Abstract
Dynamic rearrangement of actin filament networks is critical for cell motility, phagocytosis and endocytosis. Coronins facilitate these processes, in part, by their ability to bind F-actin (filamentous actin). We previously identified a conserved surface-exposed arginine (Arg30) in the β-propeller of Coronin 1B required for F-actin binding in vitro and in vivo. However, whether this finding translates to other coronins has not been well defined. Using quantitative actin-binding assays, we show that mutating the equivalent residue abolishes F-actin binding in Coronin 1A, but not Coronin 1C. By mutagenesis and biochemical competition, we have identified a second actin-binding site in the unique region of Coronin 1C. Interestingly, leading-edge localization of Coronin 1C in fibroblasts requires the conserved site in the β-propeller, but not the site in the unique region. Furthermore, in contrast with Coronin 1A and Coronin 1B, Coronin 1C displays highly co-operative binding to actin filaments. In the present study, we highlight a novel mode of coronin regulation, which has implications for how coronins orchestrate cytoskeletal dynamics.
Keywords: actin, co-operative binding, coronin, lamellipodia
Abbreviations: Arp2/3, actin-related protein 2/3; Coro1, Type I coronin; F-actin, filamentous actin; GFP, green fluorescent protein; IA32, Ink/Arf-null; NS, non-targeting; shRNA, short hairpin RNA
INTRODUCTION
Coronins, a family of highly conserved actin-binding proteins originally identified in Dictyostelium [1], have emerged as key regulators of actin dynamics via effects on assembly by the Arp2/3 (actin-related protein 2/3) complex and disassembly by cofilin [2]. Mammalian genomes contain at least six coronin genes organized into three classifications [3]. Most functional studies have focused on Coro1s (Type I coronins) (Coro1A, Coro1B and Coro1C). Coro1B and Coro1C are ubiquitously expressed isoforms, whereas Coro1A has a primarily haematopoietic expression pattern. Genetic deletion of Coro1A in mice results in severe defects in T-lymphocyte trafficking and chemotaxis [4]. Coro1B localizes to the leading edge of migrating fibroblasts [5] and its depletion alters lamellipodial architecture and whole-cell motility [6]. Like Coro1B, Coro1C localizes to lamellipodia and interacts with the Arp2/3 complex [7–9]; however, Coro1C may have additional roles in membrane trafficking [10] and tumour metastasis [11].
The crystal structure of Coro1A revealed that the most prominent structural characteristic is a seven-bladed WD40 repeat-containing β-propeller [12]. The β-propeller, a scaffold for protein–protein interactions [13], is flanked by N- and C-terminal extensions that contribute to its stability, followed by a highly variable unique region and a coiled-coil domain. Although the structure of the unique region has not been resolved, it has been shown to participate in coronin function. Yeast coronin Crn1p was initially identified as a microtubule-binding protein via the unique region [14]. In addition, a CA-like sequence, conserved in WASP (Wiskott–Aldrich syndrome protein)/SCAR (suppressor of cAMP receptor) proteins, is present in the unique region of yeast coronin and regulates the activity of the Arp2/3 complex [15]. The primary role of the coiled-coil domain is in coronin oligomerization [12,16,17].
Coronins were first identified by their ability to bind to F-actin (filamentous actin), but their actin-binding function is incompletely understood. Early studies attempted to address this by using truncation mutants [9,18–21]. The resolution of the crystal structure of Coro1A [12] suggested that the resulting proteins from truncations or deletions of portions of the β-propeller are likely to adopt unstable conformations. We provided the first direct demonstration that F-actin binding is essential for coronin function in vitro and in vivo by identifying a conserved arginine residue (Arg30) on the surface of the β-propeller in Coro1B, which when mutated abolished F-actin binding [22]. Three-dimensional reconstructions further validated the requirement of the conserved site in the β-propeller and have also identified potential non-contiguous surfaces for the Coro1A–actin interaction [23]. A recent report on yeast coronin showed similar results that mutations in residues in a ridge extending across the side of the β-propeller disrupted F-actin binding [24]. Yeast coronin may contain a second lower-affinity actin-binding site in the coiled-coil domain that inhibits cofilin activity, but this site has not been mapped at high resolution [25]. However, another study has found no detectable actin binding for a fragment containing the coiled-coil domain [15].
In the present paper, we sought to provide further characterization of the actin-binding function of the mammalian Coro1s. In our analysis, we not only confirm our previous observations concerning the role of Arg30 in actin binding, but also reveal the unexpected findings that one member, Coro1C, contains a second actin-binding site and binds co-operatively to actin filaments.
EXPERIMENTAL
Cell culture and viral transduction
HEK (human embryonic kidney)-293FT cells (Invitrogen) were used to generate recombinant protein. IA32 (Ink/Arf-null) mouse embryonic fibroblasts were cultured as described previously [26]. Lentivirus production and infection were performed as described previously [27].
