primary antibody dilutions: mouse α-rhodopsin Rho4D2 1:20051; goat α-β-Galactosidase (Serotec) 1:400; rabbit α-blue opsin (Jeremy Nathans) 1:1000; rabbit α-Gnat1 1:200 (Santa Cruz); rabbit α-Cleaved Caspase-3 (Cell Signaling) 1:100; rabbit α-Cleaved Lamin A (Cell Signaling) 1:100; rabbit α-GLUT-1 (Alpha Diagnostics) 1:100; rabbit α-p*-mTOR (Ser2448) (Cell Signaling) 1:300; rabbit α-p*-S6 (Ser235/236) (Cell Signaling) 1:100; rabbit α-HIF-1α (R&D Systems) 1:300; rat α-LAMP-2 (clone: GL2A7, from DSHB) 1:200. Time points analyzed for the rod and cone death kinetics (P: postnatal day; PW: postnatal week): PDE-β−/−: P10–P20 daily, PW3–10 weekly, PW 12, PW15, PW18, PW45; PDE-γ-KO: P10–P20 daily, PW3–PW10 weekly, PW15, PW25, PW45; Rho-KO: PW4–PW8 weekly, PW10, PW11, PW17, PW20, PW25, PW27, PW31, PW34, PW37, PW45, PW55, PW80; P23H: PW5, PW10, PW16, PW25, PW30, PW35, PW40, PW65, PW70, PW75, PW80, PW85.
Supplementary Material
Acknowledgments
We thank Jeremy Nathans for the cone-lacZ line, and the blue cone opsin antibody. We are grateful to Steve Tsang, Muna Naash and Janis Lem for the PDE-γ-KO, the P23H and the Rho-KO respectively. The LAMP-2 antibody developed by Bruce Granger was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the HICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. We thank Jeff Trimarchi, Rahul Kanadia, Norbert Perrimon, Jonathan Zirin, Mel Feany and Clifford Tabin for critical reading of the manuscript. This work was supported by the NIH (RO1 EY014466), the Macular Research Foundation, The Foundation for Retinal Research, HHMI, Merck, and by an EMBO fellowship to CP.
References
- 1.Madreperla SA, Palmer RW, Massof RW, Finkelstein D. Visual acuity loss in retinitis pigmentosa. Relationship to visual field loss. Arch Ophthalmol. 1990;108:358–61. doi: 10.1001/archopht.1990.01070050056030. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg RH. Survival factors in retinal degenerations. Curr Opin Neurobiol. 1994;4:515–24. doi: 10.1016/0959-4388(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 3.Mohand-Said S, et al. Normal retina releases a diffusible factor stimulating cone survival in the retinal degeneration mouse. Proc Natl Acad Sci U S A. 1998;95:8357–62. doi: 10.1073/pnas.95.14.8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Streichert LC, Birnbach CD, Reh TA. A diffusible factor from normal retinal cells promotes rod photoreceptor survival in an in vitro model of retinitis pigmentosa. J Neurobiol. 1999;39:475–90. [PubMed] [Google Scholar]
- 5.Mohand-Said S, et al. Photoreceptor transplants increase host cone survival in the retinal degeneration (rd) mouse. Ophthalmic Res. 1997;29:290–7. doi: 10.1159/000268027. [DOI] [PubMed] [Google Scholar]
- 6.Mohand-Said S, Hicks D, Dreyfus H, Sahel JA. Selective transplantation of rods delays cone loss in a retinitis pigmentosa model. Arch Ophthalmol. 2000;118:807–11. doi: 10.1001/archopht.118.6.807. [DOI] [PubMed] [Google Scholar]
- 7.Leveillard T, et al. Identification and characterization of rod-derived cone viability factor. Nat Genet. 2004;36:755–9. doi: 10.1038/ng1386. [DOI] [PubMed] [Google Scholar]
- 8.Gupta N, Brown KE, Milam AH. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res. 2003;76:463–71. doi: 10.1016/s0014-4835(02)00332-9. [DOI] [PubMed] [Google Scholar]
- 9.Komeima K, Rogers BS, Lu L, Campochiaro PA. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2006;103:11300–5. doi: 10.1073/pnas.0604056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komeima K, Rogers BS, Campochiaro PA. Antioxidants slow photoreceptor cell death in mouse models of retinitis pigmentosa. J Cell Physiol. 2007 doi: 10.1002/jcp.21152. [DOI] [PubMed] [Google Scholar]
- 11.Yu DY, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005;80:745–51. doi: 10.1016/j.exer.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Bowes C, et al. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–80. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 13.Tsang SH, et al. Retinal degeneration in mice lacking the gamma subunit of the rod cGMP phosphodiesterase. Science. 1996;272:1026–9. doi: 10.1126/science.272.5264.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lem J, et al. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999;96:736–41. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naash MI, Hollyfield JG, al-Ubaidi MR, Baehr W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc Natl Acad Sci U S A. 1993;90:5499–503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Applebury ML, et al. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–23. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 17.John SK, Smith JE, Aguirre GD, Milam AH. Loss of cone molecular markers in rhodopsin-mutant human retinas with retinitis pigmentosa. Mol Vis. 2000;6:204–15. [PubMed] [Google Scholar]
