Congenital stationary night blindness is characterized by impaired night vision, decreased visual acuity, nystagmus, myopia, and strabismus. A genome-wide linkage scan was completed that localized the critical interval to the short arm of chromosome 3 and sequencing identified a novel missense mutation in GNAT1.
Abstract
Purpose.
Congenital stationary night blindness is a nonprogressive retinal disorder manifesting as impaired night vision and is generally associated with other ocular symptoms, such as nystagmus, myopia, and strabismus. This study was conducted to further investigate the genetic basis of CSNB in a consanguineous Pakistani family.
Methods.
A consanguineous family with multiple individuals manifesting cardinal symptoms of congenital stationary night blindness was ascertained. All family members underwent detailed ophthalmic examination, including fundus photographic examination and electroretinography. Blood samples were collected and genomic DNA was extracted. Exclusion and genome-wide linkage analyses were completed and two-point LOD scores were calculated. Bidirectional sequencing of GNAT1 was completed, and quantitative expression of Gnat1 transcript levels were investigated in ocular tissues at different postnatal intervals.
Results.
The results of ophthalmic examinations were suggestive of early-onset stationary night blindness with no extraocular anomalies. The genome-wide scan localized the critical interval to chromosome 3, region p22.1-p14.3, with maximum two-point LOD scores of 3.09 at θ = 0, flanked by markers D3S3522 and D3S1289. Subsequently, a missense mutation in GNAT1, p.D129G, was identified, which segregated within the family, consistent with an autosomal recessive mode of inheritance, and was not present in 192 ethnically matched control chromosomes. Expression analysis suggested that Gnat1 is expressed at approximately postnatal day (P)7 and is predominantly expressed in the retina.
Conclusions.
These data suggest that a homozygous missense mutation in GNAT1 is associated with autosomal recessive stationary night blindness.
Congenital stationary night blindness (CSNB) is a clinically heterogeneous group of nonprogressive retinal disorders, characterized by impaired night vision, decreased visual acuity, nystagmus, myopia, and strabismus.1 CSNB can be classified into two groups based on the electroretinographic recordings that exhibit waveforms in response to flashes of light that correspond to changes in the polarization of the photoreceptor and bipolar cells.2,3 The Schubert-Bornschein type is characterized by an electronegative electroretinogram (ERG) at the scotopic bright flash, in which the amplitude of the b-wave is smaller than that of the a-wave,2 while the Riggs type is defined by proportionally reduced a- and b-waves.3 The Schubert-Bornschein- and Riggs-type CSNB patients not only differ electrophysiologically, but manifest different clinical characteristics. Decreased visual acuity, myopia, and nystagmus can be associated with Schubert-Bornschein CSNB, whereas patients with Riggs-type CSNB have visual acuity within normal range with no symptoms of myopia and/or nystagmus.1
Familial cases of CSNB with autosomal dominant, autosomal recessive as well as X-linked inheritance have been reported. Mutations in RHO, PDE6B, and GNAT1 have been associated with autosomal dominant CSNB, and patients with mutations in these genes exhibit the Riggs- or Schubert-Bornschein-type ERG.4–10 The Schubert-Bornschein form can be further subdivided into complete and incomplete types. Complete CSNB is characterized by reduced rod b-wave response, whereas the incomplete CSNB is characterized by both a reduced rod b-wave and substantially reduced flicker cone responses, due to both ON and OFF bipolar cell dysfunction.11 Mutations in GRM6 and TRPM1 lead to autosomal recessive CSNB, whereas mutations in NYX to X-linked complete CSNB.12–18 Mutations in CABP4 have been associated with autosomal recessive CSNB and mutations in CACNA1F with X-linked incomplete CSNB.19–21 Recently, SLC24A1 was implicated in the pathogenesis of autosomal recessive CSNB with Riggs-type ERG response.22
Transducin is a trimeric G protein that is expressed in the retina and consists of three subunits.23 The α-subunit binds GTP and activates cyclic GMP phosphodiesterase (PDE), whereas the β and γ subunits form a complex that is necessary to interact with rhodopsin.24 The α-subunit expressed in the rod cells is encoded by GNAT1, whereas α-subunit expressed in the cone cells is encoded by GNAT2.25,26 Transducin is activated by conformational changes in rhodopsin due to the absorption of light, which causes a GDP bound α subunit to be exchanged with GTP and subsequent dissociation from the β-gamma complex.27,28
Previously, a missense mutation c.113G>A was identified in the Nougaret form of autosomal dominant CSNB.9 This mutation (Gly38Arg) affects part of small loop of α subunit of rod transducin and is believed to affect GTPase activity.9 Recently, Szabo et al.10 identified a heterozygous mutation; c.598C>G, in exon 6 of GNAT1 in individuals affected with autosomal dominant CSNB. This mutation (Gln200Glu) resides in a domain that plays an important role in binding and hydrolyzing GTP, and the ERG recordings were suggestive of a nonprogressive presynaptic defect in rod phototransduction.10
Here, we report a consanguineous family with multiple individuals diagnosed with CSNB that segregated within the family in an autosomal recessive fashion. A genome-wide linkage scan localized the disease phenotype to chromosome 3, region p22.1-p14.3, harboring GNAT1, a gene previously associated with autosomal dominant CSNB. Bidirectional sequencing identified a homozygous missense mutation in GNAT1 that segregated with the disease phenotype in the family and was not present in ethnically matched control chromosomes. To the best of our knowledge, this is the first report to implicate GNAT1 in the pathogenesis of autosomal recessive CSNB.