Anti-Coro1C antibody production
An antibody against Coro1C was generated using an immunogen containing the human C-terminus (unique region and coiled-coil) by the University of North Carolina Monoclonal Antibody Core, collected and concentrated. Anti-Coro1C antibody (clone 22.51.1.10) was used for immunoblotting experiments.
Molecular cloning
Coro1C mutations were generated by overlap extension PCR and inserted into the multiple cloning site of the pTT5SH8Q2 vector that contains the StrepTagII and His8 affinity tags.
The shRNA (short hairpin RNA) targeting mouse Coro1C was designed according to the instructions found at http://en.dogeno.us/2shrna/ and cloned into the HpaI/XhoI sites of pLL5.0(GFP) using methods described previously [6]. The 19-nt target sequence for mouse Coro1C is GACGGGACGAATTGACAAA. Wild-type and mutant versions of human Coro1C, which were not targeted by mouse Coro1C shRNA, were cloned into the multi-cloning site of pLL5.0(GFP). The NS (non-targeting) shRNA was described previously [6].
Expression and purification of recombinant coronin protein
Recombinant coronin protein was expressed and purified from a mammalian expression system as described previously [6].
Actin cosedimentation
Human platelet actin (Cytoskeleton) was depolymerized in general actin buffer [5 mM Tris/HCl (pH 8.0) and 0.2 mM CaCl2] for 1 h at room temperature (23°C) and pre-cleared by ultracentrifugation at 23 psi (100000 g for 15 min at 25°C with an A-100/18 rotor; 1 psi=6.9 kPa) in a Beckman Airfuge. Actin was polymerized by adding 1/10 vol. of polymerization buffer (500 mM KCl, 20 mM MgCl2 and 10 mM ATP) and incubating at room temperature for 1 h.
Recombinant coronin proteins were incubated with actin at room temperature for 1 h and sedimented at 100000 g for 30 min. Pellets and supernatants were run on SDS/PAGE gels and analysed by Coomassie Blue staining or immunoblotting. Coomassie Blue-stained gels were scanned using an Odyssey Infrared Imager (LI-COR Biosciences) and images were despeckled using ImageJ (http://rsbweb.nih.gov/ij/).
To determine the binding affinity of coronin to F-actin, we used an established supernatant depletion method [22,28]. Coronin bound to F-actin was determined indirectly by immunoblotting the supernatant and quantified by densitometry. Binding curves were fitted to the experimental data with Graphpad Prism software.
Immunofluorescence and image analysis
For immunofluorescent cell staining, cells were fixed, stained and mounted as described previously [29]. Cells were stained with an anti-cortactin antibody (1:400 dilution; Millipore) and Alexa Fluor® 647-phalloidin (1:500 dilution; Molecular Probes). Cells were imaged using an Olympus FV1000 confocal microscope equipped with a 40× (1.3 numerical aperture) oil-immersion objective. A maximum intensity projection was generated from a z-stack (7 slices, 1.28 μm/slice). To plot the distribution of a protein around the cell edge, a previously published contour-based ImageJ macro was used [6]. Briefly, the images from GFP (green fluorescent protein) fluorescence, cortactin and phalloidin staining were merged into a single image, which was converted to a binary mask by thresholding the cell based on the actin image. A hand-drawn polygon mask was used to exclude those regions lacking peripheral cortactin staining. Dilation or erosion operations were iteratively performed to create a series of contour lines both inside and outside the perimeter of the cell (http://www.unc.edu/~cail/code/edge2.txt). Pixel intensities from −6 μm to 0 μm were extracted from each channel and plotted as a function of distance from the edge of the cell. Extracted pixel intensities were exported to Excel for analysis and Prism (GraphPad) for graphing.