- 18.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–83. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 19.Dekanty A, Lavista-Llanos S, Irisarri M, Oldham S, Wappner P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima. J Cell Sci. 2005;118:5431–41. doi: 10.1242/jcs.02648. [DOI] [PubMed] [Google Scholar]
- 20.Hudson CC, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–14. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–81. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 22.Zhong H, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–5. [PubMed] [Google Scholar]
- 23.Thomas GV, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–7. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 24.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083–9. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- 26.Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–34. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–44. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12 (Suppl 2):1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 29.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–9. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 30.Kunchithapautham K, Rohrer B. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy. 2007;3:433–41. doi: 10.4161/auto.4294. [DOI] [PubMed] [Google Scholar]
- 31.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punzo C, Cepko CL. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev Dyn. 2008;237:1034–42. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–83. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 35.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(Pt 24):4441–50. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 37.Corrochano S, et al. Attenuation of vision loss and delay in apoptosis of photoreceptors induced by proinsulin in a mouse model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2008;49:4188–94. doi: 10.1167/iovs.08-2182. [DOI] [PubMed] [Google Scholar]
- 38.Poitry-Yamate CL, Poitry S, Tsacopoulos M. Lactate released by Muller glial cells is metabolized by photoreceptors from mammalian retina. J Neurosci. 1995;15:5179–91. doi: 10.1523/JNEUROSCI.15-07-05179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsacopoulos M, Poitry-Yamate CL, MacLeish PR, Poitry S. Trafficking of molecules and metabolic signals in the retina. Prog Retin Eye Res. 1998;17:429–42. doi: 10.1016/s1350-9462(98)00010-x. [DOI] [PubMed] [Google Scholar]
- 40.Snodderly DM, Sandstrom MM, Leung IY, Zucker CL, Neuringer M. Retinal pigment epithelial cell distribution in central retina of rhesus monkeys. Invest Ophthalmol Vis Sci. 2002;43:2815–8. [PubMed] [Google Scholar]
- 41.Young RW. The renewal of rod and cone outer segments in the rhesus monkey. Journal of Cell Biology. 1971;49:303–318. doi: 10.1083/jcb.49.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biel M, et al. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci U S A. 1999;96:7553–7. doi: 10.1073/pnas.96.13.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang RB, et al. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. J Neurosci. 1999;19:5889–97. doi: 10.1523/JNEUROSCI.19-14-05889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stearns G, Evangelista M, Fadool JM, Brockerhoff SE. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J Neurosci. 2007;27:13866–74. doi: 10.1523/JNEUROSCI.3136-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouras P, Kjeldbye H, Zack DJ. Reporter gene expression in cones in transgenic mice carrying bovine rhodopsin promoter/lacZ transgenes. Vis Neurosci. 1994;11:1227–31. doi: 10.1017/s0952523800007021. [DOI] [PubMed] [Google Scholar]
- 46.Woodford BJ, Chen J, Simon MI. Expression of rhodopsin promoter transgene product in both rods and cones. Exp Eye Res. 1994;58:631–5. doi: 10.1006/exer.1994.1058. [DOI] [PubMed] [Google Scholar]
- 47.al-Ubaidi MR, et al. Mouse opsin. Gene structure and molecular basis of multiple transcripts. J Biol Chem. 1990;265:20563–9. [PubMed] [Google Scholar]
- 48.Wang Y, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–40. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 49.Punzo C, Cepko C. Cellular responses to photoreceptor death in the rd1 mouse model of retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48:849–57. doi: 10.1167/iovs.05-1555. [DOI] [PubMed] [Google Scholar]
- 50.Harding EF. An Efficient, Minimal-Storage Procedure for Calculating the Mann-Whitney U, Generalized U and Similar Distributions. Applied Statistics. 1984;33:1–6. [Google Scholar]
- 51.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–60. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.