Materials and Methods
Clinical Assessment
A consanguineous Pakistani family with four affected individuals with a history of night blindness was recruited to participate in study to investigate autosomal recessive CSNB. Institutional review board (IRB) approval for this study was obtained from the National Centre of Excellence in Molecular Biology and the National Eye Institute. The participating subjects gave informed written consent consistent with the tenets of the Declaration of Helsinki. Funduscopy was performed at Layton Rehmatullah Benevolent Trust (LRBT) Hospital (Lahore, Pakistan). Electroretinography (ERG) measurements were recorded by using equipment manufactured by LKC (Gaithersburg, MD). Rod responses were determined through incident flash attenuated by −25 dB, and the rod–cone response was measured at 0 dB. Isolated cone responses were recorded at 0 dB with a 30-Hz flicker to a background illumination of 17 to 34 cd/m2. Color vision was assessed through Ishihara charts. Blood samples were collected from affected and unaffected family members and genomic DNA was extracted by the nonorganic method as described previously.29
Genotype Analysis
A genome-wide scan was performed with 382 highly polymorphic fluorescent markers from a linkage mapping with (PRISM MD-10; Applied Biosystems, Inc. [ABI], Foster City, CA) having an average spacing of 10 centimorgans (cM). Multiplex polymerase chain reactions were performed, as follows (9700 PCR System; ABI): Each reaction was performed in a 5-μL mixture containing 40 ng genomic DNA, various combinations of 10 μM dye-labeled primers pairs, 0.5 μL 10× PCR buffer (GeneAmp; ABI), 1 mM dNTP mix, 2.5 mM MgCl2, and 0.2 U of Taq DNA polymerase. Initial denaturation was performed for 5 minutes at 95°C, followed by 10 cycles of 15 seconds at 94°C, 15 seconds at 55°C, and 30 seconds at 72°C and then 20 cycles of 15 seconds at 89°C, 15 seconds at 55°C, and 30 seconds at 72°C. The final extension was performed for 10 minutes at 72°C, followed by a final hold at 4°C. PCR products from each DNA sample were pooled and mixed with size standards (HD-400; ABI). The resulting PCR products were separated in a DNA analyzer (model 3100; ABI), and alleles were assigned (GeneMapper Software ver. 4.0; ABI).
Linkage Analysis
Two-point linkage analyses were performed using the FASTLINK version of MLINK, and the maximum LOD score was calculated with ILINK from the LINKAGE program.30,31 Autosomal recessive CSNB was analyzed as a fully penetrant trait with an affected allele frequency of 0.001. The marker order and distances between the markers were obtained from the Marshfield database and the National Center for Biotechnology Information (NCBI) chromosome 3 sequence maps. For the initial genome scan, equal allele frequencies were assumed, whereas for fine mapping, allele frequencies were estimated from 96 unrelated and unaffected individuals from the Punjab province of Pakistan.
Mutation Screening
Primer pairs for individual exons were designed with Primer3 software. The sequences and annealing temperatures are available on request. Amplifications were performed in 25 μL reactions containing 50 ng of genomic DNA, 8 picomoles of each primer, 250 μM dNTP, 2.5 mM MgCl2, and 0.2U Taq DNA polymerase in the standard 1× PCR buffer provided by the manufacturer (ABI). PCR amplification consisted of a denaturation step at 96°C for 5 minutes, followed by 40 cycles, each consisting of 96°C for 30 seconds followed by 57°C (or specific annealing temperature of the primer pair) for 30 seconds and at 72°C for 1 minute. PCR products were analyzed on 2% agarose gel, precipitated, and purified by ethanol precipitation. The PCR primers for each exon were used for bidirectional sequencing by means of dye termination chemistry, according to the manufacturer's instructions (Big Dye Terminator Ready Reaction Mix; ABI). Sequencing products were resuspended in 10 μL of formamide (ABI) and denatured at 95°C for 5 minutes. Sequencing was performed on an automated system (PRISM 3100; ABI). The sequencing results were assembled with sequencing software and analyzed (SeqScape software, ver. 3.7; ABI).
Prediction Analysis
The degree of evolutionary conservation of positions at Asp129 and the possible impact of an amino acid substitution on the structure of GNAT1 protein was examined with the SIFT and PolyPhen tools, available online. Evolutionary conservation of the mutated amino acids in other GNAT1orthologs was examined using the UCSC genome browser.