RESULTS
Mutation in a conserved arginine residue blocks F-actin binding in Coro1A and Coro1B, but not Coro1C
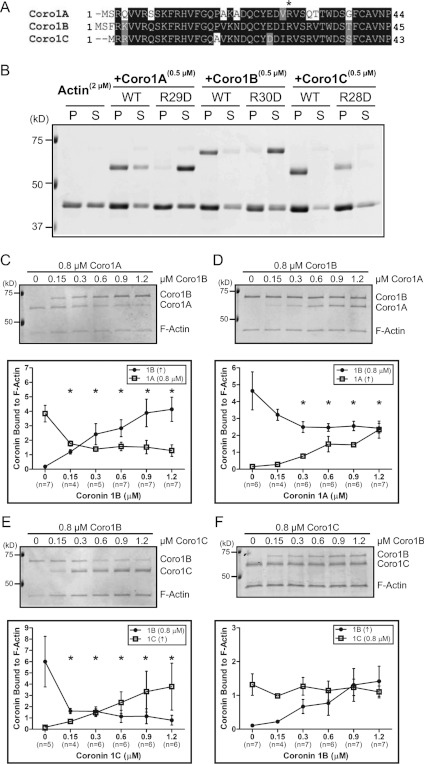
We previously identified a conserved arginine residue (Arg30) within the β-propeller domain of Coro1B, which when mutated to aspartate (R30D) abolished F-actin binding [22]. This residue is present in other Coro1s (Figure 1A) and subsequent studies have confirmed the requirement for the equivalent residue in Coro1A [30,31], but this has not been directly tested for Coro1C. In order to address this, we utilized a mammalian expression system to produce recombinant wild-type and mutant coronin proteins with >99% purity (Supplementary Figure S1 at http://www.BiochemJ.org/bj/444/bj4440089add.htm). As expected, cosedimentation assays demonstrated that all Coro1s sediment efficiently with F-actin (Figure 1B). Conversely, the arginine-to-aspartic acid mutations in Coro1A and Coro1B abolished F-actin binding. To our surprise, the R28D mutation in Coro1C failed to block F-actin binding. We verified further that this effect was not due to Coro1C sedimenting independently of its ability to bind F-actin (Supplementary Figure S2 at http://www.BiochemJ.org/bj/444/bj4440089add.htm).
Figure 1. Mutation of a conserved arginine residue abolishes F-actin binding in Coro1A and Coro1B, but not Coro1C.
(A) Protein sequence alignment of human Coro1 N-terminus. *Conserved arginine residue on the β-propeller shown previously to be involved in F-actin binding for Coro1B. (B) Actin cosedimentation was performed using 2 μM actin alone or with 0.5 μM Coro1A, Coro1B, Coro1C and their respective Arg→Asp mutants (R29D, R30D and R28D). Pellet (P) and supernatant (S) fractions were separated by SDS/PAGE. Representative Coomassie Blue-stained gel is from at least three independent experiments. Coro1A (C), Coro1B (D and E), or Coro1C (F) (0.8 μM) was incubated with 0.3 μM actin followed by incubation with increasing concentrations of the indicated coronin. Actin cosedimentation was performed and pellet fractions were separated by SDS/PAGE. Representative Coomassie Blue-stained gels are from at least four independent experiments. Quantification is shown as amount of coronin bound to F-actin, which was normalized to the amount of actin in the pellet. Results are means±S.E.M. from the indicated number of experiments (n). Asterisk indicates statistical significance compared with initial concentration of coronin by one-way ANOVA; *P<0.05. WT, wild-type. In all gels, molecular mass markers are shown in kDa on the left-hand side.
If Coro1s share a common binding site through the β-propeller, we postulated that they should compete for binding to F-actin. The primary sequence of Coro1B is longer than Coro1C and Coro1A by 10 and 23 amino acids respectively; however, Coro1B migrates approximately 10 kDa slower on SDS/PAGE gels. This property allowed us to distinguish between Coro1A and Coro1C from Coro1B and perform biochemical competition experiments. We performed cosedimentation assays under equilibrium conditions to test for competition. Coro1B was able to compete with Coro1A (Figure 1C), and conversely Coro1A was able to compete with Coro1B (Figure 1D). Increasing concentrations of Coro1C also effectively competed with Coro1B (Figure 1E). In contrast, increasing concentrations of Coro1B (within the concentration range tested) were unable to compete with Coro1C for binding to F-actin (Figure 1F). These data indicate that Coro1A and Coro1B bind F-actin through a synonymous site on the β-propeller. However, the failure of the R28D mutation on Coro1C to block actin binding and the inability of Coro1B to effectively compete with Coro1C for actin binding raise the intriguing possibility that Coro1C may harbour an additional actin-binding site.
