Abstract
Context:
Polycystic ovary syndrome (PCOS) is one of the most common endocrine disorders in women.
Objective:
Our objective was to compare gene expression pattern in sc abdominal adipose tissue in nonobese PCOS patients vs. body mass index-matched controls.
Research Design and Methods:
Eleven PCOS subjects and 12 controls (body mass index 20–28 kg/m2) were recruited. Total RNA was isolated, and gene expression profiling was performed using Affymetrix Human Genome U133 arrays. Differentially expressed genes were classified by gene ontology. Microarray results for selected genes were confirmed by quantitative real-time PCR (RT-qPCR). Frequently sampled iv glucose tolerance tests were used to assess dynamic insulin sensitivity.
Results:
Ninety-six genes were identified with altered expression of at least 2-fold in nonobese PCOS adipose tissues. Inflammatory response genes were significantly down-regulated. RT-qPCR confirmed decreases in expression of IL6 (12.3-fold), CXCL2 (18.3-fold), and SOCS3 (22.6-fold). Lipid metabolism genes associated with insulin resistance were significantly up-regulated, with confirmed increases in DHRS9 (2.5-fold), UCLH1 (2.6-fold), and FADS1 (2.8-fold) expression. Wnt signaling genes (DKK2, JUN, and FOSB) were differentially expressed. RT-qPCR confirmed significant expression changes in DKK2 (1.9-fold increase), JUN (4.1-fold decrease), and FOSB (60-fold decrease).
Conclusions:
Genes involved in inflammation, lipid metabolism, and Wnt signaling are differentially expressed in nonobese PCOS adipose tissue. Because these genes are known to affect adipogenesis and insulin resistance, we hypothesize that their dysregulation may contribute to the metabolic abnormalities observed in women with PCOS.
Polycystic ovary syndrome (PCOS) is one of the most common endocrine disorders in women, affecting at least 7–9% of women of reproductive age (1). Approximately 65% of patients with PCOS demonstrate insulin resistance above and beyond that predicted by body mass, race, or age (2), resulting in compensatory hyperinsulinemia and increased risk for metabolic syndrome, diabetes, and cardiovascular disease (3).
Adipose tissue is an important endocrine organ, with the ability to modulate lipid metabolism and peripheral inflammation. The mechanisms underlying the insulin resistance of PCOS remain unclear; however, it appears that sc adipocyte including the stimulation of glucose transport, insulin responsive glucose transporter type 4 production, and the inhibition of lipolysis are defective in the disorder (4). Furthermore, paracrine regulation of adiponectin production appears to be abnormal in PCOS, favoring the development of insulin resistance (5). The association between glucose intolerance in women with PCOS and transcription factor 7-like 2 (TCF7L2), a Wnt signaling pathway component, suggests that Wnt signaling, a powerful regulator of adipogenesis, may also be altered in PCOS (6).
We hypothesized that genes related to the regulation of chronic inflammation would be abnormally expressed in the adipose tissue of lean women with PCOS, potentially denoting a primary defect in adipose tissue function in this disorder. Although levels of visceral fat have been correlated with insulin resistance in women with PCOS (7), sc abdominal fat is also metabolically active, is more readily obtainable, and may be as important as visceral fat in contributing to insulin resistance (8). Our results demonstrated significant differences in adipose tissue expression of genes involved in inflammation, lipid metabolism, and Wnt signaling-related adipogenesis, which may directly affect the pathophysiology of PCOS, independent of obesity.
Materials and Methods
Clinical studies
Detailed descriptions of PCOS and control subjects, diagnostic and exclusion criteria, metabolic assessment, hormonal analyses, tissue processing, and quantitative real-time PCR (RT-qPCR) are presented in Supplemental Methods (published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). Briefly, 11 women with PCOS diagnosed according to the National Institutes of Health 1990 criteria with body mass index (BMI) ranging from 20–28 kg/m2, and 12 age- and BMI-matched controls were recruited. Clinical characteristics of subjects are presented in Supplemental Table 1. An additional 20 controls were used to establish endocrine normative ranges.