Comparative Homology Modeling
Comparative homology modeling was performed using crystal coordinates for the human GNAT1. The wild and mutated protein sequences were loaded on to the Swiss PDB Viewer, and homology models were created. Hydrogen bonding was incorporated in the Swiss PDB Viewer.
Quantitative Expression of Gnat1 in the Eye
C57BL/6 mice were maintained in a 12 hour dark–light cycle according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and with protocols approved by UCSD Institutional Animal Care and Use Committee. The eye tissues were collected from C57BL/6 mice that were euthanized at predetermined time points at the end of the dark cycle by CO2 asphyxiation followed by cervical dislocation. Quantitative RT-PCR was performed, and data were analyzed according to previously published protocols.32 The expression of Gnat1 was determined using GACGCTGTCACCGACATTATCATC and GGCTGCAGGGCTACAGAATCTC and analyzing Rpl19 as a housekeeping gene to normalize Gnat1 gene expression, as described.33 Expression levels (±SEM) were calculated by analyzing at least three independent samples with replica reactions and presented on an arbitrary scale that represents the expression over the housekeeping gene. Rhodopsin mRNA levels served as a molecular marker for photoreceptor development and were measured in the same set of samples.
Internet Sources
The following web sites were used in the study: ILINK from LINKAGE, Rockefeller University, New York, NY, http://linkage.rockefeller.edu/soft; the FASTLINK version of MLINK and ILINK, NCBI, National Institutes of Health, Bethesda MD, http://www.ncbi.nlm.nih.gov; Marshfield Database, Marshfield Clinic, Marshfield, WI, http://research.marshfieldclinic.org; Primer3 PCR Primer Design Tool; http://primer3.sourceforge.net; PolyPhen: Prediction of Functional Effect of Human nsSNPs, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, http://genetics.bwh.harvard.edu/pph; SIFT database, J. Craig Venter Institute, Rockville, MD, http://sift.jcvi.org; Human Genome Browser, University of California, Santa Cruz, Santa Cruz, CA, http://genome.ucsc.edu; and Swiss PDB-Viewer, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://us.expasy.org/spdbv.
Results
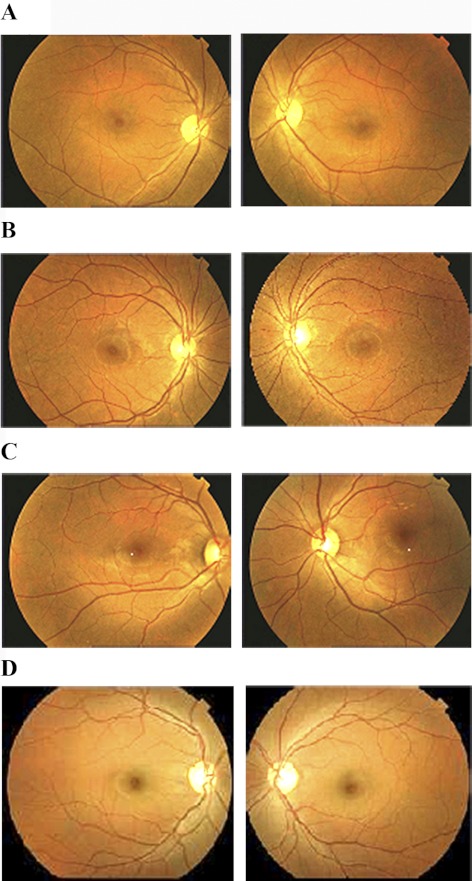
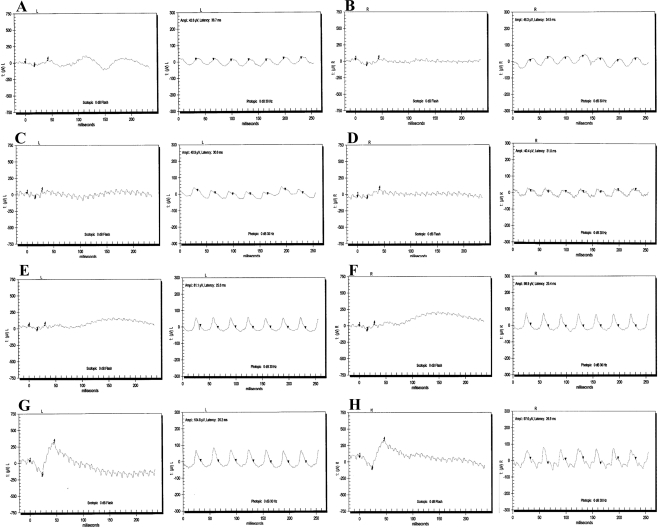
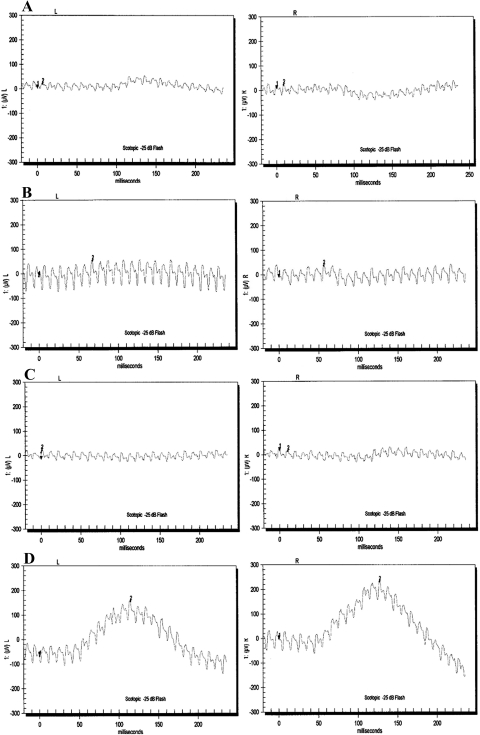