Coro1C harbours an additional actin-binding site within the unique region
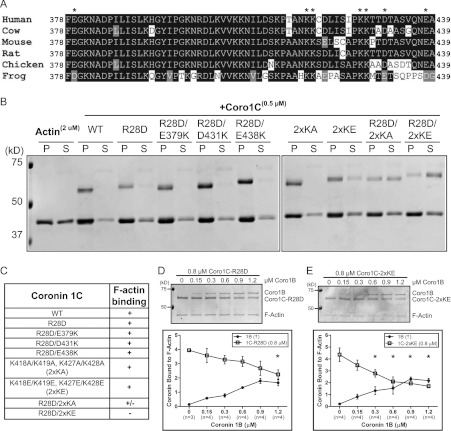
A previous study using truncation mutants of Coro1C suggested that an actin-binding site might reside within the unique region of the protein [9]; however, this site has not been clearly defined. We compared the sequences of the unique regions between the Coro1s and noted a patch of charged residues within the Coro1C unique region that was not present in other Coro1s. These residues were highly conserved amongst all vertebrate Coro1C orthologues (Figure 2A). To test whether the unique region is involved in F-actin binding, we mutated several of these conserved charged residues and performed actin cosedimentation assays using purified (full-length) mutant proteins (Figure 2B). Charge-reversal mutations in acidic residues (E379K, D431K and E438K) in combination with the R28D mutation did not affect Coro1C binding to F-actin. In addition, neither conservative alanine residue substitutions in a positive lysine patch [K418A, K419A/K427A and K428A (2×KA)] nor charge-reversal mutations [K418E, K419E/K427E and K428E (2×KE)] markedly affected F-actin binding. Interestingly, the R28D together with the 2×KA mutation reduced binding. Strikingly, only the 2×KE in conjunction with the R28D mutation abolished Coro1C binding to F-actin (summarized in Figure 2C). To further confirm these results, we repeated biochemical competition experiments using actin cosedimentation by pre-binding Coro1C–R28D (Figure 2D) or Coro1C–2×KE (Figure 2E) to actin filaments and adding increasing concentrations of Coro1B. Mutations at either of these sites allowed Coro1B to compete for binding to F-actin. Moreover, both Coro1C–R28D and Coro1C–2×KE were able to compete with Coro1B (Supplementary Figure S3 at http://www.BiochemJ.org/bj/444/bj4440089add.htm). Together, these data suggest that Coro1C has two sites that function collaboratively to promote binding to F-actin.
Figure 2. Coro1C harbours a second actin-binding site.
(A) Protein sequence alignment of the vertebrate Coro1C unique region. *Charged residues mutated for assays in (B). (B) Actin cosedimentation was performed using 2 μM actin alone or with 0.5 μM wild-type (WT) Coro1C or the following mutants: R28D; R28D/E379K; R28D/D431K; R28D/E438K; K418A, K419A/K427A, K428A (2xKA); K418E, K419E/K427E, K428E (2×KE); R28D/2xKA; or R28D/2xKE. Pellet (P) and supernatant (S) fractions were separated by SDS/PAGE. Representative Coomassie Blue-stained gel is from at least three independent experiments. (C) Table summarizing actin cosedimentation results from (B). Coro1C–R28D (D) or Coro1C–2xKE (E) (0.8 μM) was incubated with 0.3 μM actin followed by incubation with increasing concentrations of Coro1B. Actin cosedimentation was performed and pellet fractions were separated by SDS/PAGE. Representative Coomassie Blue-stained gels are from four independent experiments. Quantification is shown as amount of coronin bound to F-actin, which was normalized to the amount of actin in the pellet. Results are means±S.E.M. from the indicated number of experiments (n). Asterisk indicates statistical significance compared with initial concentration of coronin by one-way ANOVA; *P<0.05. In all gels, molecular mass markers are shown in kDa on the left-hand side.
Coro1C binds F-actin co-operatively and with high affinity
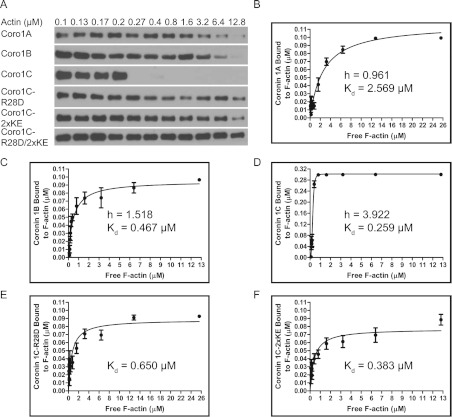
We reasoned that the presence of two actin-binding sites should impart Coro1C with the ability to bind F-actin with higher affinity relative to other Coro1s. In the present study, we systematically compared Coro1 binding to preformed filaments from human platelet actin, which contains 85% β-actin and 15% γ-actin [32]. To calculate the binding affinity, we used a method to analyse the amount of coronin (0.1 μM input) depleted from the supernatant with increasing concentrations of actin in cosedimentation assays (Figure 3A). Among the Coro1s, Coro1A bound to platelet actin with the lowest affinity with Kd=2.569 μM (Figure 3B), whereas Coro1B displayed a substantially higher affinity with a Kd=0.467 μM (Figure 3C). Strikingly, in initial experiments using the same concentration of Coro1C, we observed rapid depletion even at concentrations just above the critical concentration of actin (results not shown), indicating that Coro1C binds F-actin with very high affinity. Therefore, to more accurately measure the affinity, we used a higher input concentration of Coro1C (0.3 μM). We consistently observed a steep depletion of Coro1C from the supernatant at concentrations of actin between 0.2–0.8 μM (Figures 3A and 3D), which was not apparent for either Coro1A or Coro1B.
Figure 3. Coro1C binds F-actin co-operatively and with high affinity.