DNA microarray and gene expression data analysis
DNA microarray gene expression profiling was carried out using the Affymetrix genechip Human Genome U133 plus 2.0 arrays (Affymetrix, Inc., Santa Clara, CA), using a previously described protocol (9). The criteria for selecting differentially expressed genes was preset as at least 2-fold difference in either direction plus statistical significance (P < 0.05, unpaired t test).
Statistical analysis
Comparisons between PCOS and control subjects were carried out parametrically using paired t tests. All values were presented as mean and se. Due to limitations in the amount of adipose tissue isolated, not all subjects contributed to each of the experiments performed.
Results
Insulin sensitivity in PCOS and control subjects
To determine and compare insulin sensitivity in nonobese PCOS, all PCOS patients studied molecularly underwent a frequently sampled iv glucose tolerance test (FSIVGTT), which was compared with a group of 20 healthy BMI-matched controls who had previously undergone an FSIVGTT. There were no significant differences in BMI, age, waist to hip ratio, or blood pressure between the groups (Supplemental Table 1). PCOS subjects had significantly higher modified Ferriman-Gallwey scores, free testosterone levels, dehydroepiandrosterone sulfate than controls. They also had higher homeostasis model assessment of insulin resistance levels than controls, although there were no detectable differences between the groups in insulin sensitivity assessed by the FSIVGTT. All subjects had normal TSH and prolactin levels (Supplemental Table 1).
Determination of differentially expressed genes in adipose tissues of nonobese PCOS and control subjects
To identify differentially expressed genes, adipose tissue samples from 11 nonobese PCOS subjects (75% White) and 12 BMI-matched controls (72% White) were studied. We performed microarray analysis using adipose tissues from nonobese PCOS subjects (n = 3) and BMI-matched controls (n = 4) and used RT-qPCR to confirm differential expression in an additional independent sample of eight PCOS and eight controls. Principal component analysis visualized global gene expression profiles and sample distributions, separating both groups using only a one-dimensional level (PC = 1; 71.6%), with a 76.8% variance (Supplemental Fig. 1).
Identification and functional classification of differentially expressed genes
Differential expression of at least 2-fold change was observed in 96 genes (Supplemental Tables 2 and 3). The greatest up- or down-regulation in PCOS was observed in genes involved in inflammation, lipid metabolism, and Wnt signaling, any of which may contribute to the molecular mechanisms underlying insulin resistance in PCOS.
Genes involved in inflammation
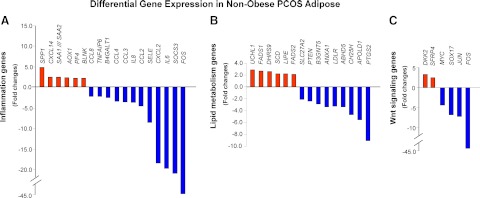
Microarray analysis identified six up-regulated and 12 down-regulated inflammation-related genes in PCOS adipose tissue (Fig. 1A and Supplemental Tables 2 and 3). Genes encoding cytokines (IL-6 and IL-8) and chemokines (CCL2, CCL3, CCL4, and CXCL2) were found to be significantly down-regulated in PCOS. Differential expression of IL6 (12.3-fold), CXCL2 (18.3-fold), and SOCS3 (22.6-fold) was confirmed by RT-qPCR and independently validated (Supplemental Fig. 2, A–C).
Fig. 1.
Inflammatory response, lipid metabolism, and Wnt signaling genes are differentially expressed in the adipose tissue from nonobese PCOS subjects (n = 4) vs. matched controls (n = 3). A, Histogram analysis of microarray data indicates inflammation-related genes with at least 2-fold change in expression; B, histogram analysis of microarray data indicates lipid metabolism-related genes with at least 2-fold change in expression; C, histogram analysis of microarray data indicates Wnt-related genes with at least 2-fold change in expression.