The family PKRP130 was recruited from the Punjab province of Pakistan (Fig. 1). The medical records available to us suggested that all four affected individuals of the family had reported an inability to see at night from the early years of their lives. Fundus photographs of affected individuals 10, 12, and 14 reveal no signs of retinal artery attenuation or bone spicule pigmentation (Fig. 2). The ERGs of affected individuals, 10, 12, and 14 show reduced or nearly absent a- and b-waves, even after 4 hours of dark adaptation (Fig. 3). The 30-Hz flicker recordings representing isolated cone responses were normal, although the amplitude of these recordings were reduced for affected individuals 10 and 12, when compared with those in unaffected individual 9 (Fig. 3). Similarly, the scotopic ERG recordings measured at −25 dB were absent in all three affected individuals, when compared with those in unaffected individual 9 (Fig 4). Taken together, these clinical data are suggestive of stationary night blindness in PKRP130 (Table 1).
Figure 1.
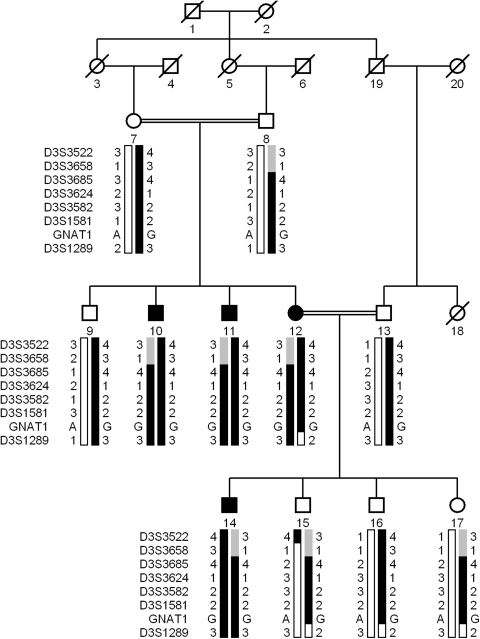
Pedigree of PKRP130 with a haplotype formed from alleles of the short arm of chromosome 3 microsatellite markers. Alleles forming the risk haplotype (black), alleles cosegregating with CSNB but not showing homozygosity (gray), and alleles not cosegregating with CSNB (white) are shown. Squares: males; circles: females; filled symbols: affected individuals; double line between individuals: consanguinity; and diagonal line through a symbol: deceased member.
Figure 2.
Fundus photographs of family PKRP130. (A) OD and OS of affected individual 10, (B) OD and OS of affected individual 12, (C) OD and OS of affected individual 14, and (D) OD and OS of unaffected individual 9. Fundus photographs of affected individuals 10, 12, and 14 revealed no signs of retinal arteries attenuation or bone spicule pigmentations in the retina.
Figure 3.
ERG recordings of family PKRP130. Combined rod and cone response recorded at 0 dB and cone responses recorded with 30-Hz flicker of affected individual 10: (A) OS and (B) OD; affected individual 12: (C) OS and (D) OD; affected individual 14: (E) OS and (F) OD; and unaffected individual 9: (G) OS and (H) OD; respectively. All affected individuals showed reduced amplitude and shortened implicit times in response to white flashes, suggesting an absence of rod function, whereas the unaffected individual had normal rod response in both eyes.
Figure 4.
Scotopic ERG recordings of family PKRP130 recorded at −25 dB after 4 hours after dark adaptation. (A) OS and OD of individual 10; (B) OS and OD of individual 12; (C) OS and OD of individual 14; and (D) OS and OD of individual 19. All affected individuals show no rod response, whereas the unaffected individual has normal rod response in both eyes.
Table 1.