(A) Coro1A, Coro1B, Coro1C–R28D, Coro1C–2×KE, Coro1C–R28D/2×KE (0.1 μM) or Coro1C (0.3 μM) was incubated with increasing concentrations of actin and cosedimentation assays were performed. Supernatant fractions were separated by SDS/PAGE and analysed by immunoblotting with an anti-His antibody. Representative immunoblots are from at least four independent experiments. Quantification of the amount of Coro1A (B), Coro1B (C), Coro1C (D), Coro1C–R28D (E) and Coro1C–2×KE (F) bound to F-actin as a function of the free F-actin concentration. Each data point is shown as mean±S.E.M. from at least four independent experiments. Binding curves were fitted to the experimental data and used to generate a Hill coefficient (h) and dissociation constant (Kd), which are displayed on the plots.
We surmised that these effects might be due to co-operative effects of Coro1C binding to enhance the ability of additional Coro1C to bind F-actin. To further analyse this phenomenon, we attempted to fit binding curves to our experimental data using Hill slopes. Consistent with our hypothesis, Coro1A and Coro1B bound F-actin non-co-operatively with a Hill coefficient of ~1. In contrast, Coro1C showed a high degree of positive co-operativity with a Hill coefficient of 3.922. We also examined the effects of the R28D or 2×KE mutation on Coro1C binding affinity (Figures 3D and 3E). Intriguingly, although either mutation only modestly impaired binding affinity, both were sufficient to block Coro1C-positive co-operative binding to F-actin (results not shown). In line with our actin cosedimentation data, mutation of both sites in combination severely decreased F-actin binding affinity (~Kd>25 μM, results not shown). Overall, our data demonstrate that binding of Coro1C to F-actin requires both residues in the β-propeller and in the unique region. Furthermore, co-operative binding is a property that distinguishes Coro1C from other Coro1s.
Arg28 is the key determinant for efficient leading-edge localization of Coro1C, but both sites are required for actin binding in vivo
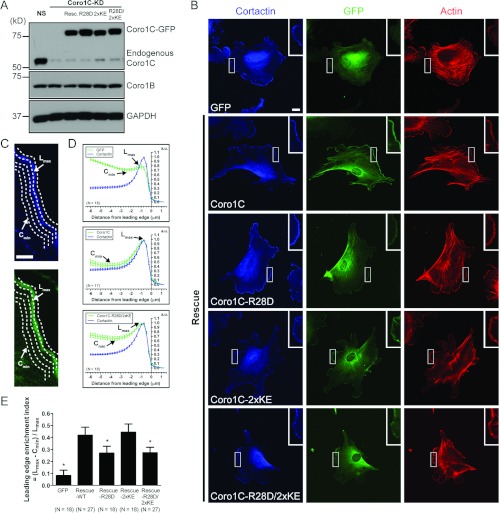
We previously demonstrated that efficient leading edge lamellipodia localization of Coro1B requires its F-actin-binding function [22]. The generation of an actin-binding-deficient mutant of Coro1C allowed us to address the specific effect of F-actin binding on its leading-edge localization in vivo. We first generated a monoclonal antibody that specifically recognized Coro1C, but not other coronins (Supplementary Figure S4 at http://www.BiochemJ.org/bj/444/bj4440089add.htm). We then simultaneously depleted endogenous Coro1C and re-expressed shRNA-resistant Coro1C–GFP and the R28D, 2×KE and R28D/2×KE mutants in IA32 mouse fibroblasts using a system we had developed previously (Figure 4A; [33]). The localization of wild-type and mutant Coro1C–GFP was examined in cells stained for cortactin (a leading-edge marker) and F-actin (Figure 4B). We analysed leading-edge localization of Coro1C using a contour-based method for determining the average fluorescence intensity of all the pixels along a contour edge (Figure 4C; [6]). We plotted the normalized cortactin and GFP fluorescence intensity as a function of the distance from the leading edge of the cell (Figure 4D). To quantify the amount of coronin at leading edge lamellipodia relative to the cytoplasm, we used a previously developed index of leadingedge enrichment [22]. This is defined as the ratio of the lamellipodia maximum intensity and the minimum cytoplasmic intensity (Figure 4E). As expected, Coro1C–GFP localized efficiently to the leading edge. In contrast, whereas the leading-edge enrichment of cortactin remained unchanged (results not shown), the localization of the actin-binding mutant Coro1C–GFP–R28D/2×KE was significantly impaired. The R28D mutation impaired leading edge enrichment of Coro1C almost to the same extent as the R28D/2×KE mutation, suggesting that whereas the R28D mutation failed to block actin binding in vitro, it was sufficient to impair leading-edge enrichment of Coro1C in vivo. Interestingly, the 2×KE mutant did not affect the leading-edge enrichment of Coro1C. These data demonstrate that, like Coro1B, efficient leading-edge localization of Coro1C requires the conserved site in the β-propeller.
Figure 4. Arg28 is the key determinant for efficient leading-edge localization of Coro1C, but both sites are required for actin binding in vivo.