Genes involved in lipid metabolism
Microarray analyses indicated that seven up-regulated genes and 10 down-regulated genes in PCOS were involved in lipid metabolism (Fig. 1B and Supplemental Tables 2 and 3). Gene expression of DHRS9 (2.5-fold), UCLH1 (2.6-fold), and FADS1 (2.8-fold) was confirmed by RT-qPCR and independently validated (Supplemental Fig. 2, D–F).
Genes involved in Wnt signaling
Microarray analyses revealed significant changes in the expression of multiple Wnt signaling genes, including DKK2 (1.9-fold increase), JUN (4.1-fold decrease), and FOSB (60-fold decrease) (Fig. 1C and Supplemental Tables 2 and 3). Microarray data were confirmed and independently validated (Supplemental Fig. 2, G–I).
Discussion
Our study demonstrates significant differences in sc adipose tissue gene expression between nonobese PCOS patients and BMI-matched controls. First, microarray and RT-qPCR data demonstrated significant differences in inflammatory gene expression. Cytokine and chemokine expression was significantly decreased in PCOS. A possible explanation for this paradoxical finding is that there may exist altered distribution in the resident macrophage subtypes (M1 vs. M2) in PCOS adipose tissue. M1 macrophages are classically activated proinflammatory macrophages, whereas M2 macrophages are alternatively activated noninflammatory macrophages, and an increase in the ratio of M2 to M1 macrophages would lead to reduced inflammatory response (10). Although this hypothesis remains to be tested, we have previously demonstrated that our adipose samples have two inherent populations of CD14-positive macrophages, and these populations shift in response to coculture with adipocytes (11).
Among the different inflammatory genes that were found to be dysregulated in nonobese PCOS adipose tissue, IL6 and SOCS3 demonstrated a lower gene expression in comparison with controls (Supplemental Table 2), consistent with microarray findings in cumulus cells of nonobese PCOS subjects (12). Due to the reduction in IL6 expression, SOCS3 expression is predicted to be reduced. IL-6 binds (IL6ST), activating Janus kinase 2 (JAK2). JAK2 then phosphorylates signal transducer and activator of transcription 3 (STAT3), which dimerizes and translocates to the nucleus activating the transcription of numerous genes, including SOCS3. In turn, suppressor of cytokine signaling 3 (SOCS3) negatively regulates IL-6 signaling by directly binding GP130 and deactivating JAK2 (13). Additionally, SOCS3 has the ability to selectively mediate inflammation, blocking IL-6-induced STAT3 signaling, while allowing the antiinflammatory IL-10 activation to proceed through STAT3 (13) (Fig. 2).
Fig. 2.
Putative molecular mechanisms of Wnt signaling, lipid metabolism, and IL-6 effects on insulin signaling in nonobese PCOS adipose tissue. Note that IL-6 signaling acts on both the phosphatidylinositol-3 kinase (PI3K) and the MAPK insulin signaling pathways (13) and that up-regulation of components of the MAPK pathway [P38, c-Jun-N-terminal kinase (JNK), and MAPK kinase (MEK)/ERK] further reduces insulin receptor substrate-1 (IRS-1)/PI3K activity. Also, note that free fatty acids (FFA) activate protein kinase Cθ (PKCθ), which leads to inactivation of IRS-1 (17). Furthermore, up-regulation of one of the genes involved in lipid metabolism, DHRS9, increases MEK/ERK, which further reduces IRS-1/PI3K activity (15, 16). Note that inactivation of Wnt signaling stimulates glycogen synthase kinase-3β (GSK3β) in the active form, which may result in decreased glycogenesis and GLUT4 translocation, leading to insulin resistance (22). Our preliminary data indicate up or down regulation of key components of both the Wnt and insulin signaling pathways (see legend). Changes in RNA expression are indicated by green and red ovals. APC, Adenomatous polyposis coli; ATF, activating transcription factor 1; CKI, casein kinase I; DAG, dystroglycan 1; DKK, dickkopf-1; GRB2, growth factor receptor-bound protein 2, Groucho, transducin-like enhancer of split (E(sp1) homolog, Drosophila); GS, glycogen synthase; HDAC1, histone deacetylase 1; JUN, jun proto-oncogene; LIPE, hormone-sensitive lipase; LRP, low density lipoprotein receptor-related protein 1; MYC, v-myc myelocytomatosis viral oncogene homolog (avian); NR5A2, nuclear receptor subfamily 5, group A, member 2; OCT4, octamer-binding transcription factor 4; PUFA, polyunsaturated fatty acid; sFRP, secreted Frizzled-related proteins; SHP2, protein tyrosine phosphatase, non-receptor type 11; SOX, SRY (sex determining region Y)-box.