Clinical Characteristics of Affected Individuals of Pedigree PKRP130 Diagnosed with Autosomal Recessive CSNB
| Individual ID | First Symptoms | Visual Acuity |
Color Vision | Fundus Findings | ERG Characteristics | |
|---|---|---|---|---|---|---|
| OD | OS | |||||
| 10 | Night blindness since early childhood | 6/6 | 6/6 | Normal | No macular atrophy, no pigment deposition, and no vascular attenuation. | The a- and b-waves are absent in the scotopic condition, whereas the cone responses are somewhat reduced in the photopic condition. |
| 11 | Night blindness since early childhood | 6/6 | 6/6 | Normal | ND | ND |
| 12 | Night blindness since early childhood | 6/6 | 6/6 | Normal | No macular atrophy, no pigment deposition, and no vascular attenuation. | The a- and b-waves are absent in the scotopic condition with normal cone responses in the photopic condition. |
| 14 | Night blindness since early childhood | 6/6 | 6/6 | Normal | No macular atrophy, no pigment deposition, and no vascular attenuation. | The a- and b-waves are absent in the scotopic condition with normal cone responses in the photopic condition. |
ND, not determined.
Initially, CABP4, GRM6, and SLC24A1 loci were excluded with closely spaced STR markers (data not shown). A genome-wide scan with four affected individuals (10, 11, 12, and 14) and five unaffected individuals (9, 13, 15, 16, and 17) was completed. A LOD score of 3.09 was obtained at D3S3582 during the genome-wide scan. Additional markers from the Marshfield database were selected to refine the critical interval, and two-point LOD scores of 3.07, 3.12, and 2.03 were obtained with markers D3S3685, D3S3624, and D3S1581, respectively, at θ = 0 (Table 2). No suggestive or significant two-point LOD scores were obtained during the genome-wide scan other than D3S3582.
Table 2.
Two-Point LOD Scores of 3p Markers for Pedigree PKRP130
| Marker | cM | Mb | 0 | 0.01 | 0.05 | 0.09 | 0.1 | 0.2 | 0.3 | zmax | θmax |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D3S3522* | 65.26 | 40.78 | −1.32 | 0.08 | 0.65 | 0.79 | 0.72 | 0.69 | 0.39 | 0.79 | 0.09 |
| D3S3658 | 65.26 | 40.90 | −1.54 | 0.07 | 0.60 | 0.74 | 0.71 | 0.59 | 0.35 | 0.74 | 0.09 |
| D3S3685 | 67.94 | 42.46 | 3.07 | 3.03 | 2.77 | 2.51 | 2.44 | 1.76 | 1.08 | 3.07 | 0.0 |
| D3S3624 | 68.47 | 44.61 | 3.12 | 3.08 | 2.81 | 2.56 | 2.48 | 1.81 | 1.12 | 3.12 | 0.0 |
| D3S3582* | 69.19 | 45.39 | 3.09 | 3.06 | 2.78 | 2.53 | 2.46 | 1.78 | 1.10 | 3.09 | 0.0 |
| D3S1581 | 70.61 | 48.59 | 2.03 | 1.98 | 1.79 | 1.6 | 1.56 | 1.08 | 0.64 | 2.03 | 0.0 |
| D3S1289* | 71.41 | 54.47 | −∞ | −0.01 | 0.54 | 0.64 | 0.65 | 0.58 | 0.4 | 0.65 | 0.1 |
Markers used in the genome-wide scan.
Haplotype analysis confirmed the linkage results and localized the critical interval to chromosome 3, region p22.1-p14.3. Recombination in unaffected individual 15 at marker D3S3522 defines the proximal boundary, whereas recombination in affected individual 12 at marker D3S1289 defines the distal boundary (Fig. 1). In addition, lack of homozygosity in affected individuals for alleles of markers D3S3522 and D3S3658 suggest that the pathogenic mutation lies distal to D3S3658 (Fig. 1). Taken together, this places the critical interval in a 6.15 cM (13.57 Mb) region flanked by markers D3S3658 proximally and D3S1289 distally.
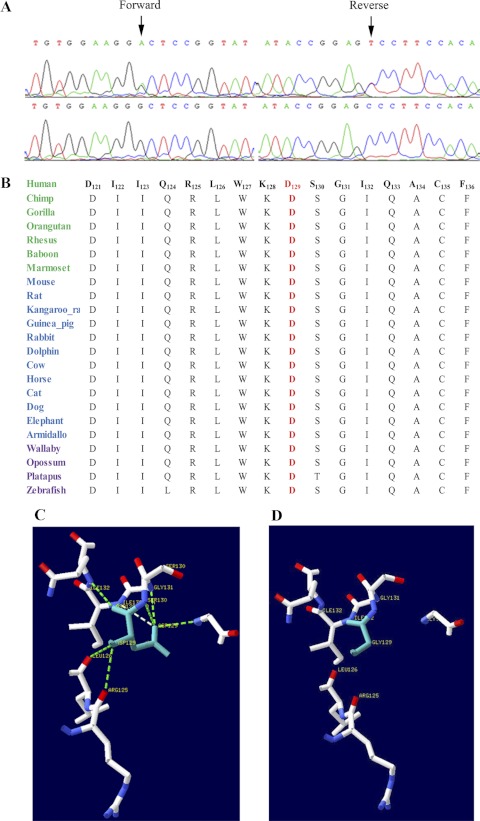
The critical interval harbors GNAT1 which has been associated with autosomal dominant CSNB. Sequencing of all the coding exons of GNAT1 in one affected and an unaffected individual of PKRP130 identified a novel homozygous missense mutation; c.386A>G that results in aspartic acid-to-glycine substitution; p.D129G (Fig. 5A). All affected individuals were homozygous for this substitution, whereas the unaffected individuals were heterozygous carriers of the mutant allele (Fig. 1). This mutation was not present in 192 ethnically matched control chromosomes.