(A) IA32 mouse fibroblasts were infected with lentivirus expressing NS shRNA (GFP), Coro1C shRNA (Coro1C-KD) or Coro1C shRNA and re-expressing Coro1C–GFP (resc.) or R28D, 2×KE, R28D/2×KE mutants. Lysates were separated on SDS/PAGE and analysed by immunoblotting with anti-Coro1C and anti-Coro1B antibodies. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was probed as a loading control. (B) IA32 mouse fibroblasts expressing GFP or depleted of endogenous Coro1C and re-expressing (rescue) Coro1C–GFP, Coro1C–GFP–R28D, Coro1C–GFP–2×KE or Coro1C–GFP–R28D/2×KE were stained with an anti-cortactin antibody and phalloidin to label actin. Scale bar, 20 μm. Insets show magnified regions of the lamellipodia. (C) Magnified region of the lamellipodia in IA32 mouse fibroblasts re-expressing Coro1C–GFP depicts an example schematic of leading edge contour analysis. Scale bar, 5 μm. The average fluorescence intensity of cortactin and GFP for all the pixels along a contour edge was measured. Lmax is the maximum fluorescence intensity in the lamellipodia. Cmin is the minimum fluorescence intensity in the cytoplasm. (D) Fluorescence intensities from contour edge analysis were normalized. Representative plots were generated from normalized fluorescence intensities as a function of distance from the leading edge for cortactin and GFP combined from indicated number of cells (N). Results are means±S.E.M. a.u., absorbance units. (E) Quantification of relative leading-edge enrichment index from indicated number of cells (N), which is defined as (Lmax−Cmin)/Lmax. *P<0.01 compared with wild-type Coro1C–GFP rescue by one-way ANOVA with Dunnett post-test. Results are means±95% confidence interval.
Since the coronin at the leading edge lamellipodia represents only a subset of the total coronin in the cell, we further analysed the global Coro1C distribution by separating cell lysates into supernatant and Triton-insoluble fractions (Supplementary Figure S5 at http://www.BiochemJ.org/bj/444/bj4440089add.htm). As anticipated, wild-type Coro1C–GFP was associated with the Triton-insoluble cytoskeletal fraction. Coro1C–R28D and Coro1C–2×KE were also found in the Triton-insoluble compartment. Interestingly, a greater amount of the R28D mutant was present in the supernatant. In line with our actin cosedimentation data, the fully defective R28D/2×KE actin-binding mutant was exclusively detected in the supernatant. Together, these data suggest that, although the conserved arginine residue in the β-propeller is a key determinant in leading-edge localization, both sites in the β-propeller and the unique region of Coro1C are required for global association with the actin cytoskeleton in vivo.
DISCUSSION
On the basis of our findings, we provide additional insight into the F-actin-binding function of the mammalian Coro1s. We have substantial evidence to support the idea that Coro1C harbours a second actin-binding site. First, a mutation in a conserved arginine residue in Coro1A and Coro1B that abrogated F-actin binding did not block F-actin binding in Coro1C. Secondly, charge-reversal mutations in the positive patch in the unique region blocked F-actin binding only when in combination with the R28D mutation. Thirdly, the F-actin-binding affinity of Coro1C with either of these mutations was not dramatically affected. Moreover, we demonstrated that an actin-binding mutant of Coro1C was not enriched in leading-edge lamellipodia in fibroblasts, reinforcing the general notion that F-actin binding is essential for coronin function.
One initial interpretation of our coronin competition results was that they might only reflect differences in actin-binding affinity. However, our measurements of the binding affinities suggest that this is not the case. Coro1A has a substantially lower affinity for F-actin than Coro1B, but it was able to compete with Coro1B for binding. On the other hand, Coro1B and Coro1C share similar binding affinities, but Coro1B was unable to compete with Coro1C. It is possible that inability of Coro1B to compete with Coro1C may also be related to the ability of Coro1C to bind F-actin co-operatively. Mutations in the site in β-propeller or in the unique region had only modest effects on Coro1C-binding affinity, but allowed Coro1B to compete for binding to actin, suggesting that the location of the these binding sites may be in close proximity on the actin filament.
To validate our in vitro results, we examined the localization of Coro1C and found that actin binding is required for efficient leading-edge localization. However, one distinction from our in vitro results is that the R28D mutation was sufficient to block leading-edge localization in vivo, whereas the 2×KE mutant Coro1C still retained leading edge localization. This raises the possibility that the conserved site in the β-propeller has a function in addition to its role in actin binding. Indeed, a previous study demonstrated that mutation of this site in Coro1A abolished phospholipid binding [30]. Alternatively, either the 2×KE mutation impairs Coro1C localization to other actin-based structures or alters a behaviour not evident in our leading-edge localization studies. Indeed, we confirmed this effect by finding that the R28D/2×KE mutant was completely excluded from the Triton-insoluble cytoskeleton. In the future it will be interesting to distinguish between the differential effects of both actin-binding sites on Coro1C localization and function.