In contrast, microarray and RT-qPCR data demonstrated significant increases in the expression levels of several lipid metabolism genes, including DHRS9, UCHL1, and FADS1. Dehydrogenase/reductase (SDR family) member 9 (DHRS9), an oxidoreductase/decarboxylase, activates lipid metabolism to promote insulin resistance (14). DHRS9 is also involved in androgen and progesterone steroidogenesis, increasing 17-α hydroxyl-dehydrosteroid reductase activity, which leads to increased androgen production (15). Additionally, DHRS9 may be associated with other hormones, because PCOS patients had significantly higher levels of dehydroepiandrosterone sulfate (P < 0.05) (Supplemental Table 1). UCHL1 encodes a ubiquitin protein that alters the lipid composition of the plasma membrane, vesicles, and endosomes (16). Although the role of ubiquitin carboxyl-terminal esterase L1 in adipose tissue has yet to be elucidated, we hypothesize that alterations in membrane lipid composition could contribute to insulin resistance in nonobese PCOS adipose tissue.
FADS1 is a member of the fatty acid desaturase (FADS) gene family, which catalyzes the biosynthesis of highly unsaturated fatty acids from precursor essential polyunsaturated fatty acids. Microarray studies between sc adipose tissues from insulin-sensitive and insulin-resistant patients found a negative correlation between FADS1 expression levels and insulin resistance (17), whereas we observed an association between FADS1 expression in PCOS and increased homeostasis model assessment for insulin resistance values (Supplemental Table 1). Additionally, FADS1 was found to be decreased in sc adipose tissue of postmenopausal women after a 3-month treatment with estradiol valerate (18). One possible explanation for these results could stem from the synergistic effect of FADS1 and androgens in exacerbating the insulin resistance of PCOS patients (Fig. 2).
Our microarray results also identified decreased expression of selected genes involved in lipid metabolism in nonobese PCOS adipose tissue, including SLC27A2, annexin A1 (ANXA1), cholesterol 25-hydroxylase (CH25H), and low-density lipoprotein (LDL) receptor (LDLR) (Supplemental Table 3). SLC27A2 encodes the fatty-acid-coenzyme A ligase, which plays a key role in lipid biosynthesis and fatty acid degradation (19), and annexin A1 modulates lipolysis and the release of the inflammatory factor IL-6 (20), which we also found to be down-regulated in nonobese PCOS adipose tissue. Cholesterol 25-hydroxylase increases the concentration of cholesterol in the blood flow, and LDL receptor is involved in the uptake of LDL from the blood, thereby affecting the lipid profile. Altered expression of genes involved in lipid metabolism may contribute to insulin resistance in nonobese PCOS patients (Supplemental Fig. 3). This is consistent with PCOS studies that identified altered expression of genes involved in lipolysis (21).