Figure 5.
Sequence chromatograms illustrating the missense mutation, conservation Asp129 in GNAT1 orthologs and the proposed model illustrating hydrogen bond interactions in wild-type and mutant GNAT1 proteins. (A) Forward and reverse sequence chromatograms of unaffected individual 9 (top traces) and affected individual 10 (bottom traces) illustrating the c.386A>G substitution. (B) Sequence alignment of amino acids 121-136 of GNAT1 illustrating the conservation of the amino acid Asp129 in red (blue, primates; green, placental mammals; and purple, vertebrates). The model illustrating molecular interactions for (C) the wild-type and (D) the mutant GNAT1 proteins. In the wild-type GNAT1, Asp129 forms hydrogen bonds with the surrounding amino acids, which are compromised in the p.D129G mutant protein.
To investigate the possible impact of p.D129G substitution on the GNAT1 protein, we used SIFT and PolyPhen software. SIFT prediction based on evolutionary conservation suggested that p.D129G substitution will not be tolerated by the native three dimensional structure of GNAT1 protein. Likewise, the position-specific score difference of 2.742 obtained from PolyPhen strongly suggested that the D129G substitution could have a deleterious effect on GNAT1 protein structure. We further analyzed the evolutionary conservation of the amino acids around the missense mutation (121–136 amino acids) by aligning GNAT1 orthologs. The results strongly suggest that not only Asp129 but amino acids in the immediate neighborhood are well conserved among GNAT1 orthologs (Fig. 5B). To further understand the effect of the p.D129G substitution on GNAT1 structure, we compared the three-dimensional models of wild-type and mutant protein. Our results suggest that the wild-type sequence of D129 forms hydrogen bonds with the surrounding amino acids, but these interactions are compromised with G129 substitution (Figs. 5C, 5D).
Next, we investigated the quantitative expression of Gnat1 transcript levels in ocular tissues at different postnatal intervals. Rhodopsin mRNA levels were measured in the same set of samples and served as a molecular marker for photoreceptor development. Both Gnat1 and rhodopsin expression were not detected before P5; however, they increased exponentially from P7 indicating that Gnat1 is present in the photoreceptors (Fig. 6A). The expression profile of Gnat1 is similar to Slc24a1, a gene recently implicated in the pathogenesis of CSNB.22 The level of rhodopsin expression in the retina showed a steady increase with a maximum level at P30 and is in agreement with the profile of rhodopsin expression reported previously.32,33 Comparison of the expression levels of Gnat1 in different eye tissues strongly suggested that GNAT1 is highly expressed in the retina followed by the ciliary body, iris, and retinal pigment epithelium (Fig. 6B).
Figure 6.
Expression pattern of Gnat1 mRNA in postnatal mouse eyes. (A) Expression of Gnat1 and rhodopsin postnatal mouse eyes at postnatal day (P)1, P3, P5, P10, P15, and P30. Gnat1 expression is presented as bars and rhodopsin expression is presented as a line, with the relative scale on the y-axis. (B) Quantitative expression of Gnat1 in adult mouse eye tissue (180 days) presented as an arbitrary scale. All the values are the mean (±SEM) of three independent observations after normalization with the control gene (Rpl19) expression. RPE, retinal pigment epithelium; ON, optic nerve.
Discussion
Herein, we report a consanguineous Pakistani family with CSNB that was recruited from the Punjab province of Pakistan. Clinical evaluation consisting of fundus photography and ERG responses confirmed the disease phenotype, and linkage analyses localized the critical interval to chromosome 3, region p22.1-p14.3, with statistically significant LOD scores. Sequential analyses identified a novel mutation, p.D129G, which segregated within the family, consistent with an autosomal recessive mode of inheritance, and was absent in 192 ethnically matched control chromosomes. Although the maximum two-point LOD score of 3.12 is only slightly higher than the traditional limiting value of 3.0, nevertheless, it represents the maximum value obtainable with this family with real allele frequencies. In addition, the lack of evidence for linkage during the genome-wide scan, except with markers on the short arm of chromosome 3 provides additional support for localization of the CSNB locus to 3p. Although mutations in GNAT1 have been associated with autosomal dominant CSNB, this is the first report to implicate GNAT1 in the pathogenesis of autosomal recessive CSNB.