By performing binding affinity measurements, we were able to determine that a unique function for Coro1C is that it displays highly co-operative binding to actin filaments. There are only a few other known examples of modulators that bind actin co-operatively: cofilin, tropomyosin, gelsolin and myosin subfragment-1 [34–37]. Intriguingly, cofilin co-operative binding has been postulated to propagate structural changes along the filament to lower the affinity of actin for the Arp2/3 complex [38]. At present, we cannot discern if Coro1C co-operativity is due to Coro1C–Coro1C interactions or the ability of Coro1C to alter the structure of the actin filament to enhance the affinity for additional Coro1C. It is tempting to speculate that these effects may be related to the nucleotide state of actin, as our previous results suggest that Coro1B binds preferentially to ATP- and ADP-Pi-actin over ADP-actin with approximately 50-fold higher affinity [22].
Coronins facilitate actin dynamics at the level of Arp2/3 actin assembly and cofilin-mediated disassembly. Both Coro1A and Coro1B contain a phosphorylation site by PKC (protein kinase C) at Ser2 which regulates the interaction of Coro1A and Coro1B with the Arp2/3 complex and has effects on whole-cell motility [4, 5]. Intriguingly, Coro1C lacks this phosphorylation site and can still interact with the Arp2/3 complex [8], which may contribute to the residual localization observed with the Coro1C actin-binding mutant. In contrast with a previous study suggesting that Coro1A and cofilin synergize to promote actin depolymerization [39], Coro1A and Coro1B protect actin filaments from cofilin binding. Instead, Coro1B targets the cofilin-activating phosphatase Slingshot-1 to the leading edge of migrating cells, suggesting a more indirect mechanism by which coronins facilitate cofilin activity [6]. It will be interesting to determine how Coro1C co-operative binding affects these processes.
With the generation of a completely deficient actin-binding mutant of Coro1C, we now have the ability to formally assess the actin-binding function of Coro1C in a variety of physiological contexts. A future challenge will be to determine the biophysical properties of Coro1C co-operative binding and how this feature mechanistically contributes to the regulation of actin cytoskeletal dynamics.
Online data
AUTHOR CONTRIBUTION
Keefe Chan performed the experiments. David Roadcap and Nicholas Holoweckyj generated and tested the Coro1C monoclonal antibody. Keefe Chan and James Bear designed the experiments, analysed the data and wrote the paper.
ACKNOWLEDGEMENTS
We thank Tao Bo for technical assistance and members of the Bear lab for helpful discussions.
FUNDING
This work was supported by the National Institutes of Health [grant number G-M083-35], the American Chemical Society [grant number RSG-08-157-01], and the Howard Hughes Medical Institute.
References
- 1.de Hostos E. L., Bradtke B., Lottspeich F., Guggenheim R., Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G-protein β subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan K. T., Creed S. J., Bear J. E. Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011;21:481–488. doi: 10.1016/j.tcb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uetrecht A. C., Bear J. E. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Foger N., Rangell L., Danilenko D. M., Chan A. C. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 2006;313:839–842. doi: 10.1126/science.1130563. [DOI] [PubMed] [Google Scholar]
- 5.Cai L., Holoweckyj N., Schaller M. D., Bear J. E. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J. Biol. Chem. 2005;280:31913–31923. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- 6.Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iizaka M., Han H. J., Akashi H., Furukawa Y., Nakajima Y., Sugano S., Ogawa M., Nakamura Y. Isolation and chromosomal assignment of a novel human gene, CORO1C, homologous to coronin-like actin-binding proteins. Cytogenet. Cell Genet. 2000;88:221–224. doi: 10.1159/000015555. [DOI] [PubMed] [Google Scholar]
- 8.Rosentreter A., Hofmann A., Xavier C. P., Stumpf M., Noegel A. A., Clemen C. S. Coronin 3 involvement in F-actin-dependent processes at the cell cortex. Exp. Cell Res. 2007;313:878–895. doi: 10.1016/j.yexcr.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Spoerl Z., Stumpf M., Noegel A. A., Hasse A. Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J. Biol. Chem. 2002;277:48858–48867. doi: 10.1074/jbc.M205136200. [DOI] [PubMed] [Google Scholar]
- 10.Kimura T., Kaneko Y., Yamada S., Ishihara H., Senda T., Iwamatsu A., Niki I. The GDP-dependent Rab27a effector coronin 3 controls endocytosis of secretory membrane in insulin-secreting cell lines. J. Cell Sci. 2008;121:3092–3098. doi: 10.1242/jcs.030544. [DOI] [PubMed] [Google Scholar]
- 11.Thal D., Xavier C. P., Rosentreter A., Linder S., Friedrichs B., Waha A., Pietsch T., Stumpf M., Noegel A., Clemen C. Expression of coronin-3 (coronin-1C) in diffuse gliomas is related to malignancy. J. Pathol. 2008;214:415–424. doi: 10.1002/path.2308. [DOI] [PubMed] [Google Scholar]