Lastly, altered expression was observed in several genes in the Wnt signaling pathway, which is involved in regulating adipogenesis. Studies have identified genetic associations between Wnt signaling gene TCF7L2 and PCOS (6). Nonobese PCOS adipose tissue displayed increased expression of DKK2, an antagonist of Wnt, binding to its receptor. Inhibition of Wnt signaling leads to degradation of β-catenin and increases the active form of glycogen synthase kinase-3β, which results in increased adipogenesis and insulin resistance (22). We also identified decreased expression levels of JUN (c-Jun) and FOSB. These are downstream targets of the β-catenin–T cell-factor/lymphoid-enhancer-factor (TCF/LEF) complex and are key mediators of mesenchymal stem cell differentiation and proliferation, maintaining preadipocytes in an undifferentiated state (23). It is possible that decreased expression of these two genes could also lead to inhibition of Wnt signaling, increased adipogenesis, and insulin resistance in nonobese PCOS adipose tissue (Supplemental Fig. 3). Although we did not observe any change in cytochrome P450 (CYP) enzymes, CYP17 expression is very low in adipose compared with the adrenal or ovarian tissues. However, if the reduced FBJ murine osteosarcoma viral oncogene homolog expression observed in PCOS adipose tissue is indicative of the ovary or adrenal, as a key inhibitor of CYP17, a decrease or inhibition of FOS may lead to increased CYP17 expression and androgen production in tissues (24).
In summary, our results demonstrate that there are significant differences in the expression levels of genes involved in inflammation, lipid metabolism, and Wnt signaling in the adipose tissues of nonobese PCOS patients vs. BMI-matched controls. Dysregulation of these genes and pathways, which are known to affect adipogenesis and/or insulin resistance, may contribute to the metabolic abnormalities of PCOS.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant RO1-DK073632 and an endowment from the Helping Hand of Los Angeles, Inc. (to R.A.).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- BMI
- Body mass index
- CYP
- cytochrome P450
- DHRS9
- dehydrogenase/reductase (SDR family) member 9
- FSIVGTT
- frequently sampled iv glucose tolerance test
- JAK2
- Janus kinase 2
- LDL
- low-density lipoprotein
- PCOS
- polycystic ovary syndrome
- RT-qPCR
- quantitative real-time PCR
- SOCS
- suppressor of cytokine signaling 3
- STAT3
- signal transducer and activator of transcription 3.
References
- 1. Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. 2004. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab 89:2745–2749 [DOI] [PubMed] [Google Scholar]
- 2. Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF. 2009. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril 91:456–488 [DOI] [PubMed] [Google Scholar]
- 3. Ovalle F, Azziz R. 2002. Insulin resistance, polycystic ovary syndrome, and type 2 diabetes mellitus. Fertil Steril 77:1095–1105 [DOI] [PubMed] [Google Scholar]
- 4. Ciaraldi TP, Morales AJ, Hickman MG, Odom-Ford R, Olefsky JM, Yen SS. 1997. Cellular insulin resistance in adipocytes from obese polycystic ovary syndrome subjects involves adenosine modulation of insulin sensitivity. J Clin Endocrinol Metab 82:1421–1425 [DOI] [PubMed] [Google Scholar]
- 5. Chazenbalk G, Trivax BS, Yildiz BO, Bertolotto C, Mathur R, Heneidi S, Azziz R. 2010. Regulation of adiponectin secretion by adipocytes in the polycystic ovary syndrome: role of tumor necrosis factor-α. J Clin Endocrinol Metab 95:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biyasheva A, Legro RS, Dunaif A, Urbanek M. 2009. Evidence for association between polycystic ovary syndrome (PCOS) and TCF7L2 and glucose intolerance in women with PCOS and TCF7L2. J Clin Endocrinol Metab 94:2617–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lord J, Thomas R, Fox B, Acharya U, Wilkin T. 2006. The effect of metformin on fat distribution and the metabolic syndrome in women with polycystic ovary syndrome: a randomised, double-blind, placebo-controlled trial. BJOG 113:817–824 [DOI] [PubMed] [Google Scholar]