The phenomenon of a gene associated with a certain phenotype but manifesting different modes of inheritance has been reported. Previously, we identified three homozygous pathogenic mutations in RP1, a gene implicated in the pathogenicity of autosomal dominant retinitis pigmentosa34–36 that segregated with the disease phenotype in their respective families with heterozygous carriers of these causal lesions clinically unaffected.37 Likewise, causal mutations in crystalline βB1 have been associated with both autosomal dominant and autosomal recessive congenital cataracts.38,39 The mechanistic details of some mutations manifesting an autosomal dominant mode of inheritance, while other mutations in the same gene result in a recessive phenotype, are not yet completely understood. One possible explanation is that proteins harboring dominant mutations exhibit a negative effect on their wild-type counterparts; thus, one mutant allele is sufficient to manifest the disease phenotype. In contrast recessive mutations demonstrate a local effect on the native structure of the mutant protein, without affecting the wild-type gene product that subsequently requires two mutant alleles to exhibit the affectation status.
Identification of novel mutations and new genes associated with autosomal recessive CSNB will help us better understand the pathophysiology of the disease at a molecular level and will lead to development of better treatments.
Acknowledgments
The authors thank all family members for participating in the study and the staff of LRBT hospital for identification of the families and expert clinical evaluation of the affected individuals.
Footnotes
Supported in part by the Higher Education Commission (HEC) and the Ministry of Science and Technology, Islamabad, Pakistan, and by National Eye Institute Grant R01EY021237-01 (RA, SAR).
Disclosure: M.A. Naeem, None; V.R.M. Chavali, None; S. Ali, None; M. Iqbal, None; Sa. Riazuddin, None; S.N. Khan, None; T. Husnain, None; P.A. Sieving, None; R. Ayyagari, None; Sh. Riazuddin, None; J.F. Hejtmancik, None; S.A. Riazuddin, None
References
- 1. Zeitz C. Molecular genetics and protein function involved in nocturnal vision. Expert Rev Ophthalmol. 2007; 2: 467– 485 [Google Scholar]
- 2. Schubert G, Bornschein H. Analysis of the human electroretinogram (in German). Ophthalmologica. 1952; 123: 396– 413 [DOI] [PubMed] [Google Scholar]
- 3. Riggs LA. Electroretinography in cases of night blindness. Am J Ophthalmol. 1954; 38: 70– 78 [DOI] [PubMed] [Google Scholar]
- 4. Dryja TP, Berson EL, Rao VR, Oprian DD. Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat Genet. 1993; 4: 280– 283 [DOI] [PubMed] [Google Scholar]
- 5. Gal A, Orth U, Baehr W, Schwinger E, Rosenberg T. Heterozygous missense mutation in the rod cGMP phosphodiesterase beta-subunit gene in autosomal dominant stationary night blindness. Nat Genet. 1994; 7: 551 [DOI] [PubMed] [Google Scholar]
- 6. Sandberg MA, Weigel-DiFranco C, Dryja TP, Berson EL. Clinical expression correlates with location of rhodopsin mutation in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1995; 36: 1934– 1942 [PubMed] [Google Scholar]
- 7. Rosenberg T, Haim M, Piczenik Y, Simonsen SE. Autosomal dominant stationary night-blindness: a large family rediscovered. Acta Ophthalmol (Copenh). 1991; 69: 694– 702 [DOI] [PubMed] [Google Scholar]
- 8. Zeitz C, Gross AK, Leifert D, et al. Identification and functional characterization of a novel rhodopsin mutation associated with autosomal dominant CSNB. Invest Ophthalmol Vis Sci. 2008; 49: 4105– 4114 [DOI] [PubMed] [Google Scholar]
- 9. Dryja TP, Hahn LB, Reboul T, Arnaud B. Missense mutation in the gene encoding the alpha subunit of rod transducin in the Nougaret form of congenital stationary night blindness. Nat Genet. 1996; 13: 358– 360 [DOI] [PubMed] [Google Scholar]
- 10. Szabo V, Kreienkamp HJ, Rosenberg T, Gal A. p.Gln200Glu, a putative constitutively active mutant of rod alpha-transducin (GNAT1) in autosomal dominant congenital stationary night blindness. Hum Mutat. 2007; 28: 741– 742 [DOI] [PubMed] [Google Scholar]
- 11. Audo I, Robson AG, Holder GE, Moore AT. The negative ERG: clinical phenotypes and disease mechanisms of inner retinal dysfunction. Surv Ophthalmol. 2008; 53: 16– 40 [DOI] [PubMed] [Google Scholar]
- 12. Bech-Hansen NT, Naylor MJ, Maybaum TA, et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet. 2000; 26: 319– 323 [DOI] [PubMed] [Google Scholar]
- 13. Pusch CM, Zeitz C, Brandau O, et al. The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat Genet. 2000; 26: 324– 327 [DOI] [PubMed] [Google Scholar]