- 12.Appleton B. A., Wu P., Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Stirnimann C. U., Petsalaki E., Russell R. B., Muller C. W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Goode B. L., Wong J. J., Butty A. C., Peter M., McCormack A. L., Yates J. R., Drubin D. G., Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S. L., Needham K. M., May J. R., Nolen B. J. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem. 2011;286:17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oku T., Itoh S., Ishii R., Suzuki K., Nauseef W. M., Toyoshima S., Tsuji T. Homotypic dimerization of the actin-binding protein p57/coronin-1 mediated by a leucine zipper motif in the C-terminal region. Biochem. J. 2005;387:325–331. doi: 10.1042/BJ20041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kammerer R. A., Kostrewa D., Progias P., Honnappa S., Avila D., Lustig A., Winkler F. K., Pieters J., Steinmetz M. O. A conserved trimerization motif controls the topology of short coiled coils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13891–13896. doi: 10.1073/pnas.0502390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatfield J., Albrecht I., Zanolari B., Steinmetz M. O., Pieters J. Association of the leukocyte plasma membrane with the actin cytoskeleton through coiled coil-mediated trimeric coronin 1 molecules. Mol. Biol. Cell. 2005;16:2786–2798. doi: 10.1091/mbc.E05-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C. Z., Chen Y., Sui S. F. The identification of a new actin-binding region in p57. Cell Res. 2006;16:106–112. doi: 10.1038/sj.cr.7310014. [DOI] [PubMed] [Google Scholar]
- 20.Oku T., Itoh S., Okano M., Suzuki A., Suzuki K., Nakajin S., Tsuji T., Nauseef W. M., Toyoshima S. Two regions responsible for the actin binding of p57, a mammalian coronin family actin-binding protein. Biol. Pharm. Bull. 2003;26:409–416. doi: 10.1248/bpb.26.409. [DOI] [PubMed] [Google Scholar]
- 21.Mishima M., Nishida E. Coronin localizes to leading edges and is involved in cell spreading and lamellipodium extension in vertebrate cells. J. Cell Sci. 1999;112:2833–2842. doi: 10.1242/jcs.112.17.2833. [DOI] [PubMed] [Google Scholar]
- 22.Cai L., Makhov A. M., Bear J. E. F-actin binding is essential for coronin 1B function in vivo. J. Cell Sci. 2007;120:1779–1790. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- 23.Galkin V. E., Orlova A., Brieher W., Kueh H. Y., Mitchison T. J., Egelman E. H. Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. J. Mol. Biol. 2008;376:607–613. doi: 10.1016/j.jmb.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandhi M., Jangi M., Goode B. L. Functional surfaces on the actin-binding protein coronin revealed by systematic mutagenesis. J. Biol. Chem. 2010;285:34899–34908. doi: 10.1074/jbc.M110.171496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandhi M., Achard V., Blanchoin L., Goode B. L. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai L., Makhov A. M., Schafer D. A., Bear J. E. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 28.Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 29.Bear J. E., Loureiro J. J., Libova I., Fassler R., Wehland J., Gertler F. B. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 30.Tsujita K., Itoh T., Kondo A., Oyama M., Kozuka-Hata H., Irino Y., Hasegawa J., Takenawa T. Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides. J. Biol. Chem. 2010;285:6781–6789. doi: 10.1074/jbc.M109.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foger N., Jenckel A., Orinska Z., Lee K. H., Chan A. C., Bulfone-Paus S. Differential regulation of mast cell degranulation versus cytokine secretion by the actin regulatory proteins Coronin1a and Coronin1b. J. Exp. Med. 2011;208:1777–1787. doi: 10.1084/jem.20101757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De La Cruz E. M. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol. Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 33.Vitriol E. A., Uetrecht A. C., Shen F., Jacobson K., Bear J. E. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGough A., Pope B., Chiu W., Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y. Z., Korn E. D., Eisenberg E. Cooperative binding of tropomyosin to muscle and Acanthamoeba actin. J. Biol. Chem. 1979;254:7137–7140. [PubMed] [Google Scholar]
- 36.Prochniewicz E., Zhang Q., Janmey P. A., Thomas D. D. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J. Mol. Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 37.Greene L. E., Eisenberg E. Cooperative binding of myosin subfragment-1 to the actin–troponin–tropomyosin complex. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2616–2620. doi: 10.1073/pnas.77.5.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan C., Beltzner C. C., Pollard T. D. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brieher W. M., Kueh H. Y., Ballif B. A., Mitchison T. J. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.