- 8. Maffeis C, Manfredi R, Trombetta M, Sordelli S, Storti M, Benuzzi T, Bonadonna RC. 2008. Insulin sensitivity is correlated with subcutaneous but not visceral body fat in overweight and obese prepubertal children. J Clin Endocrinol Metab 93:2122–2128 [DOI] [PubMed] [Google Scholar]
- 9. Deng X, Xu J, Wang C. 2008. Improving the power for detecting overlapping genes from multiple DNA microarray-derived gene lists. BMC Bioinformatics 9(Suppl 6):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujisaka S, Usui I, Bukhari A, Ikutani M, Oya T, Kanatani Y, Tsuneyama K, Nagai Y, Takatsu K, Urakaze M, Kobayashi M, Tobe K. 2009. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes 58:2574–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chazenbalk G, Bertolotto C, Heneidi S, Jumabay M, Trivax B, Aronowitz J, Yoshimura K, Simmons CF, Dumesic DA, Azziz R. 2011. Novel pathway of adipogenesis through cross-talk between adipose tissue macrophages, adipose stem cells and adipocytes: evidence of cell plasticity. PLoS ONE 6:e17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kenigsberg S, Bentov Y, Chalifa-Caspi V, Potashnik G, Ofir R, Birk OS. 2009. Gene expression microarray profiles of cumulus cells in lean and overweight-obese polycystic ovary syndrome patients. Mol Hum Reprod 15:89–103 [DOI] [PubMed] [Google Scholar]
- 13. Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. 2003. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature Immunol 4:551–556 [DOI] [PubMed] [Google Scholar]
- 14. Weigert J, Neumeier M, Bauer S, Mages W, Schnitzbauer AA, Obed A, Gröschl B, Hartmann A, Schäffler A, Aslanidis C, Schölmerich J, Buechler C. 2008. Small-interference RNA-mediated knock-down of aldehyde oxidase 1 in 3T3-L1 cells impairs adipogenesis and adiponectin release. FEBS Lett 582:2965–2972 [DOI] [PubMed] [Google Scholar]
- 15. Bauman DR, Steckelbroeck S, Williams MV, Peehl DM, Penning TM. 2006. Identification of the major oxidative 3α-hydroxysteroid dehydrogenase in human prostate that converts 5α-androstane-3α,17β-diol to 5α-dihydrotestosterone: a potential therapeutic target for androgen-dependent disease. Mol Endocrinol 20:444–458 [DOI] [PubMed] [Google Scholar]
- 16. Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, Kimmel AR, Londos C. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem 280:42841–42847 [DOI] [PubMed] [Google Scholar]
- 17. Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. 2011. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes 60:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lundholm L, Zang H, Hirschberg AL, Gustafsson JA, Arner P, Dahlman-Wright K. 2008. Key lipogenic gene expression can be decreased by estrogen in human adipose tissue. Fertil Steril 90:44–48 [DOI] [PubMed] [Google Scholar]
- 19. Wood JR, Nelson VL, Ho C, Jansen E, Wang CY, Urbanek M, McAllister JM, Mosselman S, Strauss JF., 3rd 2003. The molecular phenotype of polycystic ovary syndrome (PCOS) theca cells and new candidate PCOS genes defined by microarray analysis. J Biol Chem 278:26380–26390 [DOI] [PubMed] [Google Scholar]
- 20. Warne JP, John CD, Christian HC, Morris JF, Flower RJ, Sugden D, Solito E, Gillies GE, Buckingham JC. 2006. Gene deletion reveals roles for annexin A1 in the regulation of lipolysis and IL-6 release in epididymal adipose tissue. Am J Physiol 291:E1264–E1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seow KM, Tsai YL, Hwang JL, Hsu WY, Ho LT, Juan CC. 2009. Omental adipose tissue overexpression of fatty acid transporter CD36 and decreased expression of hormone-sensitive lipase in insulin-resistant women with polycystic ovary syndrome. Hum Reprod 24:1982–1988 [DOI] [PubMed] [Google Scholar]
- 22. Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. 2009. Adipogenesis and WNT signalling. Trends Endocrinol Metab 20:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacDonald BT, Tamai K, He X. 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beshay VE, Havelock JC, Sirianni R, Ye P, Suzuki T, Rainey WE, Carr BR. 2007. The mechanism for protein kinase C inhibition of androgen production and 17α-hydroxylase expression in a theca cell tumor model. J Clin Endocrinol Metab 92:4802–4809 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.