- 14. Dryja TP, McGee TL, Berson EL, et al. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci U S A. 2005; 102: 4884– 4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeitz C, van Genderen M, Neidhardt J, et al. Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15-Hz flicker electroretinogram. Invest Ophthalmol Vis Sci. 2005; 46: 4328– 4335 [DOI] [PubMed] [Google Scholar]
- 16. van Genderen MM, Bijveld MM, Claassen YB, et al. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet. 2009; 85: 730– 736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Audo I, Kohl S, Leroy BP, et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2009; 85: 720– 729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Sergouniotis PI, Michaelides M, et al. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet. 2009; 85: 711– 719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeitz C, Kloeckener-Gruissem B, Forster U, et al. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet. 2006; 79: 657– 667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strom TM, Nyakatura G, pfelstedt-Sylla E, et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet. 1998; 19: 260– 263 [DOI] [PubMed] [Google Scholar]
- 21. Bech-Hansen NT, Naylor MJ, Maybaum TA, et al. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998; 19: 264– 267 [DOI] [PubMed] [Google Scholar]
- 22. Riazuddin SA, Shahzadi A, Zeitz C, et al. A mutation in SLC24A1 implicated in autosomal-recessive congenital stationary night blindness. Am J Hum Genet. 2010; 87: 523– 531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lerea CL, Somers DE, Hurley JB, Klock IB, Bunt-Milam AH. Identification of specific transducin alpha subunits in retinal rod and cone photoreceptors. Science. 1986; 234: 77– 80 [DOI] [PubMed] [Google Scholar]
- 24. Fung BK, Hurley JB, Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci U S A. 1981; 78: 152– 156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ngo JT, Bateman JB, Klisak I, et al. Regional mapping of a human rod alpha-transducin (GNAT1) gene to chromosome 3p22. Genomics. 1993; 18: 724– 725 [DOI] [PubMed] [Google Scholar]
- 26. Morris TA, Fong SL. Characterization of the gene encoding human cone transducin alpha-subunit (GNAT2). Genomics. 1993; 17: 442– 448 [DOI] [PubMed] [Google Scholar]
- 27. Hargrave PA, Hamm HE, Hofmann KP. Interaction of rhodopsin with the G-protein, transducin. Bioessays. 1993; 15: 43– 50 [DOI] [PubMed] [Google Scholar]
- 28. Downs MA, Arimoto R, Marshall GR, Kisselev OG. G-protein alpha and beta-gamma subunits interact with conformationally distinct signaling states of rhodopsin. Vision Res. 2006; 46: 4442– 4448 [DOI] [PubMed] [Google Scholar]
- 29. Kaul H, Riazuddin SA, Shahid M, et al. Autosomal recessive congenital cataract linked to EPHA2 in a consanguineous Pakistani family. Mol Vis. 2010; 16: 511– 517 [PMC free article] [PubMed] [Google Scholar]
- 30. Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984; 36: 460– 465 [PMC free article] [PubMed] [Google Scholar]
- 31. Schaffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994; 44: 225– 237 [DOI] [PubMed] [Google Scholar]
- 32. Mandal MN, Vasireddy V, Reddy GB, et al. CTRP5 is a membrane-associated and secretory protein in the RPE and ciliary body and the S163R mutation of CTRP5 impairs its secretion. Invest Ophthalmol Vis Sci. 2006; 47: 5505– 5513 [DOI] [PubMed] [Google Scholar]
- 33. Ayyagari R, Mandal MN, Karoukis AJ, et al. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci. 2005; 46: 3363– 3371 [DOI] [PubMed] [Google Scholar]
- 34. Pierce EA, Quinn T, Meehan T, McGee TL, Berson EL, Dryja TP. Mutations in a gene encoding a new oxygen-regulated photoreceptor protein cause dominant retinitis pigmentosa. Nat Genet. 1999; 22: 248– 254 [DOI] [PubMed] [Google Scholar]
- 35. Sullivan LS, Heckenlively JR, Bowne SJ, et al. Mutations in a novel retina-specific gene cause autosomal dominant retinitis pigmentosa. Nat Genet. 1999; 22: 255– 259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guillonneau X, Piriev NI, Danciger M, et al. A nonsense mutation in a novel gene is associated with retinitis pigmentosa in a family linked to the RP1 locus. Hum Mol Genet. 1999; 8: 1541– 1546 [DOI] [PubMed] [Google Scholar]
- 37. Riazuddin SA, Zulfiqar F, Zhang Q, et al. Autosomal recessive retinitis pigmentosa is associated with mutations in RP1 in three consanguineous Pakistani families. Invest Ophthalmol Vis Sci. 2005; 46: 2264– 2270 [DOI] [PubMed] [Google Scholar]
- 38. Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002; 71: 1216– 1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen D, Bar-Yosef U, Levy J, et al. Homozygous CRYBB1 deletion mutation underlies autosomal recessive congenital cataract. Invest Ophthalmol Vis Sci. 2007; 48: 2208– 2213 [DOI] [PubMed] [Google Scholar]